SUMMARY
Cultured cells convert glucose to lactate and glutamine is the major source of tricarboxylic acid (TCA) cycle carbon, but whether the same metabolic phenotype is found in tumors is less studied. We infused mice with lung cancers with isotope-labeled glucose or glutamine and compared the fate of these nutrients in tumor and normal tissue. As expected, lung tumors exhibit increased lactate production from glucose. However, glutamine utilization by both lung tumors and normal lung was minimal, with lung tumors showing increased glucose contribution to the TCA cycle relative to normal lung tissue. Deletion of enzymes involved in glucose oxidation demonstrates that glucose carbon contribution to the TCA cycle is required for tumor formation. These data suggest that understanding nutrient utilization by tumors can predict metabolic dependencies of cancers in vivo. Furthermore, these data argue that the in vivo environment is an important determinant of the metabolic phenotype of cancer cells.
INTRODUCTION
Metabolism is altered in cancer to sustain the energetic and biosynthetic demands of uncontrolled proliferation, and increased glucose uptake is a common feature of cancer cells in culture and tumors (Cairns et al., 2011; Vander Heiden et al., 2009). Positron emission tomography detection of 18F-2-deoxyglucose uptake together with magnetic resonance spectroscopy studies have confirmed increased glucose-to-lactate conversion in tumors (Warburg effect) (Keshari et al., 2013; Rodrigues et al., 2014), but understanding the metabolic phenotype of cancer is based mostly on cell culture studies, including the idea that glucose oxidation in the TCA cycle is limited (Bonnet et al., 2007; Cairns et al., 2011). Indeed, other data suggests mitochondrial function is required for tumor growth (Weinberg et al., 2010) and oxidative glucose metabolism has been observed in tumors (Yuneva et al., 2012; Marin-Valencia et al., 2012; Sellers et al., 2015). Whether tumors require glucose to support the TCA cycle remains unknown.
Non-small cell lung cancer (NSCLC) is a leading cause of cancer mortality (Herbst et al., 2008). Mutations in Kras are frequent in NSCLC and Ras activation has specific metabolic consequences (White, 2013). Ras-driven cancer cells display increased glucose uptake and aerobic glycolysis that supports both nucleotide biosynthesis and protein glycosylation for growth signaling (Jones and Thompson, 2009; Onetti et al., 1997; Ying et al., 2012). However, recent work suggests that there is selective pressure to maintain functional mitochondria in tumors (Tan et al., 2015). Limited studies of tumor metabolism have suggested the driver mutation, tissue of origin, and microenvironment all can impact metabolic phenotypes (Davidson and Vander Heiden, 2012; Yuneva et al., 2012), highlighting the need to understand metabolism in spontaneously arising tumors.
Glutamine is an important nutrient for most cancer cells in culture (Hensley et al., 2013; Mayers and Vander Heiden, 2015), and can provide carbon for TCA cycle anaplerosis in Ras-driven cancer cells (White, 2013). Indeed, the GAC splice isoform of glutaminase (Gls1) is essential in NSCLC-derived cell lines (van den Heuvel et al., 2012). Glutamine also acts as a direct nitrogen donor for nucleotide biosynthesis and can supply alpha-ketoglutarate (αKG) to support amino acid catabolism (Commisso et al., 2013; White, 2013). Glutamine catabolism in Kras-driven pancreatic cancer cells also has been implicated in NADPH production and redox balance (Lyssiotis et al., 2013; Ying et al., 2012). However, the extent of glutamine metabolism in vivo is controversial, with evidence for net glutamine synthesis in some tumors (Yuneva et al., 2012). Analysis of NSCLC tumor metabolism in patients argues both glucose and glutamine can be important for TCA anaplerosis (Cheng et al., 2011; Sellers et al., 2015). Drugs targeting Gls1 are being explored as therapies for human cancers, highlighting the importance of understanding glutamine use by tumors (Gross et al., 2014).
To understand how lung tumors utilize nutrients relative to normal lung, we examined tissue metabolism in mouse lung cancer models. This analysis suggested glucose oxidation is important for lung tumors, a finding that was not evident from examination of lung cancer cell lines in vitro. We utilized CRISPR/Cas9-based genome editing to understand whether tumors were dependent on glucose contribution to the TCA cycle. While glucose oxidation is dispensable for lung cancer cell proliferation in culture, enzymes involved in glucose oxidation are required for tumor growth in vivo. Conversely, preferential glutamine metabolism is not observed in lung tumors, and neither genetic deletion nor pharmacological inhibition of Gls1 affected the growth of KrasG12D-induced lung tumors. Together, these data better define the metabolic landscape of NSCLC and inform pathways that are required for cancer growth in a pathophysiological tissue context.
Results
Analysis of glucose metabolism in normal tissues
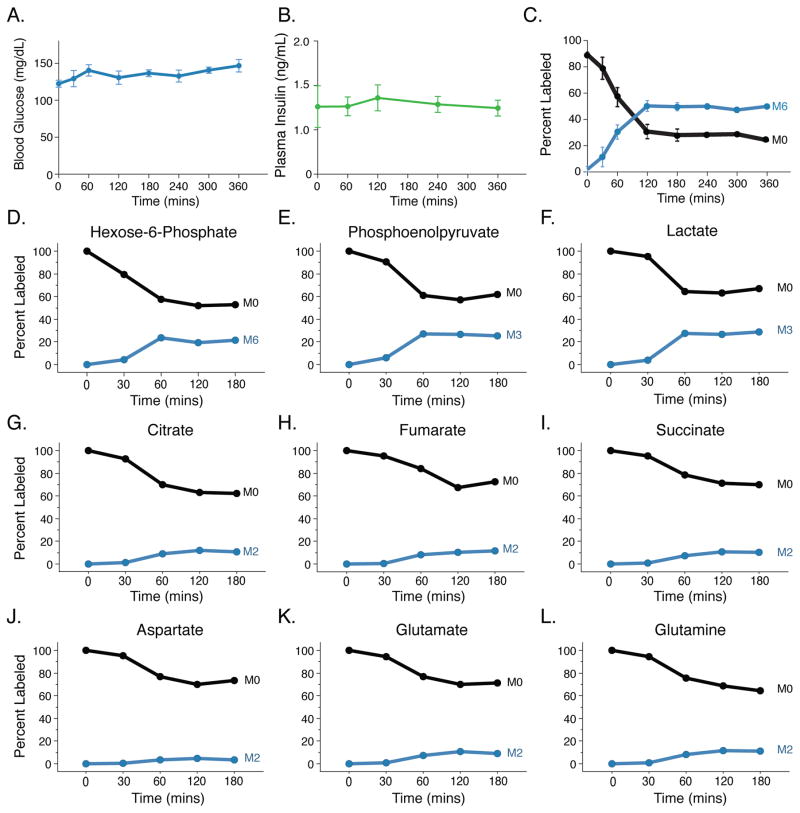
To study tissue glucose metabolism, we performed euglycemic [U-13C]glucose infusion in conscious unrestrained C57Bl6 mice (Ayala et al., 2010). Blood glucose and plasma insulin levels remained within the normal range throughout (Figures 1A, B), and isotopic enrichment of plasma glucose reached steady-state within 120 minutes (Figure 1C). To compare the labeling of tissues with different glucose uptake rates, we established the time required for glucose labeling of metabolites in tissues to reach isotopic steady-state. Because tissues with lower uptake will take longer to reach steady-state, this timing was established in normal tissues since most tumors, including Kras-driven lung tumors, exhibit elevated glucose uptake (Engelman et al., 2008). Tissue metabolites from wild-type [U-13C]glucose infused mice were analyzed by liquid-chromatography mass spectrometry (LC-MS) (Figures 1D–L, S1). The percentage of fully labeled lactate (M3 isotopomer) in lung tissue reached steady-state by 1-hour and was similar to the percentage of hexose-6-phosphate enrichment (Figures 1D, F). This suggests glucose is the primary source of lactate in normal lung tissue. Labeling of other glycolytic intermediates, TCA cycle metabolites, and TCA cycle-derived amino acids from glucose reached isotopic steady-state in the lung with similar kinetics (Figure 1D–L). The finding that TCA cycle intermediate labeling occurred with similar kinetics as glycolytic intermediates is consistent with glucose oxidation being a major fate of glucose in lung tissue. Labeling of glutamate and glutamine from glucose is also observed (Figures 1K, L).
Figure 1. Steady state labeling of metabolites in tissues.
