Abstract
Alkyne-modified nucleoside analogs are useful for nucleic acid localization as well as functional and structural studies because of their ability to participate in copper-catalyzed azide/alkyne cycloaddition (CuAAC) reactions. Here we describe the synthesis of the triphosphate of 7-ethynyl-8-aza-7-deazaadenosine (7-EAATP) and the enzymatic incorporation of 7-EAA into RNA. The free nucleoside of 7-EAA is taken up by HeLa cells and incorporated into cellular RNA by endogenous RNA polymerases. In addition, 7-EAATP is a substrate for both T7 RNA polymerase and poly (A) polymerase from E. coli in vitro, albeit at lower efficiencies than with ATP. This work adds to the toolbox of nucleoside analogs useful for RNA labeling.
Keywords: ATP analogs, T7 transcription, Polyadenylation, Cellular RNA labeling, Click chemistry
Graphical Abstract
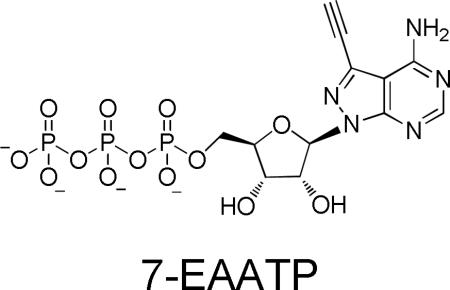
Nucleoside analogs are important for studying nucleic acid structure and function.1 Derivatives bearing terminal alkynes are particularly useful since they allow for further modification by copper-catalyzed azide/alkyne cycloaddition (CuAAC) reactions.2-7 Nucleoside analogs can be incorporated into oligonucleotides enzymatically or by chemical synthesis.2-11 Previously, our lab introduced 7-ethynyl-8-aza-7-deazaadenosine (7-EAA) into RNA using chemical synthesis with a 7-EAA phosphoramidite.2 X-ray crystallography of RNA duplexes bearing 7-EAA showed this analog forms a stable base pair with uridine and does not perturb the helical geometry of the duplex while projecting the 7-ethynyl group into the RNA duplex major groove (Fig. 1). 3 Furthermore, the high CuAAC reactivity of 7-EAA in RNA suggested it could be a useful probe of RNA structure and localization.3 Other alkyne-modified adenosine derivatives have been reported, but less is known about their effects on RNA structure.4, 11 Enzymatic incorporation of 7-EAA using RNA polymerases would allow for the synthesis of longer RNAs with more complex structures. In addition, since 7-EAA is an adenosine analog, its triphosphate is a mimic of ATP and could function as a substrate (or co-substrate) for other ATP-dependent enzymes.13-15 For these reasons, we prepared 7-EAATP and evaluated it as a substrate for two different ATP-utilizing enzymes: T7 RNA polymerase and E. coli poly (A) polymerase. In addition, we showed that HeLa cells treated with the 7-EAA nucleoside produce RNAs reactive in CuAAC reactions indicating 7-EAA uptake and intracellular conversion to the triphosphate in human cells.
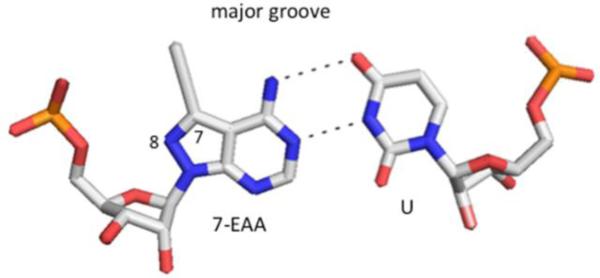
Figure 1.
