Abstract
The applicability of the recombinant LipL32 for serodiagnosis of leptospiral infection in field rodents was assessed in this study. An immunodominant region of LipL32 was determined by monoclonal antibodies, and then, truncated LipL32 (tLipL32) was designed to contain the region (87–188th amino acid). The tLipL32 was compared between two recombinant expression hosts Escherichia coli and Pichia pastoris in ELISA. With field rat sera, tLipL32 expressed by P. pastoris (tLipL32p) had high antigenicity without background reactions, while tLipL32 expressed by E. coli (tLipL32e) showed high background reactions, which were reduced by pre-adsorption of sera with E. coli. To evaluate tLipL32-ELISA, field rat sera were tentatively divided into a Leptospira infection positive (12 sera) and a negative group (12 sera) based on the results from flaB gene PCR of kidney samples and WB with whole Leptospira cell. Consequently, the sensitivity of tLipL32p-ELISA for field rat sera was 83% . A similar result was obtained from tLipL32e-ELISA with adsorbed sera, (92%). However, sensitivity of tLipL32e-ELISA using sera without an adsorption treatment was 50%. Regardless of the expression host, tLipL32-ELISA had 100% specificity and sensitivity in experimentally infected laboratory rats. These results suggest that recombinant LipL32 expressed by P. pastoris is more applicable for serodiagnosis in field rats due to a lack of background reaction.
Keywords: ELISA, Leptospira interrogans, Rattus norvegicus, wild animal, zoonosis
Leptospirosis is a bacterial zoonosis caused by pathogenic Leptospira spp (Leptospira interrogans sensu lato). The pathogenic species have a wide range of reservoirs including companion animals, livestock and wildlife. Rodents play an important role as a source of human infection as they have a persistent asymptomatic infection with leptospires and shed them in the environment throughout their life [1, 6, 24, 26]. Leptospirosis patients develop a wide range of symptoms including high fever, headache, muscular pain, abdominal pain, intense jaundice, bleeding, renal and pulmonary dysfunction, and neurologic alterations. Severe cases are also known as Weil’s disease or leptospirosis pulmonary hemorrhage syndrome (LPHS) [7, 17], and fatality rates of those cases are >10% and >74%, respectively [16]. Human leptospirosis used to be recognized as an occupational disease among agricultural and forestry workers. On the other hand, emerging outbreaks of leptospirosis have been reported after natural disasters and severe weather, such as typhoon, hurricane and heavy rainfall in tropical and subtropical regions [2, 11]. Individual cases and localized outbreaks have also been increasing in recent years after various outdoor activities, such as swimming, hiking and rafting in endemic areas of leptospirosis [3, 31]. Therefore, it is essential to obtain information on reservoir animals of Leptospira spp. and Leptospira-contaminated environments from a preventive public health perspective [1, 6].
Laboratory diagnosis of leptospirosis has been carried out using PCR, the microscopic agglutination test (MAT), ELISA, the lateral flow assay (LFA) and culture methods. Mostly, the same techniques have been applied to reservoir animals for prevalence research and veterinary practice. MAT has been a gold standard for serological diagnosis, but MAT requires live cultures of a panel of reference serovars and it is therefore difficult to conduct MAT without proper biosecurity infrastructures [34]. Whilst, several recombinant leptospiral outer membrane proteins have been developed for diagnostic antigens, including LipL32, LipL21, LipL41, Omp1, LigA and LigB, which are convenient compared to live cultures [14, 28, 30, 36].
LipL32 is the most abundant lipoprotein expressed on the bacterial membrane, and it would thus be highly immunogenic and induce immunoreactions in the early stage of infection [18]. Additionally, LipL32 is highly conserved among pathogenic leptospires and would be a distinguishing marker for leptospirosis [27]. Applications of recombinant LipL32 for serodiagnosis have already been studied using canine, equine, bovine and human sera [5, 10, 15, 36]. Periodomestic rodents are recognized as the main source of human leptospirosis worldwide, and the carriage of leptospires in rodents has recently been studied mainly by using PCR [6]. However, an application of a recombinant leptospiral antigen for rodent serological surveillance is limited.
In this study, recombinant LipL32 was applied to a serological screening test for rodent sera. Initially, the antigenic property of recombinant LipL32 was evaluated by competitive ELISA using monoclonal antibodies and sera of laboratory rats inoculated with L. interrogans. The results indicated that immunodominant area of LipL32 was located in the intermediate region. Secondly, the intermediate region was expressed by Escherichia coli and Pichia pastoris, and then, the utility of the recombinant LipL32 was evaluated for serological screening of rat sera by ELISA.
MATERIALS AND METHODS
Bacteria and yeast strains and culture media: Eight pathogenic Leptospira serovars (L. interrogans serovar Hebdomadis strains OP84 and Akiyami B, L. interrogans serovar Batavie strain Viet16, L. interrogans serovar Manilae strain UP-MMC-NIID, L. interrogans serovar Australis strains Akiyami C, L. interrogans serovar Autumnalis strain Akiyami A, L. interrogans serovar Icterohaemorrhagiae strain RGA, L. interrogans serovar Canicola strain Hond Utrecht IV) and a saprophytic Leptospira serovar (L. biflexa serovar Patoc strain Patoc I) were cultured at 28°C in modified Korthof’s medium (DENKA Seiken Co., LTD.,Tokyo, Japan).
Escherichia coli strains BL21 (DE3) (9126, TaKaRa, Otsu, Japan) and JM109 (9052, TaKaRa) were grown at 37°C in CIRCLEGROW (#3000-121, MP Biomedicals, Santa Ana, CA, U.S.A.) supplemented with ampicillin at 50 µg/ml. E. coli strain TOP10 (Invitrogen C4040, Life Technologies Co., Carlsbad, CA, U.S.A.) was grown at 37°C in Low Salt Luria-Bertani (LB) medium (1% tryptone, 0.5% yeast extract and 0.5% NaCl, pH 7.5) supplemented with Zeocin (Invitrogen, Life Technologies Co.) at 25 µg/ml.
Pichia pastoris strain KM71H (K1740-01, Invitrogen) was grown at 30°C in the following culture media: yeast extract peptone dextrose (YPD) medium (1% yeast extract, 2% peptone and 2% dextrose), YPDS plate (1 M sorbitol, 2% agar and 100 µg/ ml Zeocin (Life Technologies Co.) in YPD medium), buffered glycerol-complex (BMGY) medium (1% yeast extract, 2% peptone, 100 mM potassium phosphate [pH 6.0], 1.34% yeast nitrogen base, 4 × 10−5% biotin and 1% glycerol) and buffered methanol-complex (BMMY) medium (0.5% methanol in BMGY medium).