(A) Blood glucose levels over time in mice infused with [U-13C]glucose. Values shown are mean +/− SEM, n = 4.
(B) Plasma insulin levels over time in mice infused with [U-13C]glucose. Values shown are mean +/− SEM, n = 4.
(C) Plasma enrichment of fully labeled glucose (M6) in animals infused with [U-13C]glucose over time. Values shown are mean +/− SEM, n = 4.
(D–F) Representative serial sacrifice (n=1) of WT animals after infusion of [U-13C]glucose. Labeling of glycolytic intermediates in lung tissue from wild type mice infused with [U-13C]glucose for the indicated time at 20mg/kg/min. The unlabeled (M0) and fully-labeled (M3 or M6) isotopomer is shown for each species.
(G–I) Representative serial sacrifice (n=1) of WT animals after infusion of [U-13C]glucose. Labeling of TCA intermediates in lung tissue from wild type mice infused with [U-13C]glucose for the indicated time at 20mg/kg/min. The unlabeled (M0) and prominent M2 labeled isotopomer are shown for each species.
(J–L) Representative serial sacrifice (n=1) of WT animals after infusion of [U-13C]glucose. Labeling of aspartate, glutamate, and glutamine in in lung tissue from wild type mice infused with [U-13C]glucose for the indicated time at 20mg/kg/min. The unlabeled (M0) and prominent M2 labeled isotopomer are shown for each species.
Glucose metabolism in lung tumor tissue
To examine metabolic differences between lung tumor and normal tissue we analyzed three mouse models of KrasG12D-driven NSCLC. The mouse models initiate lung tumors through different mechanisms and result in lesions of different grades and invasiveness. We utilized two autochthonous mouse models of NSCLC: 1) The KrasLA2/+(LA2) model involves spontaneous recombination of an oncogenic KrasG12D allele in somatic cells and initiates primarily low grade lung adenomas that progress to invasive adenocarcinomas over the period of 6–8 months (Johnson et al., 2001), 2) Intratracheal delivery of adenoviral Cre (Ad5-CMV-Cre) to the lungs of animals harboring conditional KrasLSL-G12D/+; p53loxP/loxP (KP) alleles results in higher grade, metastatic tumors that progress rapidly and cause death within 12–16 weeks (DuPage et al., 2009; Jackson et al., 2001; Jonkers et al., 2001; Tuveson et al., 2004). Cell lines derived from lung tumors arising in C57/Bl6 KP mice were introduced into the lungs of syngeneic animals via intratracheal injections (KPS), with lung tumor formation observed with even faster kinetics (Curtis et al., 2010). We also sought to determine how metabolism in these KrasG12D-driven tumors compared to xenograft tumors derived from two human lung cancer cell lines: 1) A549 cells with a KRAS mutation and 2) H1975 cells with an EGFR mutation.
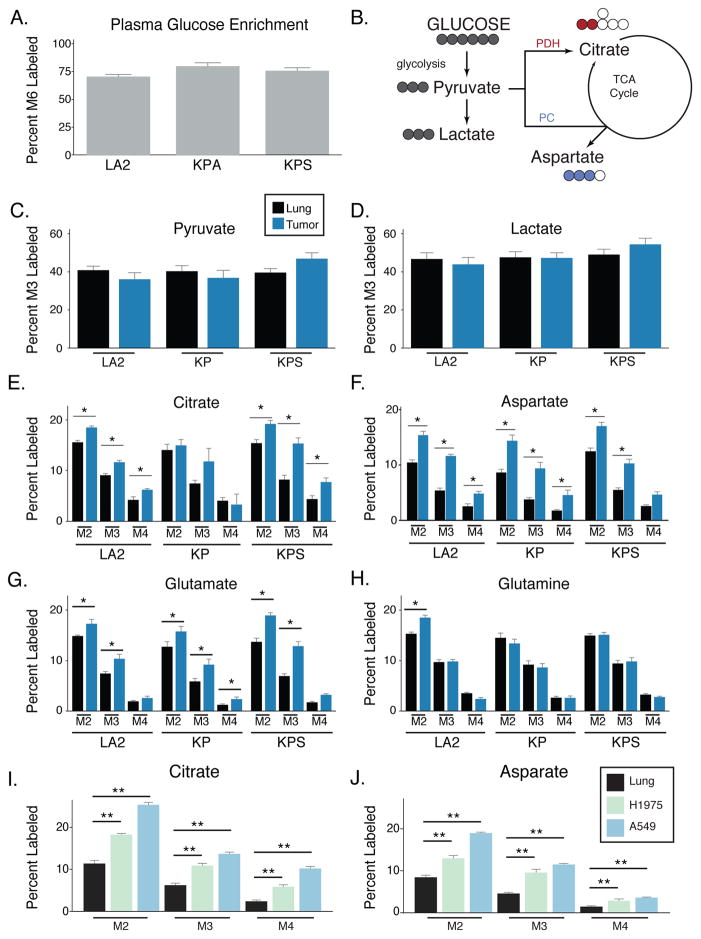
LA2, KP, and KPS mice underwent micro-computed tomography (μCT) scanning to select mice with tumors for analysis, and [U-13C]glucose was infused to reach a plasma enrichment of >65% (Figure 2A). To ensure isotopic steady-state was reached in both tumor and normal lung tissue, tissues were harvested after a 6-hour infusion and metabolites from tumor and normal lung tissue were analyzed using gas-chromatography mass-spectrometry (GC-MS) (Tables S1, S2). Labeling of lactate and pyruvate from glucose carbon was similar in tumors and adjacent lung tissue in all three models (Figures 2B, 2C, 2D), consistent with glucose acting as a source of lactate in these tissues. Lactate pool sizes were higher in KP and KPS tumors while levels of many other metabolites were similar to normal lung, suggesting that these tumors may be more glycolytic than LA2 tumors (Figure S2A, S2B). Despite no differences in percent lactate labeling, the increased uptake of glucose in tumors and the increased amount of lactate present in KP and KPS tumors argues relative glucose metabolism to lactate is increased in these NSCLC models compared to normal lung.
Figure 2. Increased glucose carbon contribution to the TCA in autochthonous K-ras driven lung tumors and xenografted lung cancer cells compared to adjacent lung.
(A) Enrichment of fully labeled glucose (M6) in plasma from mice after a 6-hour [U-13C]glucose infusion. (LA2, n = 4; KP, n = 6; KPS, n = 5).
(B) Schematic showing major isotopomer transitions from glucose to label glycolytic and TCA cycle intermediates. The dominant TCA cycle isotopomers derived from the oxidation of glucose-derived pyruvate by the PDH complex has two labels (as shown in red for citrate), while the dominant isotopomer derived from pyruvate carboxylase (PC) retains three labeled carbon (as shown in blue for aspartate).
(C, D) The percent M3 labeled pyruvate and lactate in lung (black) and lung tumors (blue) from mice following a 6-hour [U-13C]glucose infusion. (LA2, n = 4; KP, n = 6; KPS, n = 5).
(E–H) The percent labeling of citrate, aspartate, glutamate, and glutamine in lung (black) and lung tumors (blue) from mice following a 6-hour [U-13C]glucose infusion. The M2, M3 and M4 isotopomers are shown for each metabolite. (LA2, n = 4; KP, n = 6; KPS, n = 5 for tumor and adjacent lung).
For all panels, values represent the mean ± SEM. * Difference is statistically significant by two-tailed paired T-test, * p < 0.05.
(I, J) The percent labeling of citrate and aspartate in normal lung (black) and xenografts derived from H1975 cells (EGFR-driven human lung cancer cells, green) or A549 cells (KRAS-driven human lung cancer cells, blue) from mice following a 6-hour [U-13C]glucose infusion. The M2, M3 and M4 isotopomers are shown for each metabolite. (Lung, n = 8; H1975 tumor, n = 8; A549 tumor n = 8).
For all panels, values represent the mean ± SEM. * Difference is statistically significant by two-tailed paired T-test, * p < 0.05 or ** p < 0.01.