7-Ethynyl-8-aza-7-deazaadenosine (7-EAA) base pairs with uridine (U) in duplex RNA and projects a terminal alkyne into the major groove.3
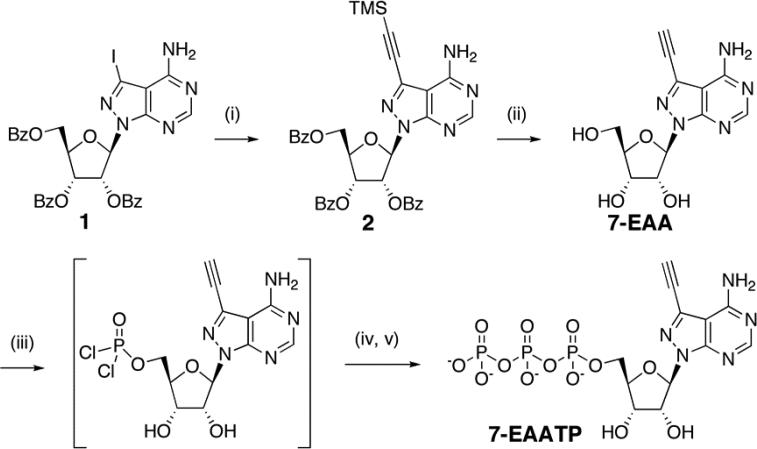
Enzymatic reactions of 7-EAA-containing RNAs showed that certain enzymes (e.g. AMV reverse transcriptase and an active site mutant of human editing enzyme ADAR2) recognized 7-EAA in RNA as adenosine.3 Given the similarity between 7-EAA and toyocamycin, a natural product known to be incorporated into cellular RNA,16 we believed 7-EAA could be taken up by human cells, converted to the triphosphate and incorporated into cellular RNAs via RNA polymerases.4-6 In order to test this idea, the 7-EAA nucleoside was required. This compound was originally synthesized by Seela and coworkers.17 Here we prepared the molecule with an optimized two step synthesis from 2’,3’,5’-tri-O-benzoyl-7-iodo-8-aza-7-deazaadenosine (1) (Scheme 1).18 The unprotected nucleoside product was then added to tissue culture media supporting the growth of HeLa cells and these cells were incubated for an additional 12 h followed by RNA isolation. We then performed a CuAAC reaction on the isolated RNA using a biotin azide. The resulting RNAs were resolved on a denaturing agarose gel, transferred to a nylon membrane and probed for the presence of biotin (Fig. 2). Biotin signal was observed from RNAs isolated from the cell sample grown in the presence of 7-EAA. Various lengths of RNAs were observed on the biotin detection image, suggesting different lengths of RNA were labeled with 7-EAA within the cell. In contrast, RNAs isolated from the cell sample grown in the absence of 7-EAA showed no biotin signal. To establish that absence of signal was not caused by absence of material, the same membrane was imaged using ethidium bromide detection. RNAs isolated from the cell sample grown without 7-EAA showed an intense signal with ethidium bromide detection. In addition, to confirm the signal observed was indeed from modified RNA, we treated the isolated RNA samples with ribonuclease V1 prior to the Northern blot detection. The results indicated the signal was generated by RNA. Together these data show that 7-EAA was taken up by HeLa cells and incorporated into cellular RNA. We also carried out a pulse/chase experiment to assess the turnover of 7-EAA-modified RNA in HeLa cells and compared this to EU-modified RNA turnover. These experiments indicate 7-EAA-modified RNA is longer lived in these cells with a t1/2 approximately 2-fold greater that EU-modified RNA (Supporting Information).
Scheme 1.
Synthesis of 7-EAATP. i) TMS-ethyne, Pd(PPh3)4, CuI, Et3N, THF, 87%; ii) DBU, MeOH, 60%; iii) (MeO)3PO, proton sponge, POCl3, −15 °C; iv) (HNBu3)2H2P2O7, NBu3, DMF, −15 °C; v) TEAB, rt, 11% (three steps).
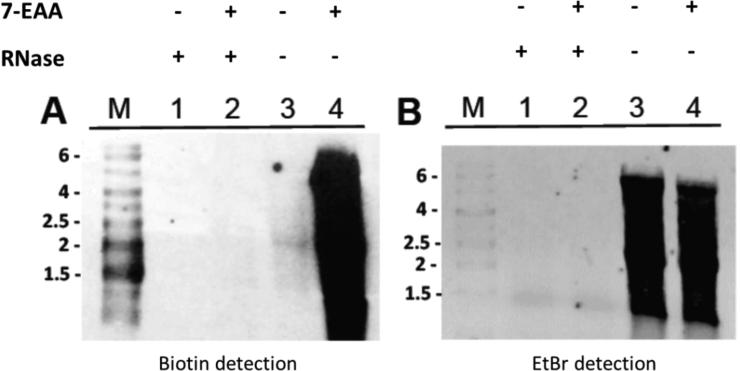
Figure 2.