Serum and kidney samples: Laboratory rats. Twenty-four female WKAH/hkm rats (6 weeks old, SLC, Hamamatsu, Japan) were inoculated intraperitoneally (i.p.) with 1 × 108L. interrogans serovar Manilae strain UP-MMC-NIID. The number of leptospires in the culture medium was counted in a counting chamber (C-Chip, AR BROWN Co., Ltd., Tokyo, Japan) under a dark field microscope. Serum and kidney specimens were collected at days 3, 6, 8, 12, 14, 21, 30, 45 and 60 after inoculation. Eight female WKAH/hkm rats (6 weeks old, SLC) were used for a control.
Field rats. A total of 33 field rats (31 Rattus norvegicus and 2 Rattus tanezumi) were captured at Hai Phong Port, Vietnam in July 2011. Serum and kidney specimens were collected and stored at −80°C until use.
Detection of the flaB gene by PCR. A portion of each kidney was comminuted, and DNA was extracted by using DNAzol reagent according to the instructions of the manufacturer (Life Technologies Co.). The extracted DNA fraction was used for nested PCR targeting the flaB gene as described previously [21]. The genomic DNA of L. interrogans serovar Manilae strain UP-MMC-NIID was used as a positive control.
Serological analysis: Enzyme-linked immunosorbent assay (ELISA). Ninety-six-well plates (3590, Corning Inc. Life Sciences, Lowell, MA, U.S.A.) were coated with recombinant LipL32 expressed by E. coli or P. pastoris (described below) or with formalin-treated leptospires at 37°C for 1 hr. After being washed three times with Dulbecco’s phosphate buffered saline (PBS) containing 0.05% Tween 20 (PBS-T) by a microplate washer (Immuno Wash Model 1575, BioRad, Hercules, CA, U.S.A.), the plates were blocked with PBS containing 3% bovine serum albumin (BSA; Sigma-Aldrich Inc., St. Louis, MO, U.S.A.) at 4°C overnight. After washing the plates, diluted rodent sera with ELISA buffer (EB; PBS containing 0.05% Tween 20 and 0.5% BSA) or culture supernatant of hybridomas were added to the plates. After incubation for 1 hr at 25°C, the plates were washed with PBS-T as described above. Bound antibodies were detected with horseradish peroxidase (HRP)-labeled secondary antibody for 1 hr at 25°C. After being washed as described above, color reactions were performed with 100 µl of 0.17% o-phenylenediamine dihydrochloride (OPD) (Sigma-Aldrich Inc.) and 0.02% H2O2 substrate solution and were allowed to develop for 10 min at 25°C. The reaction was stopped by adding 30 µl of 10% sulfonic acid. Absorbance was measured at 490/650 nm by using a SpectraMax 340 microplate spectrophotometer® (Molecular Devices, Sunnyvale, CA, U.S.A.). The results of ELISA optical density (OD) values in duplicates were averaged for the following analyses. Following HRP-conjugated secondary antibodies, goat anti-rat IgG (Kirkegaard & Perry Laboratories, Inc. (KPL), Gaithersburg, MD, U.S.A.), goat anti-mouse IgG (KPL) and mouse anti-rat IgG (KPL) were used for the detection of antibodies from laboratory rat and field rat sera, mouse monoclonal antibodies (MAbs), and antibodies from laboratory rat sera competed with MAbs, respectively.
SDS-PAGE and Western blotting (WB). An antigen was fractionated by 10–20% SDS-PAGE (ePAGEL T1020, ATTO, Tokyo, Japan) and stained with SimplyBlueTM SafeStain (Life Technologies Co.). For WB analysis, antigens were transferred to a polyvinylidene difluoride (PVDF) membrane (ATTO). The membrane was incubated overnight with 3% BSA (Sigma-Aldrich Inc.) in PBS at 4°C. After washing with PBS, the membrane was reacted with a solution containing the first antibody diluted in PBS or culture supernatant of hybridomas at room temperature for 1 hr. Binding antibodies were detected using HRP-conjugated secondary antibodies, and 4-chloro-1-naphthol (Sigma-Aldrich Inc.) was used as a peroxidase substrate. Two kinds of HRP-conjugated secondary antibodies, goat anti-rat IgG (KPL) and Protein A (Zymed-Invitrogen, Life technologies Co.), were used to detect antibodies of laboratory rats and field rats and mouse MAb, respectively.
Preparation of monoclonal antibodies. Five-week-old female BALB/c/slc mice were injected i.p. with 100 µg of formalin-treated whole cells of L. interrogans serovar Australis strain Akiyami C that was mixed with an equal volume of Freund’s complete adjuvant (Difco, Detroit, MI, U.S.A.). The mice were given booster doses using the same route. Two weeks later, the mice were injected with the same antigen mixed with Freund’s incomplete adjuvant (Difco). Four weeks after the second immunization, a final injection without the adjuvant was administered by the intravenous route. Three days later, spleens were removed and used for fusion to Sp2/O-Ag14 myeloma cells using polyethylene glycol 1500 (PEG 1500; Boehringer, Ingelheim am Rhein, Germany) as described before [20]. First, hybridoma culture fluids were screened for the presence of Leptospira-specific antibodies using the immunofluorescence assay (IFA) [29]. Next, each of the hybridomas secreting MAbs against LipL32 was examined by ELISA using recombinant LipL32 antigen as described above. Supernatants containing MAbs were recovered by centrifugation, filtered with a 0.45 µm Minisart filter (Sartorius, Goettingen, Germany) and stored at −80°C until use.
Monoclonal antibody characterization: Western blotting (WB). Reactivities of MAbs against L. interrogans serovar Hebdomadis strain OP84, serovar Batavie strain Viet16 and serovar Manilae strain UP-MMC-NIID, L. biflexa serovar Patoc strain Patoc I and recombinant LipL32 in WB were determined as described above. Leptospira was lysed with 1% SDS and 1% 2-mercaptoethanol. The lysate was boiled for 5 min at 98°C and used as the antigen at 1 × 106 leptospires/lane. Culture media of hybridoma cells were used as the first antibody, and HRP-conjugated Protein A (Zymed) at 1:200 dilution was used as the secondary antibody.
Microscopic agglutination test (MAT). MAT was conducted essentially by the same method as that described by Koizumi et al. against L. interrogans serovar Australis strain Akiyami C [21].
