Abstract
During surveillance for bovine diarrhea of unknown causes in Japanese black cattle in Kagoshima Prefecture, Japan, we found two types of novel kobu-like viruses in fecal samples of calves. Sequence analyses revealed that they had L protein and 2A protein with H-box/NC sequence motif, which are present in kobuviruses. Phylogenetic analysis revealed that they were related to kobuviruses; however, they clustered apart from other kobuviruses. In the prevalence study of two types of novel kobu-like viruses, 16.9% and 10.4% prevalence of these viruses were observed in the feces of diarrheal calves in this area.
Keywords: candidate new species, cattle, Japan, kobuvirus
Kobuviruses are members of the family Picornaviridae, which currently comprises 29 genera [4]. Picornaviruses are nonenveloped small RNA viruses with a positive-sense, single-stranded RNA genome that encodes a single polyprotein [14]. The genus Kobuvirus currently comprises three species (Aichivirus A, formerly aichi virus; Aichivirus B, formerly bovine kobuvirus; and Aichivirus C, formerly porcine kobuvirus) in addition to two unassigned viruses: caprine kobuvirus 1 and European roller kobuvirus 1 [4]. Aichivirus A and Aichivirus B are constituted by four and three types of viruses (i.e., Aichivirus A: aichi virus 1, feline kobuvirus 1, canine kobuvirus 1 and murine kobuvirus 1; Aichivirus B: bovine kobuvirus 1, ferret kobuvirus 1 and sheep kobuvirus 1), respectively, while Aichivirus C comprises only porcine kobuvirus 1.
Aichi virus and bovine kobuvirus were first recognized in Aichi Prefecture, Japan, in 1989 and 2003, respectively [17, 18]. Aichi virus 1 is a causative agent of gastroenteritis in humans [14], while porcine kobuvirus 1 was detected with a high frequency in non-diarrheic healthy pigs [12, 19]. Bovine kobuvirus 1 is thought to be associated with diarrhea in cattle; however, it can be detected in both healthy and diarrheic animals [5, 14].
Kagoshima Prefecture is located at the south end of Kyushu, and around 312,000 Japanese black breeding cattle are raised for beef in this area, which is the largest population of cattle in Japan. In the course of an epidemiological investigation of bovine diarrhea in Kagoshima Prefecture, we identified novel kobuviruses in fecal samples of Japanese black calves.
Thirteen fecal samples, collected during December 2014 to January 2015, from calves with diarrhea were investigated using a metagenomics approach. Viral RNA extraction, cDNA library construction and deep sequencing were performed as described previously [9]. We found large sequence contigs, both of which had an approximate length of 8,000 nucleotides (nt) from two calves (8 and 14 days old) raised on two distinct farms. These two samples also contained the sequence reads of nebovirus and bovine astrovirus. Using nucleotide BLAST searches, the contigs, which had about 8,000 nt length, showed similarity to kobuvirus genomes. We determined the complete genome sequence of one strain, named Kagoshima-2-24-KoV/2015/JPN (Kago-2-24), using the rapid amplification cDNA end method (5′-Full RACE Core Set; TaKaRa Bio, Otsu, Japan). The complete genome length of Kago-2-24 was 8,466 nucleotides (nt) with G+C content of 0.56, excluding the poly (A) tail. The Kago-2-24 genome included a single large open reading frame (ORF) containing 7,809 nt, which encoded a polyprotein precursor of 2,502 amino acids (aa), flanked by the 5′UTR (803 nt) and the 3′UTR (154 nt [excluding poly (A) tail]), and had a standard picornavirus genome organization, i.e. 5′UTR-L- P1 (VP0- VP3- VP1) -P2 (2A- 2B- 2C) -P3 (3A- 3B- 3C-3D) -3′UTR- poly (A). The other contig containing nearly complete genome sequences, including the complete open reading frames, was named Kagoshima-1-22-KoV/2014/JPN (Kago-1-22). The ORF of Kago-1-22 comprised 7,455 nt and encoded a polyprotein precursor of 2,484 aa. The 2A proteins of Kago-2-24 and Kago-1-22 contained H1051W-box/N1113C and H1062W-box/N1124C motifs, which are present in kobuviruses and bat picornaviruses (data not shown). Nucleotide sequences were deposited in the DNA Data Bank of Japan. The DDBJ/EMBL/GenBank accession numbers for the nucleotide sequences of Kago-2-24 and Kago-1-22 are LC 055960 and LC 055961, respectively.