7-EAA is taken up by HeLa cells and incorporated into cellular RNA. A) Detection of biotin-labeled RNA after click reaction with biotin-azide (M) biotinylated molecular weight markers in kb, (1) Sample isolated from cells grown in absence of 7-EAA, treated with RNase V1; (2) Sample isolated from cells grown in the presence of 100 μM 7EAA, treated with RNase V1; (3) Sample isolated from cells grown in the absence of 7-EAA, no RNase V1 treatment; (4) Sample isolated from cells grown in presence of 100 μM 7-EAA, no RNase V1 treatment. B) Same as in A) with ethidium bromide (EtBr) detection instead of biotin detection.
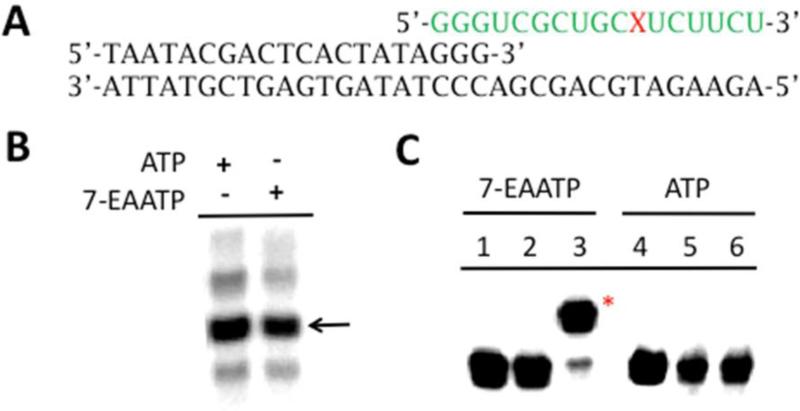
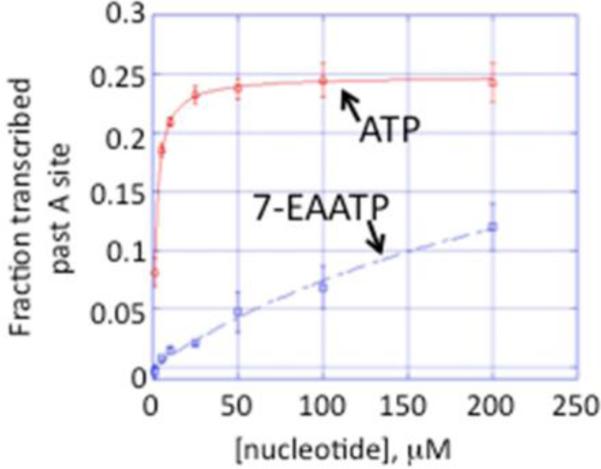
To test directly the efficiency of transcription with the triphosphate of 7-EAA, we synthesized this compound (Scheme 1, 7-EAATP). Different approaches have been reported for the synthesis of nucleoside triphosphates chemically and enzymatically.9, 19-21 Compared with other methods, the one-pot synthesis from unprotected nucleoside to nucleoside triphosphate has the advantages of minimizing functional group protection, simplifying the synthetic procedure and decreasing the number of purification steps.19, 20 Therefore we synthesized 7-EAATP from 7-EAA using the one-pot synthesis procedure shown in Scheme 1. The purified 7-EAATP was later used in transcription reactions in vitro. T7 RNA polymerase was chosen for these experiments since it is readily available and widely used in biochemistry laboratories. A 34 nucleotide (nt) DNA template was designed for in vitro transcription reactions bearing a T7 promoter sequence and a template for the synthesis of a 17 nt RNA transcript containing one A (Fig. 3A). The transcription reaction was performed in the presence of α-[32P]GTP enabling the detection of the final transcription product using storage phosphor autoradiography (Fig. 3B). To evaluate the extent of 7-EAA incorporation into the T7 transcript, we isolated the labeled 17 nt RNA product and performed a CuAAC reaction followed by analysis using polyacrylamide gel electrophoresis (Fig. 3C). RNA generated from a transcription reaction containing 7-EAATP was clearly shifted to a new position in the gel after the CuAAC reaction. Indeed, 87% of the RNA was shifted (Fig. 3C, lane 3), suggesting that at least this amount of the RNA sample contained 7-EAA. Importantly, no shift was observed for RNA generated from a transcription reaction containing ATP, supporting the idea that the shift is specific for the 7-EAA containing RNA. We then compared the efficiency of transcription beyond the 7-EAA/A position in the transcript at different 7-EAATP or ATP concentrations. The amount of product transcribed beyond the 7-EAA/A site was plotted against the concentration of 7-EAATP or ATP used in the transcription reaction (Fig. 4). These values were then used to calculate kinetic constants for each substrate as previously described.22 The results suggest that ATP has over 100-fold higher affinity for T7 RNA polymerase than 7-EAATP (7-EAA: KM=340 ± 90 μM; ATP: KM=1.90 ± 0.1 μM) (Fig. 4).
