Abstract
Mycoplasma bovis has spread widely throughout the world via animal movement and has become an important pathogen of bovine respiratory disease. However, the minimum inhibitory concentrations of antimicrobials for Mycoplasma bovis have not been studied in China. The objective of this study was to determine the prevalence and antibiotic resistance of Mycoplasma bovis isolated from young cattle with respiratory infection in China. Mycoplasma bovis was detected in 32/45 bovine respiratory infection outbreaks at beef farms in 8 provinces in China. The isolates were susceptible or had medium sensitivity to ciprofloxacin, enrofloxacin and doxycycline, but were frequently resistant to macrolides (13/32, 41%). An A2058G (Escherichia coli Numbering) mutation located in the rrnA operon in domain V of 23S rRNA was observed in strains that were resistant to macrolides. This single mutations at the rrnA operon in domain V of 23S rRNA may play an important role in the resistance of Mycoplasma bovis strains to macrolides.
Keywords: antimicrobial susceptibility, bovine respiratory infection, macrolides, mycoplasma bovis, target mutation
Mycoplasma bovis (M. bovis) was first isolated from a severe case of mastitis in cattle in the United States in 1961 [7]. Subsequently, the results of some studies have suggested that M. bovis can cause respiratory infection, arthritis and tenosynovitis in feedlot cattle [2]. In recent years, M. bovis has spread widely to all parts of the world via animal movement and has become an important pathogen of bovine respiratory disease (BRD) in China and other countries [5]. The BRD caused by M. bovis is mainly treated with antibiotics, including veterinary macrolide antibiotics and fluoroquinolones in China, but treatment of BRD with macrolides often fails, leading to important economical losses in China.
Macrolide resistance has been described for pathogens of BRD, including M. bovis, Pasteurella multocida and Mannheimia haemolytica, in different countries. However, we found that the macrolide resistance of these pathogens is quite different in different countries. The genes msr(E) mph(E) and erm(42) have been shown to confer resistance to macrolides in Pasteurella multocida and Mannheimia haemolytica in Germany [11]. However, high-level macrolide resistance of Pasteurella multocida and Mannheimia haemolytica isolated in Europe can be due to 23S rRNA mutations [12]. One study of M. bovis isolated in Israel found that a combination of mutations in two domains of 23S rRNA is necessary to achieve higher minimum inhibitory concentrations of macrolides (MICs, ≥128 µg/ml) [8]. However, little research on this topic has been conducted in China. Therefore, systematic monitoring of antibiotic susceptibility and determination of the macrolide resistance mechanism of M. bovis strains in China are important.