Enzyme-linked immunosorbent assay (ELISA). ELISA was carried out according to the procedure described above. L. interrogans serovar Icterohaemorrhagiae strain RGA, L. interrogans serovar Canicola strain Hond Utrecht IV, L. interrogans serovar Autumnalis strain Akiyami A, L. interrogans serovar Hebdomadis strain Akiyami B and L. biflexa serovar Patoc strain Patoc I were treated with formalin according to the WHO standard reference [35]. Formalin-treated leptospires at 1 × 108 /ml and 1 µg /ml of recombinant LipL32 expressed in E. coli were used as antigens. Culture supernatants of hybridoma cells were used as the first antibody, and HRP-conjugated goat anti-mouse IgG (Zymed) at 1:10,000 dilution was used as the secondary antibody. The individual MAbs were isotyped in cell culture supernatants by ELISA followed by peroxidase-conjugated rabbit anti-mouse IgG1, IgG2a, IgG2b, IgG3 or IgM (Zymed), as described above.
Epitope mapping by using synthetic peptides: Peptides were synthesized and analyzed by Sigma-Aldrich Tokyo, Japan (Custom SPOTs®, 96 SPOTs). Briefly, various 10-mer peptides of 8-mer overlapping each based on the deduced amino acid sequence of LipL32 of strain Akiyami A (GenBank accession number AB094435) were spotted on a membrane. The spotted membrane was reacted with hybridoma culture supernatants and a secondary antibody against mouse IgG.
Epitope mapping by competitive binding assay. A competitive binding assay to whole LipL32 expressed by E. coli (wLipL32, described below) in ELISA was performed. As described above, 96-well plates were coated with wLipL32 and blocked with BSA. The plates were washed three times with PBS-T. Culture fluid of MAbs was added and incubated for 1 hr at room temperature, and the plates were washed as described above. Then, 1:200 dilutions of immunized rat sera prepared as described above were added and incubated for 1 hr at room temperature. After washing as described above, HRP-conjugated mouse anti-rat IgG antibody (Zymed) was added to the wells and incubated for 1 hr at room temperature. Color development was performed as described above by using OPD reagent.
Expression of whole and truncated LipL32 by E. coli and P. pastoris: Amplification and subcloning. The full cording region of lipL32 gene was amplified from genomic DNA of strain UP-MMC-NIID prepared by DNAzol reagent as described above. Primers were designed from the LipL32 sequences of serovar Manilae strain UP-MMC-NIID (GenBank accession number JQ013519) and serovar Lai strain (GenBank accession number AY568679). Primers were also designed with an additional sequence recognized by restriction enzymes (EcoRI, XhoI and NotI sites shown by underlines). To amplify the entire coding region of LipL32, forward primer lipL32_F1 (5′- CGCTCGAGATGATGAAAAAACTTCGATT (XhoI)) and reverse primer lipL32_R819 (5′- GGAATTCTCATTACTTAGTCGCGTGAG (EcoRI)) were used. After amplification, the DNA fragment was subcloned into pRSET A plasmid vector (Invitrogen) digested with XhoI/EcoRI.
To amplify truncated LipL32 of 87–188 amino acids corresponding to the nucleotide sequence from 259 to 564, forward primer tlipL32e_F259 (5′- CGCTCGAGCCTGCCGTAATCGCT (XhoI)) and reverse primer tlipL32e_R564 (5′- GGAATTCTCAATTAGGGATCTTGAT (EcoRI)) were used. After amplification, the DNA fragment was subcloned into pRSET A plasmid vector (pRSET-A-tlipL32). To amplify the same fragment for P. pastoris expression, forward primer tlipL32p_F259 (5′- GGAATTCCCTGCCGTAATCGCT (EcoRI)) and reverse primer tlipL32p_R564 (5′- ATAGTTTAGCGGCCGCATTAGGGATCTTGAT (NotI)) were used. After amplification, the DNA fragment was subcloned into pPICZα A plasmid vector (PICZα-A-tlipL32) as described above. Sequences of the inserts were confirmed by DNA sequencing.
Production of recombinant antigens. Plasmid DNA constructs pRSET-A-wlipL32 and pRSET-A-tlipL32 were transformed into E. coli strain BL21 (DE3) (Invitrogen Life Technologies Co.). A single colony was inoculated into CIRCLEGROW (#3000–121, MP Biomedicals) containing ampicillin for small-scale culture incubation at 37°C overnight. The culture fluid was then centrifuged, and the collected cells were inoculated into 100 ml of fresh medium, and isopropyl-β-D-thiogalactopyranoside (IPTG) induction was performed according to the procedure for pET system expression (Novagen Takara). The cultured cells were collected by centrifugation, resuspended in 5 ml of 0.5 M NaCl binding buffer (0.5 M NaCl, 20 mM imidazole and 20 mM potassium phosphate) and sonicated four times for 15 sec each time on ice. Thereafter, the fusion protein was purified using a His-trap column (Amersham Biosciences, Piscataway, NJ, U.S.A.) according to the manufacturer’s instructions. The recombinant proteins expressed by pRSET-A-wlipL32 and pRSET-A-tlipL32 in E. coli were designated as wlipL32 and tlipL32e, respectively.
The plasmid pPICZα A-tlipL32 was propagated in E. coli strain TOP10 and linearized with restriction enzyme BstX1. Then, it was transformed into P. pastris strain KM71H by using an EasyCompTM Kit (K1740-01, Invitrogen) according to the manufacturer’s instructions. Transformed KM71H was grown in 200 ml BMGY in a 1 l baffled flask by agitating at 300 rpm until OD600 reaching at 2 to 6. The cultured P. pastoris was centrifuged at 140 × g for 5 min, and the pellet was resuspended in 40 ml BMMY in a 300 ml baffled flask for further culture. The expression of truncated LipL32 was induced by addition of 100% methanol to the final concentration of 0.5% every 24 hr. Recombinant protein fused with 6 × His was secreted from yeast into the culture fluid. Then, large-scale culture was conducted under the optimized expression conditions, and transformed KM71H was propagated in 500 ml BMGY and resuspended in l l BMMY for 120 hr under the same conditions as those for the small-scale culture. Thereafter, the truncated LipL32 protein was purified from culture supernatant by using a His-trap column (GE Healthcare Bio-Sciences AB, Uppsala, Sweden) as described above. The recombinant protein expressed by pPICZαA-tlipL32 in P. pastris was designated as tlipL32p.
Adsorption of rodent sera with E. coli. E. coli strain BL21 (DE3) was grown at 37°C in CIRCLEGROW (#3000-121, MP Biomedicals) until the OD600 reaching 0.8 and harvested by centrifugation at 3,500 × g for 10 min. The pellet was washed with PBS 2 times and then stored at 4°C until use. 100 µl of rodent sera were diluted 1:100 in EB, mixed with an equal volume of stored E. coli adjusted the concentration at OD600 2.38 with EB and incubated at room temperature for 1 hr agitating at 200 rpm. Adsorbent E. coli was separated by centrifugation at 3,500 × g for 5 min, and carefully, the supernatant was transferred to a new tube. The treated and non-treated sera were examined by ELISA as described above.