Figure 3.
Synthesis of 17 nt RNA by in vitro transcription with 7-EAATP and T7 RNA polymerase. A) T7 RNA polymerase transcription template for synthesis of 17 nt RNA containing one 7-EAA or A residue (X). B) Products of T7 polymerase transcriptions with template shown in (A) containing 5 mM ATP or 7-EAATP, α -32P GTP and allowed to proceed for 2 h at 37 °C. (arrow indicates major transcription product). C) CuAAC reaction with 17 nt RNAs. Lanes 1-3: product from transcription with 7-EAATP, Lanes 4-6: product from transcription with ATP. Lanes 1 and 4: no CuAAC reaction reagents added, Lanes 2 and 5: CuAAC reagents lacking azide, Lanes 3 and 6: CuAAC reagents plus azide. (*) indicates position of CuAAC product.
Figure 4.
Comparison of T7 RNA polymerase efficiency with template shown in Fig. 3A using varying concentrations of 7-EAATP or ATP.
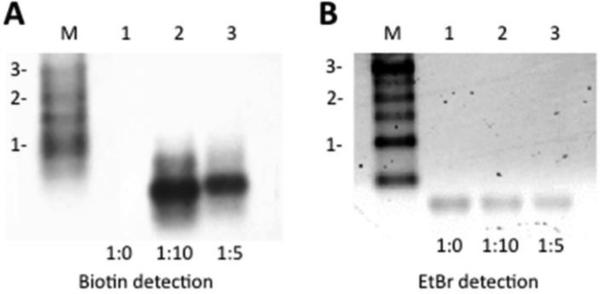
The experiments described above indicate that 7-EAATP is a substrate for T7 RNA polymerase, albeit with a substantially higher KM than ATP. To determine if multiple adenosine positions could be substituted with 7-EAA, we generated a longer RNA (335 nt) using in vitro transcription. Because of 7-EAATP's apparent low affinity for T7 RNA polymerase, we performed the transcription reaction at different ATP to 7-EAATP ratios (1:0, 1:5 and 1:10). RNA transcripts were then treated with biotin azide under CuAAC reaction conditions. The resulting RNAs underwent a Northern blotting procedure and imaged using ethidium bromide and biotin detection (Fig. 5). RNAs transcribed with the different ATP: 7-EAATP ratios showed a single band on the ethidium bromide detection image, indicating the transcription reactions generated the full-length transcript (Fig. 5B). However, no transcription product was detected for a transcription reaction carried out in the presence of 7-EAATP with no ATP added (data not shown). Importantly, a signal on the biotin detection image was observed only when 7-EAATP was present in the transcription reaction. These results indicate that RNAs transcribed in the presence of 7-EAATP were labeled with terminal alkyne and that 7-EAA was successfully incorporated into the transcript. RNA transcripts from the transcription reaction with a 1:10 ATP:7-EAATP ratio generated more signal on the biotin detection image than the RNA transcript from transcription reaction with the 1:5 ATP:7-EAATP ratio indicating a higher level of 7-EAA labeling under these conditions.
Figure 5.
Synthesis of 335 nt RNA by in vitro transcription with 7-EAATP and T7 RNA polymerase. A) Northern blot of RNA transcribed in the presence of different ratios of ATP:7-EAATP followed by click reaction modification with a biotin azide and detection for the presence of biotin. M: molecular weight markers in kb, Lane 1: no 7-EAATP in transcription, Lane 2: ATP:7-EAATP ratio is 1:10, Lane 3: ATP:7-EAATP ration is 1:5 B) Lanes are same as in A with ethidium bromide detection.