Glucose carbon can enter the TCA cycle as acetyl-CoA, with acetyl-CoA derived from oxidation of glucose-derived pyruvate via the pyruvate dehydrogenase complex (PDH) (Figure 2B). Thus, entry of PDH-derived glucose carbon into the TCA cycle results in isotopomer species with two labeled carbons (M2), and species with more labeled carbons are generated by addition of labeled acetyl-CoA to labeled oxaloacetate produced by TCA cycling (M3 and M4). When compared to the adjacent lung, the percent M2 and M4 citrate was increased in LA2 and KPS tumors, but not in KP tumors (Figure 2E). A similar pattern was observed in both glutamate and aspartate, amino acids that reflect TCA cycle labeling of αKG and oxaloacetate (OAA) (Figures 2F, 2G). Glucose label was observed in glutamine in both lung tumors and normal lung, suggesting both tissues synthesize glutamine from glucose (Figure 2H). Additionally, LA2 tumors exhibited an increase in M2 glutamine labeling, suggesting that glucose use to synthesize glutamine can be increased in some tumors (Figure 2H). To determine whether similar metabolism is observed in xenograft tumors derived from established human lung cancer cell lines, [U-13C]glucose was infused to an average final plasma enrichment of 78% (+/− 9%) in mice harboring tumors derived from A549 or H1975 cells. Analysis of the labeling patterns and metabolite pool sizes in these tumors relative to normal lung was similar to what was observed in LA2 and KPS tumors (Figures 2I, 2J, S2C, S2D, S2E and Table S2). Together, these data suggest that lung tumors oxidize glucose through PDH.
TCA cycle anaplerosis involving glucose-derived carbon and pyruvate carboxylase (Pcx) has been described as a feature of some human lung tumors (Cheng et al., 2011; Hensley and DeBerardinis, 2015; Sellers et al., 2015), and Pcx could contribute to the increased three carbon labeling (M3) of TCA cycle intermediates from glucose in all lung tumors examined (Figures 2B, 2E, 2F, 2G, 2I, 2J, S2C and S2D). To specifically examine Pcx activity in tumor tissue, we infused tumor-bearing KP animals with [1-13C]pyruvate to a final enrichment of 10.3 (+/− 2.1%) and assessed metabolite labeling in normal lung and lung tumor tissue (Figure S3A, S3B). The ratio of M1 labeled aspartate, citrate and malate to M1 labeled pyruvate was higher in tumors compared to normal lung (Figure S3B). Because the labeled carbon on [1-13C]pyruvate is lost during the PDH reaction, these data argue that pyruvate carboxylation is active in these tumors. Consistent with these studies, M3-labeled TCA cycle intermediates are observed in all models, with increased M3 species observed relative to M4 species (Figures 2E, 2F, 2G, 2I, 2J, S2C, and S2D). Furthermore, the presence of M1 labeled species in amounts that are similar to M2 labeled species, and in excess of M3 and M4 species, raises the possibility that addition of a labeled CO2, derived from glucose oxidation and pyruvate carboxylase activity contributes to the labeling patterns observed (Figures S3A, S3C). Taken together, analysis of glucose fate in lung tumors suggests that in addition to lactate production, glucose is metabolized by the PDH complex and Pcx in lung tumors, and glucose is a major source of TCA cycle carbon in these tumor models.
Glutamine metabolism in lung tumor and normal lung tissue
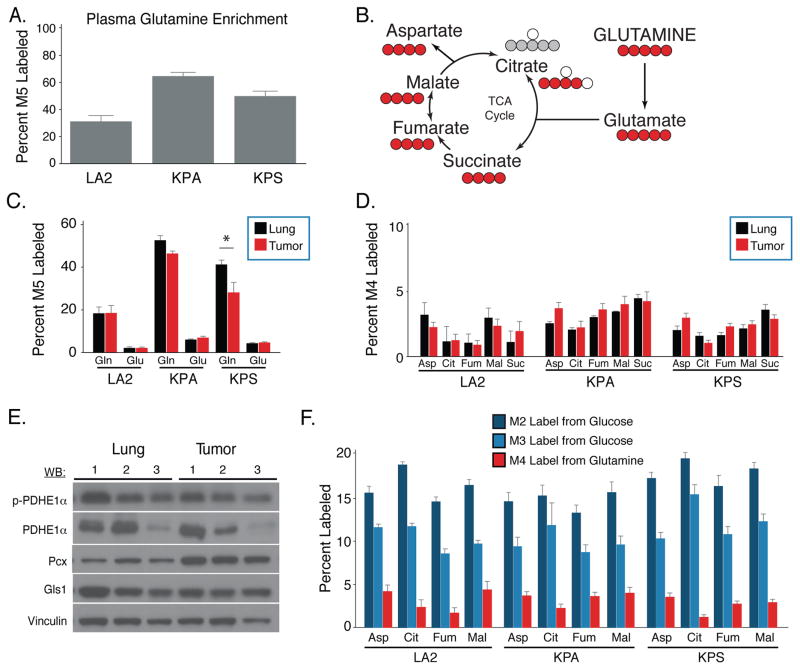
Studies of Kras-transformed cells in culture suggest glutamine can also be an important source of TCA cycle carbon. To determine the fate of glutamine in the KrasG12D-driven mouse tumor models, the human lung cancer cell line xenograft models, and normal lung tissue, [U-13C]glutamine was infused into tumor-bearing animals to a final plasma enrichment between 30–60% (Figure 3A). Surprisingly, we observed minimal labeling of glutamate and TCA intermediates in normal lung, autochthonous, syngeneic, or xenografted lung tumors despite the presence of labeled glutamine in these tissues (Figures 3B, 3C, 3D, S4A–C and Table S2). Furthermore, no significant differences in labeling of TCA cycle intermediates from glutamine was observed in tumors relative to normal lung tissue, with the exception of a small increase in M5 citrate in lung tumors relative to normal lung (Figures 3D, S4D). Cultured cells that were transplanted into the lung demonstrated decreased intracellular glutamine labeling as compared to normal lung despite comparable glutamate labeling, suggesting that these tumors may exhibit increased glutamine conversion to glutamate relative to normal lung tissue, although even in these tumors the labeling of TCA cycle intermediates was less than 5% (Figure S4A–C).
Figure 3. Glutamine carbon contributes minimally to the TCA cycle in K-ras driven lung tumors and adjacent lung.
(A) Enrichment of fully labeled glutamine (M5) in plasma from mice following 6-hour [U-13C]glutamine infusion. (LA2, n = 4; KP, n = 4; KPS, n = 4).
(B) Schematic showing major isotopomer transitions from glutamine to label TCA cycle intermediates. The dominant TCA cycle isotopomers produced by oxidative of glutamine metabolism have 4 labeled carbons for all species shown (red circles), while the dominant isotopomer of citrate from reductive glutamine metabolism is M5 (grey circles).
(C) The percent M5 labeled glutamate and glutamine in lung (black) and lung tumors (red) from mice following 6-hour [U-13C]glutamine infusion. (LA2, n = 4; KP, n = 4; KPS, n = 4).
(D) The percent M4 labeling of aspartate (Asp), citrate (Cit), fumarate (Fum), malate (Mal) and succinate (Suc) in the lung (black) and lung tumors (red) from mice following 6-hour [U-13C]glutamine infusion. (LA2, n = 4; KP, n = 4; KPS, n = 4).
(E) Western blot analysis of phospho-pyruvate dehydrogenase subunit E1α (p-PDHE1α), total PDHE1α (PDHE1α), pyruvate carboxylase (Pcx), and glutaminase (Gls1) expression in three representative KP lung tumors (Tumor) and normal lung tissue from three mice (Lung). Vinculin expression was also assessed in all samples as a loading control.
(F) The percent labeling of aspartate (Asp), citrate (Cit), fumarate (Fum), and malate (Mal) in tumor tissue from the indicated models after a 6 hour infusion of [U-13C]glucose and [U-13C]glutamine. Values were normalized to plasma enrichment of glucose or glutamine to allow comparison of the indicated isotopomers. (For [U-13C]glucose infusions, LA2, n = 4; KP, n = 6; KPS, n = 5. For [U-13C]glutamine infusions, LA2, n = 4; KP, n = 4; KPS, n = 4).
For all panels, values represent the mean ± SEM. *Difference is statistically significant by two-tailed paired T-test, p < 0.05.
To understand whether enzyme expression levels reflected the metabolic phenotypes we observe, expression of Gls1, Pcx and Pdha1 in KP tumors were determined by Western blot and/or immunohistochemistry (Figure 3E, S4E, S4F). Of those enzymes, only Pcx expression was increased in KP tumors relative to normal lung. Phosphorylation of the E1α subunit of PDH (encoded by the Pdha1 gene) can limit flux through the PDH complex to decrease glucose oxidation. In line with increased glucose labeling of TCA cycle intermediates, levels of phosphorylated PDHE1α were similar in both tumor and normal lung (Figure 3E). Furthermore, in all three KrasG12D-driven mouse cancer models labeling of TCA intermediates from glucose is observed while labeling from glutamine is minimal, suggesting that glucose rather than glutamine is a major source of carbon for the TCA cycle in these lung tumors (Figure 3F).