Statistics. ELISA data sets were analyzed in JMP software (JMP, ver. 11.0.0 SAS Institute Inc., Cary, NC, U.S.A.) and an online program for Shapiro-Wilk Normality Test (available at http://sdittami.altervista.org/shapirotest/ShapiroTest.html).
Ethics statement. Animal experimentation was performed after obtaining permission from the Institutional Animal Care and Use Committee of Hokkaido University. Experiments involving infection were performed in a BSL-2 facility, and involving experimental animals were carried out in accordance with the Guidelines approved by the National Institute of Health in the United States of America.
RESULTS
Production of hybridoma clones generating MAbs against Leptospira interrogans: A total of 123 hybridoma clones were confirmed to produce MAbs against L. interrogans serovar Australis by IFA. Clones were divided into groups A to E based on cross-reactivity against 5 strains of pathogenic Leptospira and 1 strain of nonpathogenic Leptospira by ELISA, the results of MAT against L. interrogans serovar Australis and molecular weights of the reacted fraction from L. interrogans serovar Australis in WB ( Table1 ).
Table 1. Identification of hybridomas generating MAbs against LipL32.
| Group of Hybridoma (number of clone) |
MAT | ELISA | WB | |||||
|---|---|---|---|---|---|---|---|---|
|
L. interrogans Akiyami A |
L. interrogans | L. biflexa |
L. interrogans Akiyami C |
|||||
| Akiyami C | RGA | UT IV | Akiyami A | Akiyami B | Patoc I | |||
| A (4) | – | + | + | + | + | + | – | 32 |
| B (2) | – | + | + | + | + | + | + | 40 |
| C (115) | + | + | – | – | – | – | – | 21 |
| D (1) | – | + | – | – | – | – | – | 45 |
| E (1) | – | + | – | – | – | – | – | 48 |
A total of 123 hybridoma clones were distinguished by MAT, ELISA and WB. Only Group C maintained affinity for protease K-treated antigen (data not shown).
MAbs in Group A reacted to an antigen estimated molecular weight of 32 kDa in WB, but were negative in MAT. They had cross-reactivity to all of the 5 strains of pathogenic Leptospira, but not to the strain of nonpathogenic Leptospira in ELISA. MAbs in Group B reacted to an antigen estimated molecular weight of 40 kDa fraction in WB, but were negative in MAT. They reacted with all of the pathogenic and nonpathogenic Leptospira strains. A total of 115 MAbs in Group C were positive in MAT. MAT is considered to detect reactions of antibodies against lipopolysaccharide (LPS) which defines serotype of leptospira. Therefore, MAbs in Group C might be LPS-specific antibodies. Groups D and E contained one MAb each, which recognized antigens estimated molecular weights of 45 and 48 kDa, respectively. They were positive only to homologous serovar Australis in ELISA.
Specificity of MAbs in Group A to LipL32 was expected, based on the reactivities to 32 kDa component of pathogenic leptospires. Negative result in MAT supported the MAbs in Group A bind to protein component, not to LPS. Therefore, 4 MAbs designated as D14/2, D58/3, Y22/1 and D62/1were used in further characterizations.
Characterization of MAbs in Group A: Isotype of MAbs. The immunoglobulin sub-classes of MAbs D14/2, Y22/1, D58/3 and D62/1 were determined to be IgG1, IgG2a, IgG2b and IgG2a, respectively.
Specificity of MAbs in Group A. D58/3 showed a single band around 32 kDa from whole cell antigens of L. interrogans serovars Manilae, Hebdomadis and Batavie (Fig. 1, lanes 3, 4 and 5). Next, whole LipL32 (wLipL32) expressed by E. coli was used for WB to confirm reactivity of MAbs. D58/3 reacted with wLipL32 in multiple bands (Fig. 1, lane 1).
Fig. 1.
Antigenic specificities of the MAbs against LipL32 in Western blotting assay. Monoclonal antibody D58/3 detected a 32 kDa component of L. interrogans serovars Manilae (lane 3), Hebdomadis (lane 4) and Batavie (lane 5), but no band in L. biflexa serovar Patoc strain Patoc I (lane 2). wLipL32 expressed by E.coli was detected at 33~34 kDa (lane 1). The same result was obtained by using D14/2 (not shown). Standard protein markers were developed in lane M.
No band was obtained from L. biflexa serovar Patoc (Fig. 1, lane 2), which was in accordance with the results of ELISA. The other 3 MAbs in Group A showed the same results (data not shown). Thus, the specificity of MAbs (D14/2, Y22/1, D58/3 and D62/1) to LipL32 was confirmed.
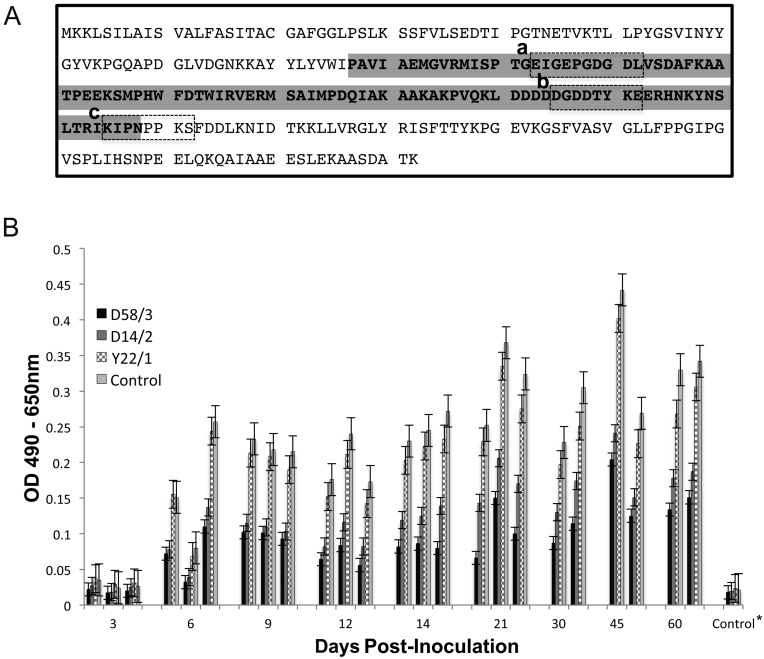
Epitope mapping. As shown in Fig. 2A, the amino acids sequences of epitopes for the MAbs were determined to be EIGEPGDGDL, DGDDTYKE and KIPNPPKS, corresponding to amino acid positions 105–114 (D14/2), 167–174 (D58/3 and D62/1) and 187–194 (Y22/1) of LipL32, respectively. Since D58/3 and D62/1 recognized the same amino acid sequence, only D58/3 was used in the following experiments.