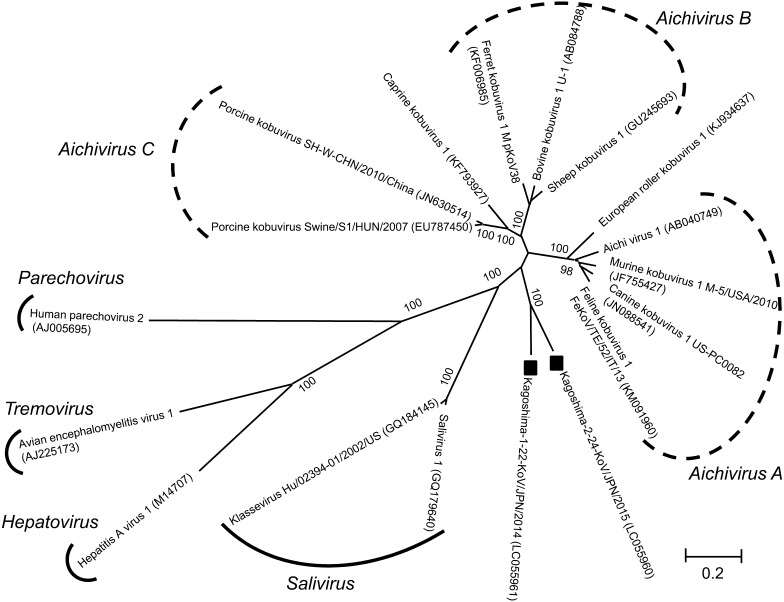
Complete amino acid sequences of Kago2-24 and Kago1-22 were aligned with representative strains of kobuviruses, salivirus-1 (Salivirus) [7], klassevirus Hu/02394-01/2002/US (Salivirus) [3], hepatitis A virus 1 (Hepatovirus) [1], human parechovirus 2 (Parechovirus) [2] and avian encephalomyelitis virus 1 (Tremovirus) [8] using ClustalW, and phylogenetic analysis was performed by the neighbor-joining method using MEGA5.22 [15]. The phylogenetic tree was statistically supported by bootstrapping with 1,000 replicates. Kago-2-24 and Kago-1-22 were related to each other and formed a distinct cluster in the tree with 100% bootstrap support. These strains are distantly related to the current official species of the genus Kobuvirus, i.e. Aichivirus A [18], B [17] and C [13, 16] and unassigned viruses: caprine kobuvirus 1 [10] and European roller kobuvirus 1 [11] (Fig. 1). Pairwise sequence identity calculations were performed using CLC Genomics Workbench 6.0 (CLC bio, Aarhus, Denmark). Pairwise alignment of the amino acid sequences of Kago-2-24 and Kago-1-22 with those of kobuviruses revealed that they shared 45.4–50.0% (complete polyprotein), 43.3–53.7% (P1), 49.4–54.8% (2C), 41.7–49.7% (P2), 56.7–60.8% (3CD) and 54.0–57.3% (P3) sequence identities (Table 1). The sequence identities between Kago-2-24 and Kago-1–22 were 65.3% (complete polyprotein), 73.6% (P1), 61.4% (2C) and 71.9% (3CD).
Fig. 1.