Environment influences the metabolic phenotype of KrasG12D lung cancer derived cells
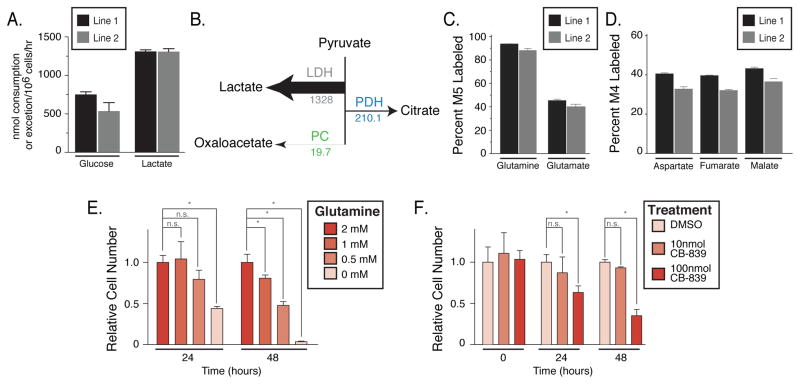
To understand whether cells derived from autochthonous tumors retained the same metabolic phenotypes in cell culture, we traced [U-13C]glucose and glutamine in cell lines derived from lung tumors arising in KP mice. Consistent with previous studies examining glucose metabolism in Ras-transformed cell lines, KP tumor-derived cell lines convert glucose to lactate at nearly a 1:2 molar ratio, suggesting the primary fate of glucose is lactate in these cells in culture (Figure 4A). To quantify the relative fate of glucose carbon under these conditions, we performed metabolic flux analysis (MFA) using stable isotope tracing of [1,2-13C]glucose and [U-13C]glutamine (Figure 4B, Tables S3–7 and Supplemental Methods). These data support that the dominant fate of glucose in these lung cancer cells in culture is lactate, while relatively little glucose-derived carbon contributes to the TCA cycle. In addition, as observed in most cultured cells, glutamine labels TCA intermediates to high levels when the lung cancer cells are cultured in either 2D or 3D culture conditions (Figures 4C, 4D, S5A). Consistent with glutamine metabolism by these cells, their ability to proliferate in culture is dependent on the concentration of glutamine in the media (Figure 4E), and they are sensitive to the glutaminase inhibitor CB-839 (Figure 4F) (Gross et al., 2014). Dose dependent reduction in M5-glutamate was observed with addition of CB-839 while M5-glutamine levels remained constant (Figure S5B, S5C), confirming that CB-839 inhibits glutaminase in these cells. These data suggest that despite a lack of preferential glutamine use by tumors in vivo, cell lines derived from KP lung tumors rely on glutamine metabolism for proliferation in vitro. Furthermore, because transplantation of the same KP cells back into the lung (KPS model) results in tumors with a metabolic phenotype similar to spontaneously arising lung cancers, the lung tissue environment must be an important determinant of lung cancer nutrient metabolism.
Figure 4. Metabolism of glucose and glutamine by cell lines derived from KP lung tumors.
(A) Rate of glucose consumption and lactate excretion by two independently derived lung cancer cell lines from KP lung tumors (Line 1 and Line 2), n = 3.
(B) Absolute fluxes downstream of pyruvate calculated from glucose and glutamine 13C labeling data in the lung cancer cell lines derived from KP lung tumors. The values shown for each flux are fmol/cell/hr, and the arrows indicating each flux are to scale.
(C) The percent M5 labeled glutamate and glutamine from [U-13C]glutamine in the lung cancer cell lines derived from KP lung tumors, n = 3.
(D) The percent M4 labeled aspartate, fumarate and malate from [U-13C]glutamine in the lung cancer cell lines derived from KP lung tumors, n = 3.
(E) Equal numbers of the lung cancer cell lines derived from KP mice were placed in media containing the indicated concentration of glutamine. Relative cell numbers present after 24 and 48 hours are shown, n=3.
(F) Equal numbers of the lung cancer cell lines derived from KP mice were plated, and then cultured in the presence of vehicle alone (DMSO) or the indicated concentration of CB-839, a glutaminase inhibitor. Relative cell numbers present at the time of vehicle or CB-839 addition or after 24 or 48 hours of exposure are shown, n=3.
For all panels, values represent mean ± SD, * denotes difference is statistically significant by two-tailed T-test, p < 0.05.
KrasG12D tumors do not preferentially rely on glutamine for tumor growth
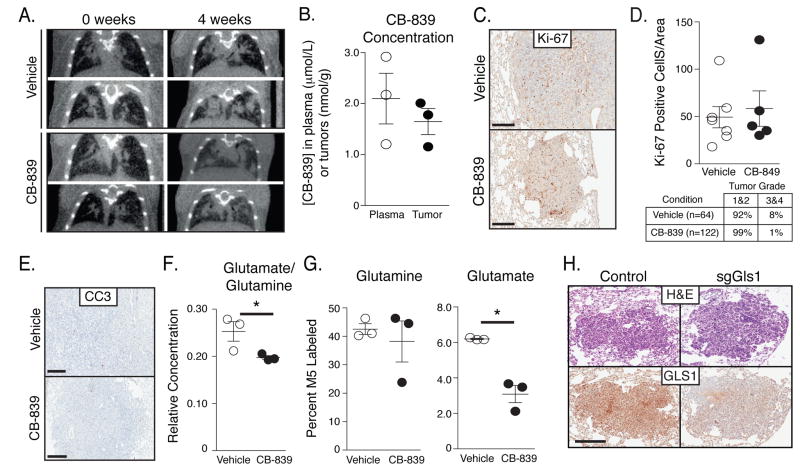
To determine whether the metabolic phenotype observed in lung tumors reflects metabolic dependencies of these tumors, we examined whether glutaminase is required for KP tumor growth in vivo. Tumor-bearing KP mice with lung tumors identified using micro-computed tomography imaging (μCT) were treated with vehicle or 200mg/kg oral CB-839 (Figure 5A). Examination of tumor size by μCT scans showed no differences after 4 weeks of continuous treatment with once daily dosing of vehicle or CB-839. The level of CB-839 in the tumor was approximately 1.5 nmol/g of tissue, an amount similar to that reported previously (Figure 5B) (Gross et al., 2014). Furthermore, when lung tumors were examined by μCT scan after 4 weeks of treatment with vehicle or CB-839, gross tumor burden was similar in both groups (Figure 5A). When tumors were harvested the histological appearance of the tumors was unchanged and no change in proliferation assessed by Ki-67 staining was observed with CB-839 treatment (Figures 5C, 5D). Additionally, unlike sensitive tumor models (Gross et al., 2014), no increase in apoptosis was observed by cleaved-caspase-3 staining (Figure 5E). To confirm CB-839 exposure in this model was adequate to inhibit glutaminase, lung tumor-bearing animals treated with vehicle or CB-839 were infused with [U-13C]glutamine prior to tumor collection. Consistent with glutaminase inhibition in CB-839 treated tumors, relative intratumoral glutamate-to-glutamine concentration ratios decreased (Figure 5F). In addition, although the percent labeled glutamine (M5) in the tumors was similar in vehicle and CB-839 treated mice, significantly less labeled glutamate was noted in tumors exposed to CB-839 (Figure 5G). These data are consistent with CB-839 inhibiting glutaminase in the tumors and suggest that this degree of glutaminase inhibition is not sufficient to slow tumor growth in this model.
Figure 5. KP-lung tumors are not dependent on high glutaminase activity.
(A) Coronal μCT scan sections showing the lungs of mice prior to treatment, and after treatment for 4 weeks with vehicle or the glutaminase inhibitor CB-839 treated mice. The images are representative of those obtained from 5 vehicle- and 6 CB-839-treated mice after 4 weeks of CB-839 treatment.
(B) Concentration of glutaminase inhibitor CB-839 in plasma and tumor, with each circle representing a value in a single animal. The mean value and SD are also shown.
(C) Ki-67 staining of lung tumor sections obtained from KP mice treated with vehicle or CB-839 for 4 weeks. The staining shown is representative of that observed in tissue from 5 vehicle- and 6 CB-839-treated mice. Scale bar: 200μM.