Fig. 2.
Epitopes of MAbs and competitive binding assay of MAbs with immune rat sera. A: Epitopes of MAbs are shown in the dashed-line boxes: (a) D14/2, (b) D58/3 and D62/1, and (c) Y22/1. Gray shaded region indicates amino acid sequence of tLipL32. B: Rat sera experimentally inoculated with L. interrogans serovar Manilae were examined by wLipl32 ELISA. MAbs D58/3, D14/2, Y22/1 and control (invalid MAb against hantavirus) competed with polyclonal antibodies to bind to wLipL32. *Results from 8 control rat sera are shown as averaged ODs.
Determination of immunodominant epitopes on LipL32. A competitive assay between MAbs and polyclonal immune sera was carried out to evaluate immunodominant epitopes on LipL32. A total of 24 sera from rats experimentally inoculated with L. interrogans serovar Manilae were tested in ELISA. As shown in Fig. 2B, ELISA ODs with polyclonal immune sera were reduced by blocking epitopes with the MAb D14/2, D58/3 or Y22/1. The average reduction rate at 6 to 60 days after inoculation was highest with D58/3 (58%) and next highest with D14/2 (46%). On the other hand, Y22/1 showed an average reduction of 13%. Thus, the antigenic region covering the two epitopes of D14/2 and D58/3 is considered to be an immunodominant region.
Preparation of truncated recombinant LipL32. In our previous experiment, wLipL32 expressed by E. coli showed rapidly reduced antigenic efficacy for ELISA. This was probably due to the temperature instability of wLipL32 related to the cycles of freeze and thaw (data not shown). To solve this problem, we developed truncated LipL32 in E. coli (tLipL32e) and in P. pastoris (tLipL32p).
wLipL32 and tLipL32e were expressed by E .coli BL21 (DE3) as soluble proteins. tLipL32p was expressed by P. pastoris KM71H as a soluble extracellular secreted protein. The expressed proteins were purified and subjected to WB analysis using MAbs. As shown in Fig. 1, wLipL32 was detected between 21~35 kDa in a ladder band. The major band was found at molecular weight of 33~34 kDa (Fig. 1, lane 1). tLipL32e was detected at molecular weight of 15~16 kDa (Fig. 3, lane 2). tLipL32p was confirmed to be a protein with an estimated molecular weight of 12~13 kDa protein (Fig. 3, lane 1). Therefore, both tLipL32e and tLipL32e were successfully expressed in expected size.
Fig. 3.
SDS-PAGE and WB analysis of truncated LipL32 expressed by E. coli and P. pastoris. Monoclonal antibody D58/3 detected tLipL32p (lane 1) and tLipL32e (lane 2). The same result was obtained by using D14/2 (not shown). Standard protein markers were developed in lane M.
Reactivity of LipL32 with rat sera: Comparison of reactivities of tLipL32 expressed by E. coli and P. pastris in ELISA with rat sera. Antigenic efficacies of tLipL32e and by tLipL32p were examined by comparing reactivities with sera from field rats that were defined as following. A field rat serum that showed the highest OD value in ELISA and positive results in flaB-nested PCR of kidney samples and WB was defined as Leptospira infection positive serum. A field rat serum that showed the lowest OD value in ELISA and negative results in flaB-nested PCR of kidney samples and WB was defined as Leptospira infection negative serum. The optimized condition of field rat sera was at a dilution 1:200, and that of goat anti-rat IgG HRP conjugate was at a dilution 1:10,000. As shown in Fig. 4, the antigen concentrations that gave plateau OD values were 1 µg/ml tLipL32p and 64 µg/ ml tLipL32e. tLipL32p showed high antigenic efficacy in ELISA compared to tLipL32e. Consequently, the tLipL32 ELISA condition was optimized to use 1 µg/ml tLipL32p and 8 µg/ml tLipL32e for practical antigen concentrations.
Fig. 4.
Optimization of antigen concentration of tLipL32 for ELISA. Pos.: serum from field rat defined as positive for leptospiral infection (solid line), Neg.: serum from field rat confirmed as negative for leptospiral infection (dashed line). The antigens tLipL32p (●) and tLipL32e (□) were diluted in the range of 26-2−8µg/ ml.
OD values with serum from an uninfected field rat indicated that tLipL32p had a very low nonspecific reaction even with higher concentrations of antigen. tLipL32e showed high background ODs with increasing antigen concentration (Fig. 4). These results also suggested that contaminated substances from expression host E. coli caused a nonspecific reaction in ELISA with field rat serum.
Evaluation of effects of E. coli adsorbed sera on ELISA. At first, a total of 24 sera from laboratory rats experimentally inoculated with L. interrogans serovar Manilae and 8 control sera from uninfected laboratory rats were examined by ELISA (Fig. 5A). In Fig. 5A, OD values with the two antigens tLipL32p and tLipL32e were well correlated with two exceptions of high ODs to tLipL32e, but lower to tLipL32p (R2=0.86, P<0.001). The OD values distribution of 8 control rats was found lower and was separately from rats inoculated with leptospires. This result presented laboratory rat sera showed low background reaction against compounds derived from E. coli. On the other hand, field rat sera showed different reaction profile (Fig. 5B). The OD values with field rat sera were scattered in confrontation of tLipL32p-ELISA and tLipL32e ELISA (R2=0.42, P<0.001). In addition, the most of sera showed higher OD values in tLipL32e ELISA. These results suggested that both tLipL32p and tLipL32e were basically applicable as ELISA antigens; but the non-specific reaction in field rat sera in tLipL32e-ELISA should be settled.
Fig. 5.
Comparison of tLipL32p and LipL32e antigens and effects of serum treatment by E. coli. The scatter diagrams of ELISA ODs with combinations of following antigens and field and laboratory rat sera with or without pretreatment of adsorption by E. coli. Closed squares indicate OD values of field rats. Closed circles indicate OD values of laboratory rats. Open triangles indicate OD values of sera from laboratory rats without inoculation. A: tLipL32p-ELISA and tLipL32e-ELISA with experimentally inoculated and control rat sera without pretreatment by E.coli. B: tLipL32p-ELISA and tLipL32e-ELISA with field rat sera without pretreatment by E.coli. C: tLipL32p-ELISA with field rat sera with and without pretreatment by E.coli. D: tLipL32e-ELISA with field rat sera with and without treatment by E.coli.