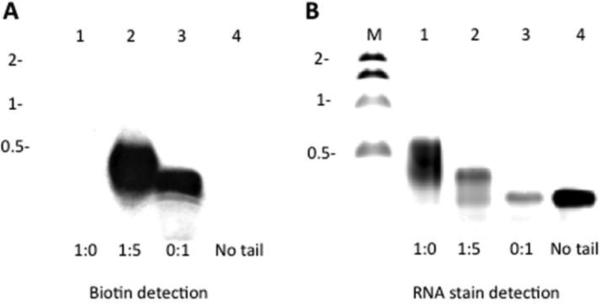
Since 7-EAATP is an ATP analog, other ATP-utilizing enzymes could accept it as well. Poly (A) polymerases use ATP as a substrate and add poly (A) tails to RNA 3’ ends.23 ATP analogs have been used to label poly (A) tails and study poly (A) related events, such as the localization, generation and the degradation of poly (A) tails.4, 11, 12 Here, we used poly (A) polymerase from E. coli to test whether 7-EAATP is suitable for labeling RNA through a polyadenylation reaction. Keeping the total concentration of both ATP and 7-EAATP the same, we carried out in vitro polyadenylation with different ratios of ATP:7-EAATP. The polyadenylation products were resolved on an agarose gel (Fig. 6). We found the RNA products generated from polyadenylation reactions with 1:0 and 1:5 ATP:7-EAATP ratios to be longer than the original RNA, indicating formation of a poly (A) tail in each case. However, RNAs generated from the polyadenylation reaction with 1:5 ATP:7-EAATP ratio were shorter than RNAs generated from the polyadenylation reaction with only ATP. Also, RNAs generated from the reaction with only 7-EAATP showed no significant change in RNA length. To determine if poly (A) tailing led to 7-EAA incorporation, we performed CuAAC reactions with the biotin azide followed by Northern blotting and biotin detection (Fig. 6A). Intense signals were observed from both the RNA polyadenylated with a 1:5 ATP:7-EAATP ratio and the RNA polyadenylated with 7-EAATP alone. Importantly, RNA polyadenylated with only ATP showed no biotin signal, supporting the idea that signals observed arise from 7-EAA modification in the poly (A) tail. When only 7-EAATP was present in the reaction, the RNA product and the RNA substrate were of similar size indicating inefficient extension of the fully modified poly (7-EAA) tail. Previous studies have shown that the presence of ATP is required for poly (A) polymerases to generate long tails in reactions containing other ATP analogs suggesting these enzymes are inefficient at extending fully modified tails.4 Nevertheless, our results show that E. coli poly(A) polymerase can recognize 7-EAATP and incorporate it into the poly(A) tail of an RNA.
Figure 6.
RNA poly(A) tailing with 7-EAATP. A) Biotin detection for RNA with poly(A) tail. B) RNA stain (SYBR® safe) detection for RNA with poly(A) tail. Lane M contains molecular marker in kb. Lanes 1 to 3 contain poly(A) tailed RNAs with different ATP:7EAATP ratios in each reaction. Lane 4 contains RNA without poly(A) tail.
In conclusion, the 7-EAA nucleoside was synthesized and added to media supporting HeLa cell growth resulting in 7-EAA incorporation into cellular RNA. 7-EAA triphosphate (7-EAATP) was also synthesized and used for modification of RNA by two different enzymes; T7 RNA polymerase and E. coli poly (A) polymerase. We found that 7-EAATP is recognized by both enzymes but low efficiencies compared to ATP require 7-EAATP/ATP mixtures for the synthesis of long modified RNAs containing multiple 7-EAA substitutions. Since 7-EAA in RNA is readily converted to triazole analogs by CuAAC reactions, this work adds to the toolbox of nucleoside analogs useful for RNA labeling.
Supplementary Material
Acknowledgments
P.A.B. acknowledges support for this research from the National Institutes of Health (R01 GM08074). Authors also acknowledge funding to the UC Davis NMR Facility (NSF DBIO-722538, NSF OSTI 97-24412 and NIH RR11973). We thank Dr. Bennett Addison for technical support and help with NMR experiments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Supplementary data associated with this article can be found.