(D) Quantification of Ki-67 staining in KP lung tumor sections obtained from mice treated with vehicle or CB-839 for 4 weeks. Each point represents data from an independently derived tumor, and the mean +/− SEM is indicated as are the % tumors graded histologically as grade 1&2 or 3&4.
(E) Representative immunohistochemical staining of Cleaved-Caspase 3 (CC3) in vehicle versus CB-839 treated mice. Scale bars indicate 200μM.
(F) Relative concentration of glutamate to glutamine in tumors from KP mice treated with vehicle or glutaminase inhibitor for 4 weeks.
(G) The percent M5 labeled glutamine and glutamate in lung tumors from KP mice infused for 6-hours with [U-13C]glutamine. Animals were treated for 4 weeks with vehicle or CB-839 as indicated. Each point represents data from an independently derived tumor, and the mean +/− SEM is indicated.
(H) Short guide RNA sequences to disrupt Gls1 or a non-targeted control were delivered along with Cas9 and Cre recombinase to induce KP lung tumors. Representative IHC images examining Gls1 expression in tumors arising in mice exposed to control sgRNAs, or sgRNAs that target Gls1 is shown. Scale bar: 200μM.
*Difference is statistically significant by two-tailed T-test, p < 0.05.
To test glutaminase dependency genetically, we attempted to delete Gls1 in KP tumor-derived cell lines using CRISPR/Cas9-based gene disruption (Figure S6A). However, in line with dependence on glutamine metabolism for proliferation in culture, we were unable to generate any Gls1 knockout cell lines (0/71 screened clones). This suggests that Gls1 is essential for KP tumor cell proliferation in culture. To assess whether Gls1 is required for the growth of autochthonous KP tumors, we utilized CRISPR/Cas9-based somatic editing to disrupt Gls1 in lung tumor tissue. A lentiviral vector containing sgRNA to Gls1, Cas9, and Cre-recombinase (pSECC) was delivered to the lungs of KP mice (Sánchez-Rivera et al., 2014), and a spectrum of tumors formed with both high and low Gls1 expression (Figure 5H, S6K). These findings argue that high Gls1 activity or expression is not required for lung tumor initiation or progression in this model and is consistent with pharmacological inhibition of glutaminase having no effect on growth of these tumors. These findings are also consistent with the metabolite tracing data showing minimal contribution of glutamine carbon to the TCA cycle in these Kras-driven lung tumors.
Mitochondrial metabolism of pyruvate is essential for tumor formation in vivo
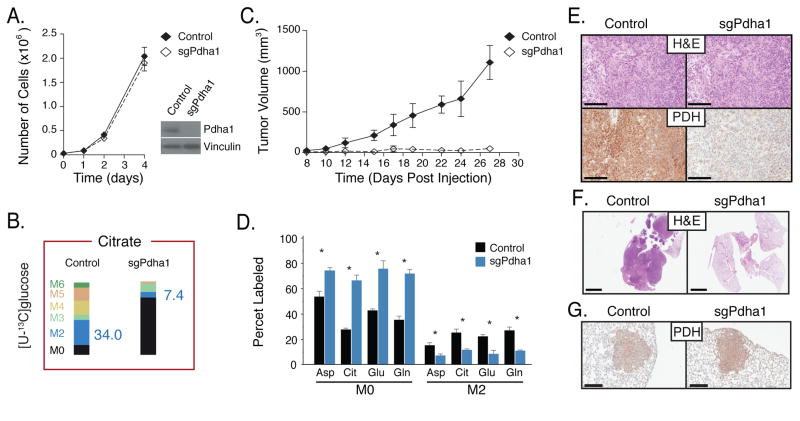
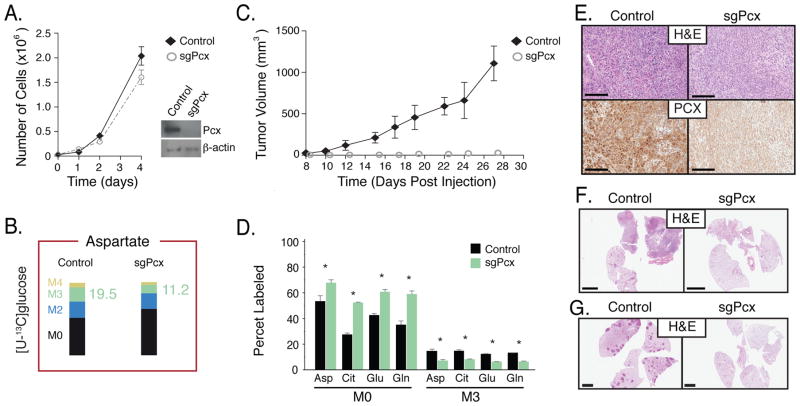
To determine whether enzymes involved in mitochondrial glucose metabolism are required for cancer cell proliferation in culture and in vivo, we deleted enzymes required for entry of glucose carbon into the TCA cycle. The PDH complex and Pcx are required for oxidative glucose metabolism, with PDH generating acetyl-CoA for the TCA cycle and Pcx required for the generation of oxaloacetate from pyruvate as a source of anaplerotic TCA cycle carbon. Using CRISPR/Cas9, both Pcx and the E1α subunit of PDH (Pdha1) were disrupted in cancer cells derived from KP lung tumors (Figures S6B, S6C). Stable cell lines lacking Pdha1 and Pcx were confirmed to harbor disruptions in these genes, and their protein products are not expressed (Figures 6A, 7A, S6B, and S6C). The ability to oxidize glucose and to utilize glucose for anaplerosis was also decreased following deletion of Pdha1 or Pcx (Figure 6B, 7B, S7). Consistent with previous studies arguing glucose oxidation is not required for proliferation of most cells in culture, we observed minimal difference in the proliferation of Pdha1- and Pcx-deleted cells relative to wild-type cells in 2D or in 3D culture conditions (Figures 6A, 7A, S6D, S6F, S6H). However, when transplanted into the flanks of syngeneic recipient mice, cells harboring mutations in either Pcx or Pdha1 fail to form tumors that grow larger than the initial mass of cells injected (Figures 6C, 7C, S6E, and S6G). We confirmed that unlike tumors that form from control cells, the Pcx- and Pdha1-deleted cells still present at the injection site as cytostatic masses did not express Pcx and Pdha1, respectively (Figures 6E, 7E), and showed changes in metabolite labeling that are consistent with enzyme deletion (Figure 6D, 7D). In addition, when transplanted orthotopically into the lungs of syngeneic mice, tumor formation from Pcx- and Pdha1-deleted cells was severely compromised (Figures 6F, 7F, S6I). Finally, disruption of Pdha1 or Pcx using pSECC-derived Lentivirus to deliver sgRNAs and Cas9 together with Cre-recombinase to the lungs of KP mice (Sánchez-Rivera et al., 2014) resulted in tumors that retained Phda1 expression (animals injected with sgPhda1, Figure 6G and S6J) or no detectable tumors (animals injected with sgPcx, Figure 7G and S6J). These data suggest that both Pcx and Pdha1 are required for tumor initiation and proliferation in vivo, and are consistent with TCA cycle metabolism being an important fate of glucose in Kras-driven lung tumors. These findings also argue that assessment of nutrient fates in tumors reflects metabolic dependencies of cancer cells in a physiological tissue environment.
Figure 6. Lung cancer cells require Pdha1 for tumor formation in vivo.
(A) sgRNAs targeting Pdha1 or control were introduced into lung cancer cells derived from KP lung tumor. Proliferation of control and Pdha1 disrupted cells in culture is shown (n=3), as is a Western blot analysis showing Pdha1 expression from isogenic clones.
(B) The same cells described in (A) were cultured in the presence of [U-13C]glucose. Percent labeling of citrate isotopomers is shown for Control and Pdha1-disrupted cell lines. The percent M2 isotopomer of citrate that can be generated from glucose via flux through Pdha1 is also presented in blue, and percent labeling of other isotopomers of citrate displayed, n = 3.
(C) The same cells described in (A) were introduced as allografts into the flanks nu/nu mice. Tumor growth over time is shown. n = 4/cell line.