Phylogenetic analyses based on the complete amino acid sequences of the polyprotein of Kago-2-24 and Kago-1-22 (with black square) with kobuviruses, salivirus-1 (Salivirus), klassevirus Hu/02394-01/2002/US (Salivirus), hepatitis A virus 1 (Hepatovirus), human parechovirus 2 (Parechovirus) and avian encephalomyelitis virus 1 (Tremovirus) obtained from DDBJ/EMBL/GenBank database. Phylogenetic tree was constructed using the neighbor-joining method in MEGA5.22 with bootstrap values (1,000 replicates).
Table 1. Pairwise amino acid identities among the predicted proteins of two Japanese bovine kobuvirses, other kobuviruses and salivirus 1.
| Amino acid identity (%) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aichivirus A (AB040749) |
Aichivirus B (AB084788) |
Aichivirus C (EU787450) |
Caprine kobuvirus (KF793927) |
European roller kobuvirus (KJ934637) |
Salivirus 1 (GQ179640) |
Kago-2-24 |
|||||||
| Kago-2-24 | Kago-1-22 | Kago-2-24 | Kago-1-22 | Kago-2-24 | Kago-1-22 | Kago-2-24 | Kago-1-22 | Kago-2-24 | Kago-1-22 | Kago-2-24 | Kago-1-22 | Kago-1-22 | |
| Complete polyprotein | 46.5 | 47.4 | 49.1 | 50.0 | 49.6 | 49.7 | 49.1 | 49.4 | 45.4 | 46.2 | 35.0 | 35.2 | 65.3 |
| P1 | 44.8 | 44.2 | 51.2 | 51.5 | 53.7 | 53.0 | 52.1 | 50.1 | 46.2 | 45.9 | 37.8 | 37.7 | 73.6 |
| 2C | 51.8 | 53.4 | 52.3 | 54.0 | 52.3 | 52.8 | 54.7 | 54.8 | 49.4 | 50.7 | 42.5 | 42.6 | 61.4 |
| 3D | 59.5 | 59.7 | 60.8 | 58.1 | 59 | 56.7 | 59.3 | 58.2 | 57.1 | 58.2 | 44.6 | 43.3 | 71.9 |
We inoculated the clarified supernatant from 10% fecal samples into the Madin-Darby Bovine Kidney cells and Vero cells. However, cytopathic effect was not observed in any inoculated cells.
To investigate the prevalence of these kobuviruses, the specific amplification primers for Kago-2-24 (forward primer: 5′-GCTTTCATGGACCATTGCGCG-3′, reverse primer: 5′-GGAGAGTCGAGAGCACCGCCGAA-3′) and Kago-1-22 (forward primer: 5′-TCTGCAAACAACTACCAGAT-3′, reverse primer: 5′-GTGGGAAGCGGTTTGTGACAGC-3′) were designed based on 5′UTR sequences of these strains, and reverse transcription PCR (RT-PCR) was performed. Of the 77 fecal samples collected from 3- to 81-day-old diarrheal calves from 34 farms in Kagoshima Prefecture from October 2014 to January 2015, 13 calves (16.9%) from two farms and 8 calves (10.4%) from five farms were positive for Kago-2-24 and Kago-1-22 specific RT-PCR, respectively. Two farms which were positive for Kago-2-24 RNA were located separately from five farms which were positive for Kago-1-22 RNA, indicating that prevalence regions of these viruses may be separated. In addition, seven and ten fecal samples of diarrheic calves from distinct regions, namely the Ishikawa Prefecture and Hokkaido Prefecture, respectively, were investigated using Kago-2-24- and Kago-1-22-specific RT-PCR. No positive sample for RNA of Kago-2-24 or Kago-1-22 was identified.