References and notes
- 1.Phelps K, Morris A, Beal PA. ACS Chem. Biol. 2012;7:100. doi: 10.1021/cb200422t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibarra-Soza JM, Morris AA, Jayalath P, Peacock H, Conrad WE, Donald MB, Kurth MJ, Beal PA. Bioorg. Med. Chem. Lett. 2012;10:6491. doi: 10.1039/c2ob25647a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phelps KJ, Ibarra-Soza JMM, Tran K, Fisher AJ, Beal PA. ACS Chem. Biol. 2014;9:1780. doi: 10.1021/cb500270x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grammel M, Hang H, Conrad NK. Chembiochem. 2012;13:1112. doi: 10.1002/cbic.201200091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jao CY, Salic A. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15779. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qu D, Zhou L, Wang W, Wang Z, Wang G, Chi W, Zhang B. Anal. Biochem. 2013;434:128. doi: 10.1016/j.ab.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 7.Guan L, Heijden GW, Bortvin A, Greenberg MM. ChemBioChem. 2011;12:2184. doi: 10.1002/cbic.201100353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cochrane JC, Strobel SA. Curr Protoc Nucleic Acid Chem. 2004 doi: 10.1002/0471142700.nc0609s17. Chapter 6. [DOI] [PubMed] [Google Scholar]
- 9.Ohtsuki T, Kimoto M, Ishikawa M, Mitsui T, Hirao I, Yokoyama S. Proc. Natl. Acad. Sci. U.S.A. 2001;98:4922. doi: 10.1073/pnas.091532698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peacock H, Maydanovych O, Beal PA. Org. Lett. 2010;12:1044. doi: 10.1021/ol100019r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curanovic D, Cohen M, Singh I, Slagle CE, Leslie CS, Jaffrey SR. Nat. Chem. Biol. 2013;9:671. doi: 10.1038/nchembio.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grammel M, Hang HC. Nat. Chem. Biol. 2013;9:475. doi: 10.1038/nchembio.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah K, Liu Y, Deirmengian C, Shokat KM. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3565. doi: 10.1073/pnas.94.8.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar V, Weng Y-CC, Geldenhuys WJ, Wang D, Han X, Messing RO, Chou W-HH. J. Biol. Chem. 2015;290:1936. doi: 10.1074/jbc.M114.598698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunamura E-I, Kamei T, Konno H, Tamaoki N, Hisabori T. Biochem. Biophys. Res. Commun. 2014;446:358. doi: 10.1016/j.bbrc.2014.02.117. [DOI] [PubMed] [Google Scholar]
- 16.Suhadolnik RJ, Uematsu T, Uematsu H. Biochim. Biophys. Acta. 1967;149:41. doi: 10.1016/0005-2787(67)90689-2. [DOI] [PubMed] [Google Scholar]
- 17.Lin W, Xu K, Seela F. Nucleosides Nucleotides Nucleic Acids. 2005;24:869. doi: 10.1081/ncn-200059218. [DOI] [PubMed] [Google Scholar]
- 18.Cottam HB, Wasson DB, Shih HC, Raychaudhuri A, Di Pasquale G, Carson DA. J. Med. Chem. 1993;36:3424. doi: 10.1021/jm00074a024. [DOI] [PubMed] [Google Scholar]
- 19.Ludwig J. Acta Biochim. Biophys. Acad. Sci. Hung. 1981;16:131. [PubMed] [Google Scholar]
- 20.Gillerman I, Fischer B. Nucleosides Nucleotides Nucleic Acids. 2010;29:245. doi: 10.1080/15257771003709569. [DOI] [PubMed] [Google Scholar]
- 21.Wu W, Bergstrom DE, Davisson VJ. J. Org. Chem. 2003;68:3860. doi: 10.1021/jo020745i. [DOI] [PubMed] [Google Scholar]
- 22.Stengel G, Urban M, Purse BW, Kuchta RD. Anal. Chem. 2010;82:1082. doi: 10.1021/ac902456n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohanty BK, Kushner SR. Wiley Interdiscip. Rev. RNA. 2011;2:256. doi: 10.1002/wrna.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.