(D) The same cells described in (A) were introduced as allografts into the flanks of mice. The percent labeling of aspartate (Asp), citrate (Cit), glutamate (Glu), and glutamine (Glu) in lung tumors derived from control cells (black) or material present at the injection site from Pdha1-disrupted cells (blue) was determined following a 6-hour [U-13C]glucose infusion. The M0 and M2 isotopomers are shown for each metabolite. All values in mean ± SEM. *Difference is statistically significant by two-tailed T-test, p < 0.05.
(E) The same cells described in (A) were introduced as allografts into the flanks of nu/nu mice. Immunohistochemistry assessing Pdha1 expression in lung tumors derived from control cells or material present at the injection site from Pdha1-disrupted cells is shown. Scale bar: 200μM.
(F) Representative H&E staining of the same cells described in (A) 4 weeks after orthotopic transplantation into the lungs of nu/nu mice. Scale bar: 200μM.
(G) Representative immunohistochemical staining for Pdha1 in tumors arising in KP mice infected with pSECC containing a control sgRNA (Control) or sgPdha1 (n = 25 tumors from ctrl and 18 tumors from sgPdha1 from 3 mice analyzed, all tumors retained Pdha1 expression). Scale bar indicates 200μm.
Figure 7. Lung cancer cells require Pcx for tumor formation in vivo.
(A) sgRNAs targeting Pcx or control were introduced into lung cancer cells derived from KP lung tumor. Proliferation of control and Pcx disrupted cells in culture is shown (n=3), as is a Western blot analysis showing Pcx expression from isogenic clones.
(B) The same cells described in (A) were cultured in the presence of [U-13C]glucose. Percent labeling of aspartate isotopomers is shown for Control and Pcx-disrupted cell lines. The percent M3 isotopomer of aspartate that can be generated from glucose via flux through Pcx is also presented in green, and percent labeling of other isotopomers of aspartate displayed, n = 3.
(C) The same cells described in (A) were introduced as allografts into the flanks of nu/nu mice. Immunohistochemistry assessing Pcx expression in lung tumors derived from control cells or material present at the injection site from Pcx-disrupted cells is shown.
(D) The same cells described in (A) were introduced as allografts into the flanks of mice. The percent labeling of aspartate (Asp), citrate (Cit), glutamate (Glu), and glutamine (Glu) in lung tumors derived from control cells (black) or material present at the injection site from Pcx-disrupted cells (green) was determined following a 6-hour [U-13C]glucose infusion. The M0 and M3 isotopomers are shown for each metabolite. All values in mean ± SEM. *Difference is statistically significant by two-tailed T-test, p < 0.05.
(E) The same cells described in (A) were introduced as allografts into the flanks of nu/nu mice. Immunohistochemistry assessing Pcx expression in lung tumors derived from control cells or material present at the injection site from Pcx-disrupted cells is shown. Scale bar: 200μM.
(F) Representative H&E staining of the same cells described in (A) 4 weeks after orthotopic transplantation into the lungs of mice. Scale bar: 200μM.
(G) Representative hematoxylin and eosin staining of KP mice infected with pSECC containing a control sgRNA (Control) and sgPcx (n = 4 mice analyzed, no tumors were observed in the sgPcx mice). Scale bar indicates 2mm.
DISCUSSION
These findings suggest increased glucose uptake in Kras-driven NSCLC is used to support both increased lactate production and increased TCA cycle metabolism, challenging the notion that tumors switch from oxidative glucose metabolism to aerobic glycolysis (Vander Heiden et al., 2009). The exclusive metabolism of glucose to lactate by tumors has also been questioned by analysis of glucose fate in glioblastoma, lung cancers and liver cancers (Maher et al., 2012; Marin-Valencia et al., 2012; Yuneva et al., 2012). The finding that oxidative metabolism of glucose is increased in vivo raises the possibility that the use of glucose to support the TCA cycle is important to support cell proliferation under the conditions present in tissues.
It has been hypothesized that high flux through glycolysis benefits tumors by allowing ATP production in the absence of oxygen and by promoting anabolic metabolism (Gatenby and Gillies, 2004; Vander Heiden et al., 2009). Increased lactate production supports NAD+ regeneration in the absence of oxygen consumption and may provide other benefits to tumor cells related to altered pH or supplying lactate to other tumor cells (Sonveaux et al., 2008). Near quantitative conversion of glucose to lactate is a phenotype observed in proliferating cells in culture, raising the possibility that a similar phenotype is present in proliferating cells of lung tumors. Because a substantial fraction of the tumor is not actively proliferating, the increase in oxidative glucose metabolism might reflect the metabolic phenotype of non-proliferating cells. In mouse breast cancer, proliferating and non-proliferating cells regulate glucose metabolism differently such that the non-proliferating cells select for increased pyruvate kinase activity, a regulatory state associated with increased oxidative glucose metabolism (Israelsen et al., 2013). Differences in lactate production are present across the NSCLC models considered for this study. Whereas the KP, KPS, and xenograft models form tumors with a higher proliferative index, the LA2 model forms lower grade tumors. Consistent with a model where increased lactate production tracks with increased proliferation, lactate levels are increased in KP, KPS, and xenograft tumors relative to normal tissue but not in LA2 tumors. However, KPS tumors proliferate faster than KP tumor, and if anything lactate levels are lower in the KPS tumors arguing that lactate levels do not always scale with proliferation. While all of the tumor models examined contain a high proportion of cancer cells relative to other cell types, differences in glucose metabolism are observed in different regions of human cancers (Hensley et al., in press). Thus, unknown factors driving tumor heterogeneity might also contribute to the metabolic phenotype differences observed in tumor tissue.
Decreased PDH flux has been described as a property of tumors, with PDH reactivation proposed as a cancer therapy (Michelakis et al., 2008). Surprisingly, glucose metabolism through PDH is increased in tumors relative to normal lung tissues and the cancer cells are dependent on this enzyme for tumor formation. Increased inhibitory phosphorylation of PDHE1α was also not observed in these tumors. One possibility is the lack of increased PDHE1α phosphorylation reflects increased oxygenation. Indeed, the lung is a relatively well-perfused organ providing access to circulating nutrients and oxygen, although as lung cancers grow vascularization can become limiting and even large human lung tumors have elevated PDH flux (Hensley et al. in press). This suggests that oxidative glucose metabolism may provide benefits to tumor cells such as the production of aspartate (Sullivan et al., 2015) regardless of tumor size.
Despite exposure to among the highest oxygen tensions in the body, normal lung uses 30–50% of consumed glucose to make lactate and 20% to make CO2 while contributing relatively minor amounts to protein and lipid biosynthesis (Fisher, 1984). Outside of cytosolic ATP production and NAD+ regeneration, the benefit for normal lung physiology of relatively high flux to lactate remains unclear. Although the flux from glucose to lactate in the lung is high relative to other tissues, glucose also supports production of TCA cycle intermediates in the lung. Notably, glucose fuels glutamate and glutamine production in lung tissue in rats and humans, and lung tissue is among the highest producers of glutamine (Hensley et al., 2013).
Glutamine is the most abundant amino acid in tissue culture media and in the blood, yet in contrast to observations in cell culture, blood glutamine contributes minimally to both normal lung and lung tumor metabolism. Labeled glutamine was abundant in both tissues suggesting ineffective glutamine delivery cannot explain why this amino acid is not used as a fuel. Instead, both normal lung and lung tumors synthesize α-ketoglutarate, glutamate and glutamine from glucose-derived carbon. This is consistent with findings in MYC-driven lung tumors and glioblastoma (Maher et al., 2012; Marin-Valencia et al., 2012; Yuneva et al., 2012) and may reflect the glutamine use to excrete excess nitrogen from amino acid catabolism (Stumvoll et al., 1999). Urea production for nitrogen excretion is restricted primarily to the liver and kidney in mammals, and glutamine carries excess nitrogen from peripheral tissues to these organs. Because the use of glutamine as a carbon source produces ammonia, glutaminase activity may be better tolerated in tissue culture where large media volumes prevent ammonia accumulation to toxic levels. However, because of an increased need for glutathione (Sellers et al., 2015), nucleotides, and amino acids; tumor nitrogen requirement is higher than that of normal tissues (Mayers and Vander Heiden, 2015). Nevertheless, the net production of glutamine by lung tumors despite the availability of this amino acid from the circulation suggests alternative sources of nitrogen are used, possibly reflecting the propensity of RAS-cancers to catabolize extracellular protein (Commisso et al., 2013; Kamphorst et al., 2015).