In summary, we identified two novel kobu-like viruses from Japanese black cattle. These viruses had a standard picornavirus genome organization and sequence homology to the genus Kobuvirus. Kago-2-24 and Kago-1-22 have L protein. The 2A proteins of Kago-2-24 and Kago-1-22 contain H-box/NC motifs which are present in kobuviruses. Thus, it is suggested that these viruses belong to the genus Kobuvirus. Phylogenetic analysis revealed that they were related to kobuviruses; however, they clustered apart from other kobuviruses. The International Committee on Taxonomy of Viruses (ICTV) has proposed that the different species within kobuviruses are defined by amino acid identities in the complete polyprotein, P1, 2C and 3CD regions being <70%, <70%, <80% and <80%, respectively [4]. Based on these criteria, Kago-2-24 and Kago-1-22 do not fall into the current proposed species of Kobuvirus and probably represent a previously unrecognized species. Furthermore, the Picornaviridae section in the 9th ICTV Report has proposed that different picornavirus genera are defined by amino acid identity in the polyprotein being <58% [6]. Based on the criteria, Kago-2-24 and Kago-1-22 may not fall into the current official genera.
These new kobu-like viruses were identified from diarrheal calves of Japanese black cattle in Kagoshima Prefecture, which is a major region of beef cattle production in Japan. In this area, 16.9% and 10.4% detection rates for RNA of Kago-2-24 and Kago-1-22, respectively, were observed in the feces of diarrheal calves. No RT-PCR-positive sample for RNA of Kago-2-24 and Kago-1-22 was identified in diarrheal calves from other distinct areas. Although these new kobu-like viruses were identified from calves with diarrhea, the pathogenic potential of these viruses in cattle is still unclear. In the metagenomics analyses of the fecal samples containing Kago-2-24 and Kago-1-22, in addition to these kobu-like viruses, the sequence reads of nebovirus and astrovirus were simultaneously identified. Nebovirus and astrovirus are bovine enteric viruses; however, their association with enteric diseases is not well documented. In future, the pathogenic potential of these kobu-like viruses including co-infection with other enteric viruses should be clarified, and their potential transmission to other domestic animals or humans should be investigated.
Acknowledgments
This work was supported by the Grants from the Ministry of Health, Labor and Welfare of Japan.
REFERENCES
- 1.Cohen J. I., Ticehurst J. R., Purcell R. H., Buckler-White A., Baroudy B. M.1987. Complete nucleotide sequence of wild-type hepatitis A virus: comparison with different strains of hepatitis A virus and other picornaviruses. J. Virol. 61: 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghazi F., Hughes P. J., Hyypiä T., Stanway G.1998. Molecular analysis of human parechovirus type 2 (formerly echovirus 23). J. Gen. Virol. 79: 2641–2650. doi: 10.1099/0022-1317-79-11-2641 [DOI] [PubMed] [Google Scholar]
- 3.Greninger A. L., Runckel C., Chiu C. Y., Haggerty T., Parsonnet J., Ganem D., DeRisi J. L.2009. The complete genome of klassevirus - a novel picornavirus in pediatric stool. Virol. J. 6: 82. doi: 10.1186/1743-422X-6-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Committee on Taxonomy of Viruses Virus taxonomy 2008. [cited 2015 June 2] http://www.ictvonline.org/virustaxonomy.asp?version=2014.