To reduce non-specific reaction in ELISA, rat sera were pretreated with E. coli to adsorb substances in sera that reacted against components of E. coli. A total of 33 field rat sera were adsorbed with E. coli as described in Materials and Methods. At first, tLipL32p was used for testing non-treated and treated sera. As shown in Fig. 5C, correlation function between non-treated and treated sera was R2=0.95 (P<0.001). This showed that the serum treatment with E. coli had almost no influence on tLipL32p-ELISA. Next, tLipL32e was used for testing non-treated and treated sera (Fig. 5D). The distribution of ELISA OD values showed a decrease of ODs with treated sera compared to those with non-treated sera. These results indicated that pretreatment of serum with E. coli was effective to reduce background reaction for an E. coli-expressed antigen, such as tLipL32e.
Evaluation of tLipL32 ELISA for field rat sera. Generally, to evaluate a novel diagnosis system, true positive and true negative groups of sera are required. However, true negative individuals were difficult to be identified among field rats. Therefore, we tentatively defined true positive sera and true negative sera groups based on the results from flaB gene PCR of kidney samples and WB with whole Leptospira cell. A total of 33 field rat sera were divided into 4 groups: PCR alone positive, WB alone positive, PCR and WB positive, and PCR and WB negative (Fig. 6). Among them, sera in the PCR and WB positive group were tentatively regarded as true positive, and sera in the PCR and WB negative group were tentatively regarded as true negative.
Fig. 6.
Distribution of ELISA ODs obtained from tLipL32 ELISA in the relevance to flaB-nested PCR of kidney samples and WB. A total of 33 field rats were divided into four groups according to the results of PCR and WB using L. interrogans serovar Manilae as an antigen. Tentatively, both positive rats were regarded as true positive, and both negative rats were defined as negative. A tentative cutoff point for antigens, which was defined as OD values for 95% of the true negative group, is shown by a broken line. A: tLipL32p-ELISA with non-treated sera, B: tLipL32e-ELISA with treated sera, C: tLipL32e-ELISA with non-treated sera.
The distribution ranges of OD values in ELISA using tLiPL32 among true positive and true negative groups were compared. A tentative cutoff point for tLipL32 ELISA was defined as 95% of the OD values among sera in the true negative group, so as to be 95% specificity [15]. Sensitivity was defined as the percentage of sera in the true positive group for which OD values were greater than the cutoff value. As shown in Fig. 6A, tLipL32p could differentiate true positive and true negative groups. With the tentative cutoff point, 83% sensitivity was obtained by tLipL32p ELISA. A similar distribution range of OD values was obtained from ELISA using tLipL32e with treated sera (Fig. 6B), and the sensitivity was 92%. However, as shown in Fig. 6C, tLipL32e with non-treated sera resulted in OD values of sera in the true positive group being overlapped with ODs of sera in the true negative group, and the sensitivity was 50%.
DISCUSSION
LipL32 is abundantly expressed on the bacterial outer membrane and is present exclusively in pathogenic Leptospira species [18, 27]. Therefore, LipL32 has been applied for serological diagnosis of Leptospira infection as a conserved antigen among pathogenic Leptospira species [15]. Despite the importance of rodents as a source of human leptospirosis, application of LipL32 for serodiagnosis of rodents has not been reported to the best of our knowledge. Hence, we aimed to establish recombinant LipL32-based ELISA for serological diagnosis in field rodents. Firstly, wLipL32 was expressed by E. coli and used as an ELISA antigen. However, wLipL32 degraded easily and showed rapid reduction of antigenicity, probably due to the size of the protein and the temperature instability (data not shown). Then, we designed tLipL32 including a major epitope region defined by MAbs. Previous studies also demonstrated that the immunodominant part was mainly the central portion of Lipl32 [9, 22, 23]. Our competitive inhibition study using MAbs and laboratory rat sera also confirmed that the immunodominat region is located in the central part.
tLipL32 was expressed by E. coli, and it was able to differentiate experimentally infected laboratory rat sera from control rat sera in ELISA, indicating that tLipL32 is applicable as a serodiagnostic antigen. Subsequently, tLipL32e was evaluated with field rat sera in ELISA. Unfortunately, high background reactions were observed in field rat sera that showed negative results in WB and PCR. These results suggested contamination of tLipL32e by components from E. coli, which caused background reaction in ELISA. In spite of SDS-PAGE and WB analysis, it was not possible to find out the causative molecule of background reaction.
A similar phenomenon occurred and was a struggle of serodiagnosis of Lyme borreliosis among patients with other bacterial infections, viral infections and autoimmune diseases [12, 13, 19, 25]. Fawcett et al. proved the reduction of false positive patients of Lyme borreliosis by the adsorption of test sera with components of E. coli [12]. We obtained a similar result using tLipL32e with E. coli adsorbed sera. The high background reaction was attributed to a reaction to contaminants of expression host E. coli in the recombinant antigen. Meanwhile, the background reaction was not a problem for testing laboratory infected rat sera. These results support the theory that experimental Leptospira infection studies cannot replicate field conditions as discussed previously [8]. The results of our experiments suggest that wildlife potentially has a high antibody titer against multiple environmental microorganisms, e.g. E. coli. This is an important reminder when applying recombinant antigens expressed by E. coli to a serodiagnosis system. A comparison of results from multiple diagnosis approaches may be appropriate for a research on prevalence in wildlife.
In this study, saccharomycetaceae P. pastori was used as another expression host to reduce background reactions against E. coli components. It was confirmed that tLipL32p caused less background reactions in ELISA with field rat sera.
Generally, the sensitivity and specificity of a novel diagnostic system are determined based on the diagnostic accuracy using true positive specimens and true negative specimens. However, definition of true positive and true negative in wildlife is almost impossible. We tentatively defined true positive and true negative field rat sera based on the results from PCR and WB, and then compared distributions of ELISA OD values between the true positive and the true negative groups to study an availability of truncated LipL32 for ELISA antigen. tLipL32p-ELISA with non-treated sera and tLipL32e-ELISA with adsorbed sera were able to divide OD distributions between positive and negative groups. On the other hand, tLipL32e-ELISA with non-treated sera could not divide OD distributions. Based on the above understanding, tLipL32p is a valuable antigen for a serological study among field rats as it does not require sera adsorption by E. coli.
Rodents are critical vectors of not only leptospirosis but also other zoonotic diseases, such as plague, hantavirus infections and hepatitis E infection [4, 24]. Therefore, monitoring of prevalence among rodents would provide valuable information for preventive medicine. However, detection of multiple pathogens may be burdensome due to the requirement of different specimens and types of test. On the other hand, serological research can provide multiple infections information at a time.