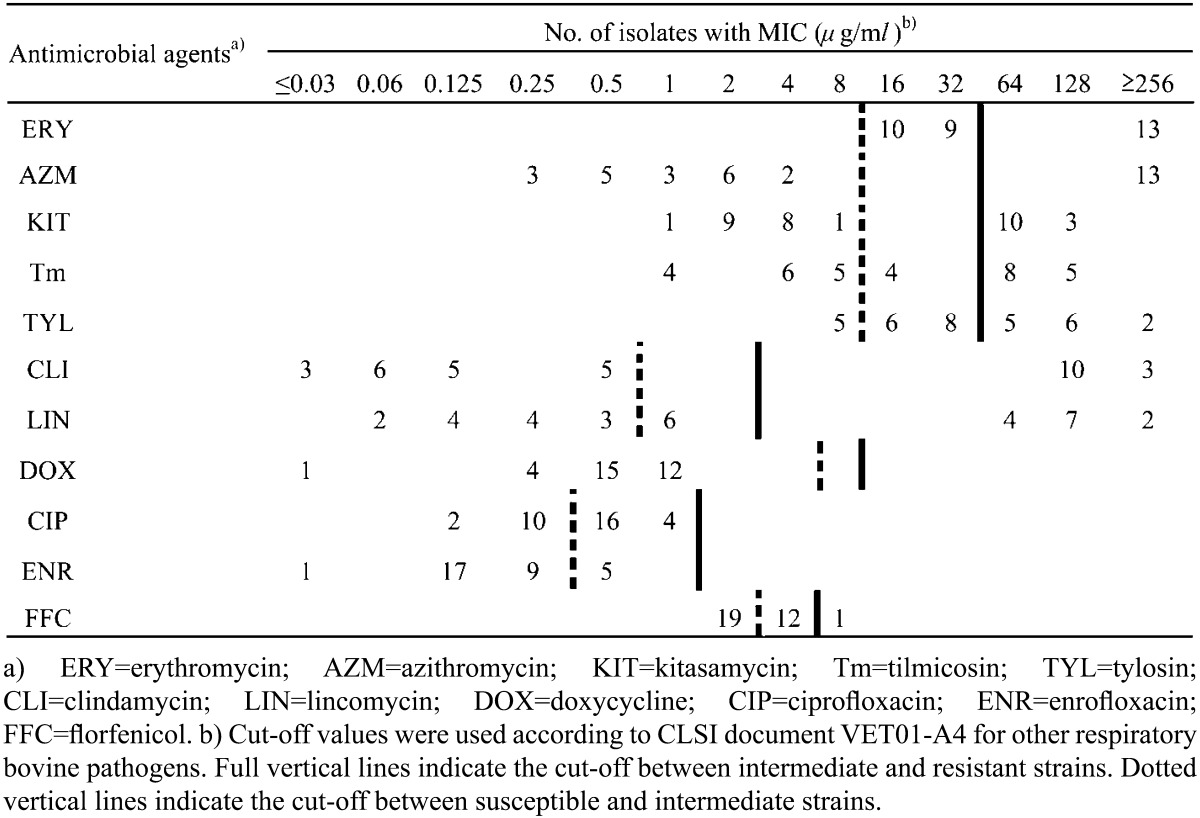
A total of 32 M. bovis strains originating from 32 feedlot cattle herds located in 8 provinces in China (Jilin, Heilongjiang, Neimenggu, Liaoning, Shandong, Hebei, Henan and Jiangsu) were tested in this study. Samples from lungs and nasal swabs were collected from distinct outbreaks between 2011 and 2013. All samples were incubated on pleuropneumonia-like organism (PPLO) agar plates. Suspected colonies with typical “fried egg” morphology were selected from each sample and identified using biochemical methods and PCR assay, as described previously [13]. The number of color changing units (CCU) was calculated, and antimicrobial susceptibility testing was performed by the microplate dilution method [4, 10]. Two-fold dilutions of antibiotics from 0.03 to 256 µg/ml were tested. Because there are no CLSI-approved MIC cut-off values for veterinary Mycoplasma species, it is difficult to interpret the impact of antimicrobial activity in vitro. CLSI-approved interpretative criteria for other respiratory bovine pathogens are frequently used to understand the implication of M. bovis sensitivity testing in vitro [6, 14]. The cut-off values for enrofloxacin and ciprofloxacin (susceptible, ≤0.25 µg/ml; resistant, ≥2 µg/ml), doxycycline (susceptible, ≤4 µg/ml; resistant, ≥16 µg/ml), clindamycin (susceptible, ≤0.5 µg/ml; resistant, ≥4 µg/ml), tulathromycin (susceptible, ≤16 µg/ml; resistant, ≥64 µg/ml) and florfenicol (susceptible, ≤2 µg/ml; resistant, ≥8 µg/ml) were defined, and the cut-off values for macrolides (susceptible, ≤16 µg/ml; resistant, ≥64 µg/ml) were defined by dichotomizing the latest CLSI criteria for veterinary pathogenic bacteria BRD [3]. The reference strains, Escherichia coli ATCC 25922 and M. bovis type strain PG45 (ATCC 25523), were used for a quality control. To identify rRNA mutations of M. bovis that confer resistance to macrolides, the two alleles that contain domain II and domain V were detected. PCR was performed according to a previous study [8]. The primers and program are shown in Table 1. PCR was performed in a 25 µl reaction volume containing 2.5 µl 10 × PCR buffer, 0.5 mmol L−1 dNTP, 0.5 µmol L−1 of each primer, 15.6 µl PCR water and 1 U LA-Taq polymerase (Takara, Otsu, Japan). The sequences were compared with the sequence of PG45, and sequences editing, consensus and alignment construction were performed by DNASTAR and ClustalW. Numbering of nucleotide and amino acid positions is based on the 23 rRNA gene or L4/L22 proteins of Escherichia coli, respectively.