- 5.Khamrin P., Maneekarn N., Peerakome S., Okitsu S., Mizuguchi M., Ushijima H.2008. Bovine kobuviruses from cattle with diarrhea. Emerg. Infect. Dis. 14: 985–986. doi: 10.3201/eid1406.070784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knowles N. J., Hovi T., Hyypiä T., King A. M. Q., Lindberg A. M., Pallansch M. A., Palmenberg A. C., Simmonds P., Skern T., Stanway G., Yamashita T., Zell R.2012. Picornaviridae. pp. 855–880. In: Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. (King, A. M. Q., Adams, M. J., Carstens, E. B. and Lefkowitz, E. J. eds.), Elsevier, San Diego. [Google Scholar]
- 7.Li L., Victoria J., Kapoor A., Blinkova O., Wang C., Babrzadeh F., Mason C. J., Pandey P., Triki H., Bahri O., Oderinde B. S., Baba M. M., Bukbuk D. N., Besser J. M., Bartkus J. M., Delwart E. L.2009. A novel picornavirus associated with gastroenteritis. J. Virol. 83: 12002–12006. doi: 10.1128/JVI.01241-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marvil P., Knowles N. J., Mockett A. P., Britton P., Brown T. D., Cavanagh D.1999. Avian encephalomyelitis virus is a picornavirus and is most closely related to hepatitis A virus. J. Gen. Virol. 80: 653–662. doi: 10.1099/0022-1317-80-3-653 [DOI] [PubMed] [Google Scholar]
- 9.Nagai M., Shimada S., Fujii Y., Moriyama H., Oba M., Katayama Y., Tsuchiaka S., Okazaki S., Omatsu T., Furuya T., Koyama S., Shirai J., Katayama K., Mizutani T.2015. H2 genotypes of G4P[6], G5P[7], and G9[23] porcine rotaviruses show super-short RNA electropherotypes. Vet. Microbiol. 176: 250–256. doi: 10.1016/j.vetmic.2015.02.002 [DOI] [PubMed] [Google Scholar]
- 10.Oem J. K., Lee M. H., Lee K. K., An D. J.2014. Novel Kobuvirus species identified from black goat with diarrhea. Vet. Microbiol. 172: 563–567. doi: 10.1016/j.vetmic.2014.06.009 [DOI] [PubMed] [Google Scholar]
- 11.Pankovics P., Boros Á., Kiss T., Reuter G.2015. Identification and complete genome analysis of kobuvirus in faecal samples of European roller (Coracias garrulus): for the first time in a bird. Arch. Virol. 160: 345–351. doi: 10.1007/s00705-014-2228-7 [DOI] [PubMed] [Google Scholar]
- 12.Reuter G., Boldizsár A., Kiss I., Pankovics P.2008. Candidate new species of Kobuvirus in porcine hosts. Emerg. Infect. Dis. 14: 1968–1970. doi: 10.3201/eid1412.080797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reuter G., Boldizsár A., Pankovics P.2009. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch. Virol. 154: 101–108. doi: 10.1007/s00705-008-0288-2 [DOI] [PubMed] [Google Scholar]
- 14.Reuter G., Boros A., Pankovics P.2011. Kobuviruses - a comprehensive review. Rev. Med. Virol. 21: 32–41. doi: 10.1002/rmv.677 [DOI] [PubMed] [Google Scholar]
- 15.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S.2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C., Lan D., Cui L., Yang Z., Yuan C., Hua X.2012. Molecular characterization of a porcine kobuvirus strain in China. Arch. Virol. 157: 573–578. doi: 10.1007/s00705-011-1205-7 [DOI] [PubMed] [Google Scholar]
- 17.Yamashita T., Ito M., Kabashima Y., Tsuzuki H., Fujiura A., Sakae K.2003. Isolation and characterization of a new species of kobuvirus associated with cattle. J. Gen. Virol. 84: 3069–3077. doi: 10.1099/vir.0.19266-0 [DOI] [PubMed] [Google Scholar]
- 18.Yamashita T., Kobayashi S., Sakae K., Nakata S., Chiba S., Ishihara Y., Isomura S.1991. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J. Infect. Dis. 164: 954–957. doi: 10.1093/infdis/164.5.954 [DOI] [PubMed] [Google Scholar]
- 19.Yu J. M., Jin M., Zhang Q., Li H. Y., Li D. D., Xu Z. Q., Li J. S., Cui S. X., Yang S. H., Liu N., Duan Z. J.2009. Candidate porcine Kobuvirus, China. Emerg. Infect. Dis. 15: 823–825. doi: 10.3201/eid1505.081518 [DOI] [PMC free article] [PubMed] [Google Scholar]