The recombinant LipL32-based ELISA that we employed in this study has advantages in laboratories with few resources. The procedures for expressing a recombinant antigen are basically standardized and are safer than handling live leptospires. Recombinant protein-based ELISA also allows easier control of antigen quality and quantity than MAT using live leptospires. Moreover, diagnostic LipL32 antigen has the most promising aspect to detect only antibodies against pathogenic Leptospira. Recently, ELISA based on combinations of leptospiral recombinant outer membrane proteins has been developed for human and equine Leptospira infection diagnosis [32, 36]. These studies suggested that combining other leptospiral antigens improve a diagnostic accuracy of Leptospira infection. Although the dynamics of Leptospira infection in Norway rats have not been studied in detail, it has been reported that Norway rats showed various pathogenesis from transient infection to chronic infection by experimental inoculation of various strains [33]. Therefore, we propose the use of both serological and molecular detection approaches, such as ELISA or WB and PCR, in parallel to provide more reliable results for research on prevalence of field rat Leptospira infection.
In conclusion, we developed recombinant LipL32-based ELISA for serodiagnosis of Leptospira infection in rodents. Truncated LipL32 expressed by P. pastoris was shown to be the most valiant recombinant antigen for ELISA in this study. A limitation of this study is that our study population was small. A further study with larger numbers of field rat sera is needed to obtain a more accurate cutoff point in tLipL32 ELISA.
Acknowledgments
This research was supported by the Japan Society for the Promotion of Science (JSPS) Fellows Grant Number 2846, Japan. This study was supported in part by the Japan Initiative for Global Research Network on Infectious Diseases (J-GRID).
REFERENCES
- 1.Adler B., de la Peña Moctezuma A.2010. Leptospira and leptospirosis. Vet. Microbiol. 140: 287–296. doi: 10.1016/j.vetmic.2009.03.012 [DOI] [PubMed] [Google Scholar]
- 2.Agampodi S. B., Dahanayaka N. J., Bandaranayaka A. K., Perera M., Priyankara S., Weerawansa P., Matthias M. A., Vinetz J. M.2014. Regional differences of leptospirosis in Sri Lanka: observations from a flood-associated outbreak in 2011. PLoS Negl. Trop. Dis. 8: e2626. doi: 10.1371/journal.pntd.0002626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agampodi S. B., Karunarathna D., Jayathilala N., Rathnayaka H., Agampodi T. C., Karunanayaka L.2014. Outbreak of leptospirosis after white-water rafting: sign of a shift from rural to recreational leptospirosis in Sri Lanka? Epidemiol. Infect. 142: 843–846. doi: 10.1017/S0950268813001465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayral F., Artois J., Zilber A. L., Widén F., Pounder K. C., Aubert D., Bicout D. J., Artois M.2015. The relationship between socioeconomic indices and potentially zoonotic pathogens carried by wild Norway rats: a survey in Rhône, France (2010-2012). Epidemiol. Infect. 143: 586–599. doi: 10.1017/S0950268814001137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bomfim M. R., Ko A., Koury M. C.2005. Evaluation of the recombinant LipL32 in enzyme-linked immunosorbent assay for the serodiagnosis of bovine leptospirosis. Vet. Microbiol. 109: 89–94. doi: 10.1016/j.vetmic.2005.05.002 [DOI] [PubMed] [Google Scholar]
- 6.Cosson J. F., Picardeau M., Mielcarek M., Tatard C., Chaval Y., Suputtamongkol Y., Buchy P., Jittapalapong S., Herbreteau V., Morand S.2014. Epidemiology of leptospira transmitted by rodents in southeast Asia. PLoS Negl. Trop. Dis. 8: e2902. doi: 10.1371/journal.pntd.0002902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa F., Porter F. H., Rodrigues G., Farias H., de Faria M. T., Wunder E. A., Osikowicz L. M., Kosoy M. Y., Reis M. G., Ko A. I., Childs J. E.2014. Infections by Leptospira interrogans, Seoul virus, and Bartonella spp. among Norway rats (Rattus norvegicus) from the urban slum environment in Brazil. Vector Borne Zoonotic Dis. 14: 33–40. doi: 10.1089/vbz.2013.1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desvars A., Michault A., Chiroleu F.2013. Influence of risk factors on renal leptospiral load in naturally infected wild black rats. Acta Trop. 125: 258–261. doi: 10.1016/j.actatropica.2012.11.011 [DOI] [PubMed] [Google Scholar]
- 9.Deveson Lucas D. S., Lo M., Bulach D. M., Quinsey N. S., Murray G. L., Allen A., Adler B.2014. Recombinant LipL32 stimulates interferon-gamma production in cattle vaccinated with a monovalent Leptospira borgpetersenii serovar Hardjo subtype Hardjobovis vaccine. Vet. Microbiol. 169: 163–170. doi: 10.1016/j.vetmic.2013.12.016 [DOI] [PubMed] [Google Scholar]
- 10.Dey S., Mohan C. M., Kumar T. M., Ramadass P., Nainar A. M., Nachimuthu K.2004. Recombinant LipL32 antigen-based single serum dilution ELISA for detection of canine leptospirosis. Vet. Microbiol. 103: 99–106. doi: 10.1016/j.vetmic.2004.07.018 [DOI] [PubMed] [Google Scholar]
- 11.Easton A.1999. Leptospirosis in Philippine floods. BMJ 319: 212. doi: 10.1136/bmj.319.7204.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fawcett P. T., Gibney K. M., Rose C. D., Klein J. D., Doughty R. A.1991. Adsorption with a soluble E. coli antigen fraction improves the specificity of ELISA tests for Lyme disease. J. Rheumatol. 18: 705–708. [PubMed] [Google Scholar]
- 13.Fawcett P. T., Rose C. D., Gibney K. M.1995. Comparative evaluation of adsorption with E. coli on ELISA tests for Lyme borreliosis. J. Rheumatol. 22: 684–688. [PubMed] [Google Scholar]
- 14.Fernandes C. P., Seixas F. K., Coutinho M. L., Vasconcellos F. A., Seyffert N., Croda J., McBride A. J., Ko A. I., Dellagostin O. A., Aleixo J. A.2007. Monoclonal antibodies against LipL32, the major outer membrane protein of pathogenic Leptospira: production, characterization, and testing in diagnostic applications. Hybridoma (Larchmt) 26: 35–41. doi: 10.1089/hyb.2006.033 [DOI] [PubMed] [Google Scholar]