Table 1. MICs distribution of M. bovis isolates.
| Amplifictaion target | Primer | Primers sequence (5′–3′) | Product | Annealing Temperature |
|---|---|---|---|---|
| (bp) | (°C) | |||
| rrnA | rrnA-F | GGATATCTAACGCCGTGTCT | 5,041 | 50 |
| rrnA-R | GTACTGGTCAGCTCAACAC | |||
| rrnB | rrnB-F | GCATGCAAGGTTAAGCAG | 2,848 | 50 |
| rrnB-R | CTAATTCCAAGTGCCACTAGCG | |||
| L4 | L4-F | TTTAGAAAAAAGAAATGAAGACAA | 603 | 49 |
| L4-R | CTACTCATATTGGCGATCTAGTT | |||
| L22 | L22-F | ATGAGTACTCAACAAGCTAAAGCA | 329 | 49 |
| L22-R | AATGCTATTGATAAATTAGATGTTC |
Then, the MIC of erythromycin was determined in the presence of the potent efflux inhibitors, carbonyl cyanide m-chlorophenyl hydrazones (CCCPs) and verapamil, at appropriate concentrations using the broth microdilution method [17]. Some previous studies have found that MICs change in the presence or absence of inhibitors due to changes in the level of activity of efflux pumps. However, a two-fold reduction is not sufficient to rule out false positives. Thus, a four-fold or greater reduction in strain MICs in the presence of inhibitors is considered to be due to the activity of efflux pumps [9, 18]. Each experiment was repeated three times.
The MIC values of the 10 antimicrobial agents obtained from the examinations of the China M. bovis isolates are shown in Table 2, and the MIC values for PG45 were as follows: erythromycin (2 µg/ml), azithromycin (2 µg/ml), kitasamycin (2 µg/ml), tylosin (0.125 µg/ml), clindamycin (0.125 µg/ml), lincomycin (0.25 µg/ml), doxycycline (0.03 µg/ml), ciprofloxacin (0.125 µg/ml), enrofloxacin (0.125 µg/ml) and florfenicol (2 µg/ml). Fluoroquinolones were found to be the most active compounds in vitro (MIC ≤1 µg/ml). For one isolate, the MIC for florfenicol was high (MIC=8 µg/ml), while the rest of the strains were inhibited by florfenicol at lower concentrations (MIC ≤4 µg/ml). Thirteen (41%) isolates were resistant to erythromycin, tylosin, azithromycin and kitasamycin, with MICs ≥64 µg/ml. The MICs of clindamycin and lincomycin were different in two distinct populations of isolates: 13 strains yielded MICs ≥128 µg/ml, while the rest yielded MICs ≤0.125 µg/ml. These results are in accordance with previous studies in other countries, which found that the most active compounds were fluoroquinolones and florfenicol [15, 16]. However, macrolides have been used traditionally and are losing their efficacy against M. bovis in many countries, which is in accordance with our results, as many M. bovis strains in China are already resistant to macrolides. Furthermore, the strains that were resistant to macrolides were also resistant to lincomycin and clindamycin, which has been previously observed in other Mycoplasma of animal and human origins. The mechanism of this resistance probably involves rRNA mutations [1, 10].
Table 2. MICs distribution of M. bovis isolates.