The use of pyruvate carboxylase appears to be an important for anaplerosis in tumors in vivo (Fan et al., 2009; Sellers et al., 2015). Consistent with these findings, genetic deletion of Pcx prevents tumor growth while genetic deletion or inhibition of glutaminase has no observable effect. The fact that opposite dependencies were suggested by cell culture experiments highlights the importance of selecting targets for cancer therapy based on nutrient use in a relevant tissue context. The possibility that tissue context is an important determinant of how tumors utilize nutrients might also explain why chemotherapies targeting nucleotide metabolism show efficacy based on cancer tissue of origin. More directly, it suggests that targeting PDH or PCX might inhibit lung tumor growth. In fact, lipoate derivatives act in part through PDH activity suppression and can be potent anticancer drugs in vivo (Zachar et al., 2011).
This study highlights the importance of model selection to identify metabolic cancer targets. Cell culture conditions are non-physiological with respect to nutrients, oxygen, and tolerance for excretion of toxic metabolites such as ammonia. Cell culture also selects for the most rapidly proliferating cells, and many tumor cells cannot be cultured in vitro. The fact that transplanted tumors exhibit a phenotype more similar to tumors arising in the lung that are never exposed to cell culture suggests that environment has a greater impact on how nutrients are utilized than genetic or epigenetic selection associated with cell line formation. The similarities between Kras-driven NSCLC, an EGFR-driven lung tumor, and published studies of genetically distinct tumors further argues that tissue environment can dictate metabolic phenotypes across tumor types. This highlights the importance of considering environmental context in addition to genetics in metabolism studies, particularly in considering how best to target cancer metabolism.
EXPERIMENTAL PROCEDURES
Mouse Cancer Models
All animal studies were approved by the MIT Committee on Animal Care. For autochthonous models, mice from a mixed 129/Sv and B6 genetic background were used. C57Bl6/J or nu/nu mice were used for allografts and xenografts respectivley. Flank tumor growth was measured with calipers. Lung tumor volumes were assessed using an eXplore CT120-whole mouse μCT (GE Healthcare) with MicroView 3D Image Viewing (Parallax-Innovations) and OsiriX (OsiriX-Viewer) used for image analysis.
Glucose and Glutamine Infusion
Arterial and venous catheters were surgically implanted into the jugular veins and/or carotid artery of animals 3–4 days prior to infusions. Infusions were performed in free-moving conscious animals after a 6 hour fast. Mice were infused for a 6-hours at the specified rate prior to terminally anesthetia with sodium pentobarbital and rapid tissue harvest. Tissue was rapidly frozen using a BioSqueezer (BioSpec Products) to quench metabolism and stored at −80°C prior to metabolite extraction. Plasma insulin levels were determined using an ultrasensitive mouse insulin ELISA kit (Crystal Chem, #90080). Blood glucose levels from the same mice were measured using a handheld glucometer (One Touch).
Cell Culture
Cell lines from KP lung tumors were established by standard protocols. Isogenic clones were selected for comparison of CRISPR cell lines. For labeling studies and MFA, cells were cultured for 24 hours in DMEM containing 17.5mmol/L [U-13C]glucose, [1,2-13C]glucose, or 4mmol/L [U-13C]glutamine (Cambridge Isotopes Laboratories) prior to metabolite extraction. For labeling in 3D culture, Adherence plates (Corning) were used. Proliferation in 3D was assessed using CellTiter-Glo (Promega).
Metabolite Measurement
For metabolite extraction, 10–40mg tissue was weighed and homogenized cryogenically (Retsch Cryomill) prior to extraction in chloroforom:methanol:water (400:600:300). Samples were centrifuged to separate aqueous and organic layers, and polar metabolites dried under nitrogen gas for subsequent analysis by mass-spectrometry. For liquid chromatography mass spectrometry (LC-MS), dried metabolites were resuspended in water based on tissue weight with valine-D8 included as an injection control. Polar metabolites were analyzed using a Nexera X2 U-HPLC (Shimadzu, Marlborough, MA) and a Q-Exactive hybrid quadrupole orbitrap mass spectrometer (Thermo Fisher Scientific; Waltham, MA). Hydrophobic interaction liquid chromatography (HILIC) was used for positive ion mode (Townsend et al., 2013; Wang et al., 2011) and negative ion mode (Avanesov et al., 2014) as previously descrbied. MS data were processed using Tracefinder (version 3.2, Thermo Fisher Scientific; Waltham, MA). For gas chromatography mass spectrometry (GC-MS) dried metabolites were dissolved in 10μL/10mg wet tissue weight of 2% methoxyamine hydrochloride in pyridine (Sigma) and held at 37C for 1.5h, norvaline was added as an injection control. Tert-butyldimethylsilyl derivatization was initiated by adding 15μL/10mg wet weight of N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide + 1% tert-butyldimethylchlorosilane (Sigma) and incubated at 37C for 1hr. GC-MS analysis was performed using an Agilent 7890 GC equipped with 30m DB-35MS capillary column connected to an Agilent 5975B MS operating under electron impact ionization at 70eV with helium as a carrier gas and the detector in scanning mode. MIDs were corrected for natural isotope abundance as previously reported (Commisso et al., 2013). Glucose, lactate, glutamine, and glutamate were measured using YSI biochemistry analyzer (Yellow Springs Instruments, Yellow Springs, OH). Exponential growth over the culture period was assumed for flux calculations.
CRISPR/Cas9 Gene Editing
sgRNAs were designed using E-CRISP (e-crisp.org) to Pdha1, Pcx, and Gls1 with restriction enzyme compatible sites and a G was added to the +1 position where not already present for U6 transcription. sgRNAs cloned in U6-sgRNA-EFS-Cas9-2A-Puro or U6-sgRNA-EFS-Cas9-2A-Cre (pSECC) were used for cell lines and in vivo experiments respectively as previously described (Sánchez-Rivera et al., 2014).
Western Blot and Immunohistochemistry
Western blotting was conducted using manufacturer’s recommended concentrations of antibodies against Vinculin (Sigma/V9131), Pcx (Thermo-Fisher/PA5-23055), Gls1 (Abcam/AB93434), p-Pdha1 (Calbiochem/AP1062), and Pdha1 (Proteintech/18068). For IHC, whole lungs and tumors were perfused with 4% PFA and fixed overnight at room temperature. Tissues were paraffin embedded and cut into 5μM sections. Following antigen retrieval, sections were stained with Ki-67 (BD Pharmingen/556003), Cleaved Caspase 3 (Cell Signaling Technology/#9661), or the above antibodies using ABC Vectastain kit (Vector Laboratories) and developed with DAB and counterstained with hematoxylin and eosin. Quantification of Ki-67 and Cleaved Caspase 3 were performed using ImageJ (http://imagej.nih.gov).
Glutaminase Inhibitor
Animals were treated with 200mg/kg CB-839 or vehicle for 4 weeks after observing initial masses by μCT in the lung. The vehicle contained 25% (w/v) hydroxypropyl-β-cyclodextrin in 10mmol/L citrate, pH 2.0 and CB-839 was formulated at 20mg/mL for a final dosing volume of 10mL/kg as previously described (Gross et al., 2014). CB-839 was quantified in tissues and plasma using LC-MS.
Statistical Analysis
Two-tailed paired and unpaired Student’s T-test were performed for all experiments unless otherwise specified. Results for independent experiments are presented as mean ± SEM; results for technical replicates are presented as mean ± SD; results for MFA are presented with 95% confidence intervals.
Supplementary Material
HIGHLIGHTS.
Metabolic phenotyping of tumors can identify essential metabolic pathways
Kras-driven lung tumors require pyruvate carboxylase and pyruvate dehydrogenase
Kras-driven lung tumors are less dependent on glutaminase than cultured cells
Tissue environment is an important determinant of tumor metabolic phenotypes
Acknowledgments
We thank Natalia Drosu for reagents and advice, the Swanson Biotechnology Center for mouse tissue processing, and the Vander Heiden Laboratory for thoughtful discussions. S.M.D. and A.L. were supported by an NSF Graduate Research Fellowship and T32GM007287. T.P. received support from Hope Funds for Cancer Research Fellowship. B.A.O. received support from K08HL119355 and Gilead Sciences Research Scholars Program. D.Y.G. received support from T32GM007753. M.G.V.H acknowledges support from the Broad Institute SPARC program, the Ludwig Center at MIT, the Burroughs Wellcome Fund and the NIH (P30CA1405141, R01CA168653).