- 15.Flannery B., Costa D., Carvalho F. P., Guerreiro H., Matsunaga J., Da Silva E. D., Ferreira A. G., Riley L. W., Reis M. G., Haake D. A., Ko A. I.2001. Evaluation of recombinant Leptospira antigen-based enzyme-linked immunosorbent assays for the serodiagnosis of leptospirosis. J. Clin. Microbiol. 39: 3303–3310. doi: 10.1128/JCM.39.9.3303-3310.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foronda P., Martin-Alonso A., Del Castillo-Figueruelo B., Feliu C., Gil H., Valladares B.2011. Pathogenic Leptospira spp. in wild rodents, Canary Islands, Spain. Emerg. Infect. Dis. 17: 1781–1782. doi: 10.3201/eid1709.101470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouveia E. L., Metcalfe J., de Carvalho A. L., Aires T. S., Villasboas-Bisneto J. C., Queirroz A., Santos A. C., Salgado K., Reis M. G., Ko A. I.2008. Leptospirosis-associated severe pulmonary hemorrhagic syndrome, Salvador, Brazil. Emerg. Infect. Dis. 14: 505–508. doi: 10.3201/eid1403.071064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haake D. A., Matsunaga J.2002. Characterization of the leptospiral outer membrane and description of three novel leptospiral membrane proteins. Infect. Immun. 70: 4936–4945. doi: 10.1128/IAI.70.9.4936-4945.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henriksen A. Z., Maeland J. A.1987. Serum antibodies to outer membrane proteins of Escherichia coli in healthy persons and patients with bacteremia. J. Clin. Microbiol. 25: 2181–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kida H., Brown L. E., Webster R. G.1982. Biological activity of monoclonal antibodies to operationally defined antigenic regions on the hemagglutinin molecule of A/Seal/Massachusetts/1/80 (H7N7) influenza virus. Virology 122: 38–47. doi: 10.1016/0042-6822(82)90375-0 [DOI] [PubMed] [Google Scholar]
- 21.Koizumi N., Muto M., Yamamoto S., Baba Y., Kudo M., Tamae Y., Shimomura K., Takatori I., Iwakiri A., Ishikawa K., Soma H., Watanabe H.2008. Investigation of reservoir animals of Leptospira in the northern part of Miyazaki Prefecture. Jpn. J. Infect. Dis. 61: 465–468. [PubMed] [Google Scholar]
- 22.Lottersberger J., Guerrero S. A., Tonarelli G. G., Frank R., Tarabla H., Vanasco N. B.2009. Epitope mapping of pathogenic Leptospira LipL32. Lett. Appl. Microbiol. 49: 641–645. doi: 10.1111/j.1472-765X.2009.02723.x [DOI] [PubMed] [Google Scholar]
- 23.Maneewatch S., Adisakwattana P., Chaisri U., Saengjaruk P., Srimanote P., Thanongsaksrikul J., Sakolvaree Y., Poungpan P., Chaicumpa W.2014. Therapeutic epitopes of Leptospira LipL32 protein and their characteristics. Protein Eng. Des. Sel. 27: 135–144. doi: 10.1093/protein/gzu006 [DOI] [PubMed] [Google Scholar]
- 24.Meerburg B. G., Singleton G. R., Kijlstra A.2009. Rodent-borne diseases and their risks for public health. Crit. Rev. Microbiol. 35: 221–270. doi: 10.1080/10408410902989837 [DOI] [PubMed] [Google Scholar]
- 25.Michael J. G., Whitby J. L., Landy M.1962. Studies on natural antibodies to gram-negative bacteria. J. Exp. Med. 115: 131–146. doi: 10.1084/jem.115.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monahan A. M., Callanan J. J., Nally J. E.2008. Proteomic analysis of Leptospira interrogans shed in urine of chronically infected hosts. Infect. Immun. 76: 4952–4958. doi: 10.1128/IAI.00511-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murray G. L.2013. The lipoprotein LipL32, an enigma of leptospiral biology. Vet. Microbiol. 162: 305–314. doi: 10.1016/j.vetmic.2012.11.005 [DOI] [PubMed] [Google Scholar]
- 28.Natarajaseenivasan K., Vijayachari P., Sharma S., Sugunan A. P., Selvin J., Sehgal S. C.2008. Serodiagnosis of severe leptospirosis: evaluation of ELISA based on the recombinant OmpL1 or LipL41 antigens of Leptospira interrogans serovar autumnalis. Ann. Trop. Med. Parasitol. 102: 699–708. doi: 10.1179/136485908X355229 [DOI] [PubMed] [Google Scholar]
- 29.Pinne M., Haake D.2011. Immuno-fluorescence assay of leptospiral surface-exposed proteins. J. Vis. Exp. 53: 2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sankar S., Harshan H. M., Somarajan S. R., Srivastava S. K.2010. Evaluation of a recombinant LigB protein of Leptospira interrogans serovar Canicola in an enzyme-linked immunosorbent assay for the serodiagnosis of bovine leptospirosis. Res. Vet. Sci. 88: 375–378. doi: 10.1016/j.rvsc.2009.11.004 [DOI] [PubMed] [Google Scholar]
- 31.Sejvar J., Bancroft E., Winthrop K., Bettinger J., Bajani M., Bragg S., Shutt K., Kaiser R., Marano N., Popovic T., Tappero J., Ashford D., Mascola L., Vugia D., Perkins B., Rosenstein N., Eco-Challenge Investigation Team.2003. Leptospirosis in “Eco-Challenge” athletes, Malaysian Borneo, 2000. Emerg. Infect. Dis. 9: 702–707. doi: 10.3201/eid0906.020751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun A., Wang Y., Du P., Wu S., Yan J.2011. A sensitive and specific IgM-ELISA for the serological diagnosis of human leptospirosis using a rLipL32/1-LipL21-OmpL1/2 fusion protein. Biomed. Environ. Sci. 24: 291–299. [DOI] [PubMed] [Google Scholar]
- 33.Thiermann A. B.1981. The Norway rat as a selective chronic carrier of Leptospira icterohaemorrhagiae. J. Wildl. Dis. 17: 39–43. doi: 10.7589/0090-3558-17.1.39 [DOI] [PubMed] [Google Scholar]
- 34.Villanueva S. Y., Ezoe H., Baterna R. A., Yanagihara Y., Muto M., Koizumi N., Fukui T., Okamoto Y., Masuzawa T., Cavinta L. L., Gloriani N. G., Yoshida S.2010. Serologic and molecular studies of Leptospira and leptospirosis among rats in the Philippines. Am. J. Trop. Med. Hyg. 82: 889–898. doi: 10.4269/ajtmh.2010.09-0711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization2003, posting date. Human Leptospirosis: Guidance for Diagnosis, Surveillance and Control. [Online]
- 36.Ye C., Yan W., McDonough P. L., McDonough S. P., Mohamed H., Divers T. J., Chang Y. F., Yang Z.2014. Serodiagnosis of equine leptospirosis by enzyme-linked immunosorbent assay using four recombinant protein markers. Clin. Vaccine Immunol. 21: 478–483. doi: 10.1128/CVI.00649-13 [DOI] [PMC free article] [PubMed] [Google Scholar]