The 23S rRNA gene sequences of susceptible strains and resistance strains were analyzed (Table 3). The macrolide-resistant strains only had one mutation type, an A2058G substitution in domain V in the rrnA operon of the 23S rRNA. None of the macrolide-resistant strains contained substitutions in the rrnB operon. Additionally, there were no significant differences in domain II of L4 or L22 ribosomal proteins between resistant and susceptibl isolates. The A2058G substitution in domain V was the most prevalent substitution in our study and a previous study [8], and this single mutation may play an important role in the resistance of M. bovis strains to macrolides. However, other point mutations were found in previous studies in one or two domains of 23SrRNA, including G748A, C752T, A2059G and A2059C, which can reduce the sensitivity of M. bovis to tylosin and tilmicosin. Differences in the genotypes of different M. bovis strains may be due to differences in their evolutionary courses in different countries or to the development of different resistance mechanisms. Our results indicate that the values of erythromycin and azithromycin were not decreased in the presence of CCCP (32 µg/ml and 64 µg/ml for both antibiotics, respectively) or verapamil (64 µg/ml and 128 µg/ml, respectively) in highly resistant strains, and Chinese macrolide-resistant strains were negative for antibiotic efflux. Whether M. bovis strains have other resistance mechanisms should be investigated further in future studies.
Table 3. Molecular characterization of macrolides-resistant M. bovis field isolates.
| Isolate types | No. of isolates | 23S rRNA |
23S rRNA |
23S rRNA |
23S rRNA |
L4 |
L22 |
|---|---|---|---|---|---|---|---|
| Domain II rrnA | Domain V rrnA | Domain II rrnB | Domain V rrnB | ribosomal protein | ribosomal protein | ||
| resistant isolates | 13 | None | A2058G | None | None | None | None |
| susceptible isolates | 19 | None | None | None | None | None | None |
This is the first report of systematic monitoring of antibiotic susceptibility of M. bovis in China. We believe that studies should be performed to evaluate changes in MIC values and genetic mutations to determine the prevalence of M. bovis strains that are resistant to different antimicrobials.
Acknowledgments
This research was funded by the National Natural Science Foundation of China (Grant No. 31272611), the National Natural Science Foundation of China (Grant No. 31140026), the World Bank Loan Project of Jilin Province (Grant No. 2011-Y05), the Science and Technology Development Plan in Jilin Province (Grant No. 20111820) and State Key Laboratory of Veterinary Biotechnology.
REFERENCES
- 1.Bébéar C., Pereyre S., Peuchant O.2011. Mycoplasma pneumoniae: susceptibility and resistance to antibiotics. Future Microbiol. 6: 423–431. doi: 10.2217/fmb.11.18 [DOI] [PubMed] [Google Scholar]
- 2.Caswell J. L., Bateman K. G., Cai H. Y., Castillo-Alcala F.2010. Mycoplasma bovis in respiratory disease of feedlot cattle. Vet. Clin. N. Am-Food A. 26: 365–379. [DOI] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute2013. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. Approved Standard, 4th ed. In: ClSI document VET01-A4, CLSI, Wayne. [Google Scholar]
- 4.Clinical and Laboratory Standards Institute2008. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved Standard, 3rd ed. In: CLSI document M31-A3, CLSI, Wayne. [Google Scholar]
- 5.Gautier-Bouchardon A. V., Ferré S., Le Grand D., Paoli A., Gay E., Poumarat F.2014. Overall decrease in the susceptibility of Mycoplasma bovis to antimicrobials over the past 30 years in France. PLoS ONE 9: e87672. doi: 10.1371/journal.pone.0087672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gerchman I., Levisohn S., Mikula I., Lysnyansky I.2009. In vitro antimicrobial susceptibility of Mycoplasma bovis isolated in Israel from local and imported cattle. Vet. Microbiol. 137: 268–275. doi: 10.1016/j.vetmic.2009.01.028 [DOI] [PubMed] [Google Scholar]