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization: S.M.D, T.P., B.A.O., M.G.V.H. Methodology: S.M.D, T.P., B.A.O., M.A.K., A.L., Formal Analysis: S.M.D., T.P., A.L. Investigation: J.E.H., A.L., J.P.O, M.R.B., A.K.J., J.P.O., K.A.P., D.Y.G., L.B.S., T.M.W., L.S., C.R.C. Writing: S.M.D., M.G.V.H. Visualization: S.M.D. Supervision: T.J., C.B.C., M.G.V.H. Funding Acquisition: S.M.D., B.A.O., M.G.V.H.
References
- Avanesov AS, Ma S, Pierce KA, Yim SH, Lee BC, Clish CB, Gladyshev VN. Age- and diet-associated metabolome remodeling characterizes the aging process driven by damage accumulation. Elife. 2014:e02077–e02077. doi: 10.7554/eLife.02077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala JE, Samuel VT, Morton GJ, Obici S, Croniger CM, Shulman GI, Wasserman DH, McGuinness OP Consortium NMMPC. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech. 2010:525–534. doi: 10.1242/dmm.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet S, Archer SL, Allalunis-Turner J, Haromy A, Beaulieu C, Thompson R, Lee CT, Lopaschuk GD, Puttagunta L, Bonnet S, et al. A Mitochondria-K+ Channel Axis Is Suppressed in Cancer and Its Normalization Promotes Apoptosis and Inhibits Cancer Growth. Cancer Cell. 2007:37–51. doi: 10.1016/j.ccr.2006.10.020. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature Reviews Cancer. 2011:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Cheng T, Sudderth J, Yang C. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells (PNAS) 2011 doi: 10.1073/pnas.1016627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013:633–637. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis SJ, Sinkevicius KW, Li D, Lau AN, Roach RR, Zamponi R, Woolfenden AE, Kirsch DG, Wong K-K, Kim CF. Primary tumor genotype is an important determinant in identification of lung cancer propagating cells. Cell Stem Cell. 2010:127–133. doi: 10.1016/j.stem.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson SM, Vander Heiden MG. METabolic adaptations in the tumor MYCroenvironment. Cell Metab. 2012:131–133. doi: 10.1016/j.cmet.2012.01.005. [DOI] [PubMed] [Google Scholar]
- DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, Maira M, McNamara K, Perera SA, Song Y, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan TWM, Lane AN, Higashi RM, Farag MA, Gao H, Bousamra M, Miller DM. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM) Mol Cancer. 2009:41–41. doi: 10.1186/1476-4598-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher AB. Intermediary metabolism of the lung. Environ Health Perspect. 1984:149–158. doi: 10.1289/ehp.8455149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature Reviews Cancer. 2004:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B, Janes JR, Laidig GJ, Lewis ER, Li J, et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther. 2014:890–901. doi: 10.1158/1535-7163.MCT-13-0870. [DOI] [PubMed] [Google Scholar]
- Hensley CT, DeBerardinis RJ. In vivo analysis of lung cancer metabolism: nothing like the real thing. J Clin Invest (American Society for Clinical Investigation) 2015:495–497. doi: 10.1172/JCI79188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Wasti AT, DeBerardinis RJ. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013:3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley CT, Faubert B, Yuan Q, Lev-Cohain N, Jin E, Kim J, Jiang L, Ko B, Skelton R, Loudat L, Wozak M, Klimko C, McMillan E, Butt Y, Ni M, Oliver D, Torrealba J, Malloy CR, Kernstine K, Lenkinski RE, Deberardinis RJ. Metabolic heterogeneity in human lung tumors. Cell. 2015 doi: 10.1016/j.cell.2015.12.034. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & Development. 2001:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Development. 2009:537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015:544–553. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshari KR, Sriram R, Van Criekinge M, Wilson DM, Wang ZJ, Vigneron DB, Peehl DM, Kurhanewicz J. Metabolic reprogramming and validation of hyperpolarized 13C lactate as a prostate cancer biomarker using a human prostate tissue slice culture bioreactor. Prostate. 2013:1171–1181. doi: 10.1002/pros.22665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, Son J, Cantley LC, Kimmelman AC. Pancreatic cancers rely on a novel glutamine metabolism pathway to maintain redox balance. Cell Cycle. 2013:1987–1988. doi: 10.4161/cc.25307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher EA, Marin-Valencia I, Bachoo RM, Mashimo T, Raisanen J, Hatanpaa KJ, Jindal A, Jeffrey FM, Choi C, Madden C, et al. Metabolism of [U-13 C]glucose in human brain tumors in vivo. NMR Biomed. 2012:1234–1244. doi: 10.1002/nbm.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Valencia I, Yang C, Mashimo T, Cho S, Baek H, Yang X-L, Rajagopalan KN, Maddie M, Vemireddy V, Zhao Z, et al. Analysis of tumor metabolism reveals mitochondrial glucose oxidation in genetically diverse human glioblastomas in the mouse brain in vivo. Cell Metab. 2012:827–837. doi: 10.1016/j.cmet.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayers JR, Vander Heiden MG. Famine versus feast: understanding the metabolism of tumors in vivo. Trends Biochem Sci. 2015:130–140. doi: 10.1016/j.tibs.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelakis ED, Webster L, Mackey JR. Dichloroacetate (DCA) as a potential metabolic-targeting therapy for cancer. Br J Cancer. 2008:989–994. doi: 10.1038/sj.bjc.6604554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onetti R, Baulida J, Bassols A. Increased glucose transport in ras-transformed fibroblasts: a possible role for N-glycosylation of GLUT1. FEBS Lett. 1997:267–270. doi: 10.1016/s0014-5793(97)00340-2. [DOI] [PubMed] [Google Scholar]
- Rodrigues TB, Serrao EM, Kennedy BWC, Hu D-E, Kettunen MI, Brindle KM. Magnetic resonance imaging of tumor glycolysis using hyperpolarized 13C-labeled glucose. Nat Med. 2014:93–97. doi: 10.1038/nm.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Rivera FJ, Papagiannakopoulos T, Romero R, Tammela T, Bauer MR, Bhutkar A, Joshi NS, Subbaraj L, Bronson RT, Xue W, et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature. 2014:428–431. doi: 10.1038/nature13906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers K, Fox MP, Bousamra M, Slone SP, Higashi RM, Miller DM, Wang Y, Yan J, Yuneva MO, Deshpande R, et al. Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J Clin Invest. 2015:687–698. doi: 10.1172/JCI72873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonveaux P, Végran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest. 2008:3930–3942. doi: 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumvoll M, Perriello G, Meyer C, Gerich J. Role of glutamine in human carbohydrate metabolism in kidney and other tissues. Kidney Int. 1999:778–792. doi: 10.1046/j.1523-1755.1999.055003778.x. [DOI] [PubMed] [Google Scholar]
- Tan AS, Baty JW, Dong L-F, Bezawork-Geleta A, Endaya B, Goodwin J, Bajzikova M, Kovarova J, Peterka M, Yan B, et al. Mitochondrial Genome Acquisition Restores Respiratory Function and Tumorigenic Potential of Cancer Cells without Mitochondrial DNA. Cell Metab. 2015:81–94. doi: 10.1016/j.cmet.2014.12.003. [DOI] [PubMed] [Google Scholar]
- Townsend MK, Clish CB, Kraft P, Wu C, Souza AL, Deik AA, Tworoger SS, Wolpin BM. Reproducibility of metabolomic profiles among men and women in 2 large cohort studies. Clin Chem. 2013:1657–1667. doi: 10.1373/clinchem.2012.199133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson DA, Shaw AT, Willis NA, Silver DP. Endogenous oncogenic K-rasG12D stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- van den Heuvel APJ, Jing J, Wooster RF, Bachman KE. Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol Ther. 2012:1185–1194. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. Exploiting the bad eating habits of Ras-driven cancers. Genes & Development. 2013:2065–2071. doi: 10.1101/gad.228122.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. CELL. 2012:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuneva MO, Fan TWM, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, Matés JM, Alonso FJ, Wang C, Seo Y, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012:157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachar Z, Marecek J, Maturo C, Gupta S, Stuart SD, Howell K, Schauble A, Lem J, Piramzadian A, Karnik S, et al. Non-redox-active lipoate derivates disrupt cancer cell mitochondrial metabolism and are potent anticancer agents in vivo. J Mol Med (Springer-Verlag) 2011:1137–1148. doi: 10.1007/s00109-011-0785-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.