- 7.Hale H. H., Helmboldt C. F., Plastridge W. N., Stula E. F.1962. Bovine mastitis caused by a Mycoplasma species. Cornell Vet. 52: 582–591. [PubMed] [Google Scholar]
- 8.Lerner U., Amram E., Ayling R. D., Mikula I., Gerchman I., Harrus S., Teff D., Yogev D., Lysnyansky I.2014. Acquired resistance to the 16-membered macrolides tylosin and tilmicosin by Mycoplasma bovis. Vet. Microbiol. 168: 365–371. doi: 10.1016/j.vetmic.2013.11.033 [DOI] [PubMed] [Google Scholar]
- 9.Kong L. C., Gao D., Gao Y. H., Liu S. M., Ma H. X.2014. Fluoroquinolone resistance mechanism of clinical isolates and selected mutants of Pasteurella multocida from bovine respiratory disease in China. J. Vet. Med. Sci. 76: 1655–1657. doi: 10.1292/jvms.14-0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lysnyansky I., Gerchman I., Flaminio B., Catania S.2015. Decreased Susceptibility to Macrolide-Lincosamide in Mycoplasma synoviae Is Associated with Mutations in 23S Ribosomal RNA. Microb. Drug Resist.doi: 10.1089/mdr.2014.0290 [DOI] [PubMed] [Google Scholar]
- 11.Michael G. B., Eidam C., Kadlec K., Meyer K., Sweeney M. T., Murray R. W., Watts J. L., Schwarz S.2012. Increased MICs of gamithromycin and tildipirosin in the presence of the genes erm(42) and msr(E)-mph(E) for bovine Pasteurella multocida and Mannheimia haemolytica. J. Antimicrob. Chemother. 67: 1555–1557. doi: 10.1093/jac/dks076 [DOI] [PubMed] [Google Scholar]
- 12.Olsen A. S., Warrass R., Douthwaite S.2015. Macrolide resistance conferred by rRNA mutations in field isolates of Mannheimia haemolytica and Pasteurella multocida. J. Antimicrob. Chemother. 70: 420–423. doi: 10.1093/jac/dku385 [DOI] [PubMed] [Google Scholar]
- 13.Rifatbegović M., Assunção P., Poveda J. B., Pasić S.2007. Isolation of Mycoplasma bovis from the respiratory tract of cattle in Bosnia and Herzegovina. Vet. Rec. 160: 484–485. doi: 10.1136/vr.160.14.484 [DOI] [PubMed] [Google Scholar]
- 14.Rosenbusch R. F., Kinyon J. M., Apley M., Funk N. D., Smith S., Hoffman L. J.2005. In vitro antimicrobial inhibition profiles of Mycoplasma bovis isolates recovered from various regions of the United States from 2002 to 2003. J. Vet. Diagn. Invest. 17: 436–441. doi: 10.1177/104063870501700505 [DOI] [PubMed] [Google Scholar]
- 15.Sato T., Okubo T., Usui M., Higuchi H., Tamura Y.2013. Amino acid substitutions in GyrA and ParC are associated with fluoroquinolone resistance in Mycoplasma bovis isolates from Japanese dairy calves. J. Vet. Med. Sci. 75: 1063–1065. doi: 10.1292/jvms.12-0508 [DOI] [PubMed] [Google Scholar]
- 16.Shin S. B., Yoo M. H., Jeong J. B., Kim Y. M., Chung J. K., Huh M. D., Komisar J. L., Jeong H. D.2005. Molecular cloning of the gyrA gene and characterization of its mutation in clinical isolates of quinolone-resistant Edwardsiella tarda. Dis. Aquat. Organ. 67: 259–266. doi: 10.3354/dao067259 [DOI] [PubMed] [Google Scholar]
- 17.Soehnlen M. K., Kunze M. E., Karunathilake K. E., Henwood B. M., Kariyawasam S., Wolfgang D. R., Jayarao B. M.2011. In vitro antimicrobial inhibition of Mycoplasma bovis isolates submitted to the Pennsylvania Animal Diagnostic Laboratory using flow cytometry and a broth microdilution method. J. Vet. Diagn. Invest. 23: 547–551. doi: 10.1177/1040638711404155 [DOI] [PubMed] [Google Scholar]
- 18.Spigaglia P., Barbanti F., Louie T., Barbut F., Mastrantonio P.2009. Molecular analysis of the gyrA and gyrB quinolone resistance-determining regions of fluoroquinolone-resistant Clostridium difficile mutants selected in vitro. Antimicrob. Agents Chemother. 53: 2463–2468. doi: 10.1128/AAC.01252-08 [DOI] [PMC free article] [PubMed] [Google Scholar]