Abstract
Genomic resource of type strains and historically important strains of genus Stenotrophomonas allowed us to reveal the existence of 18 distinct species by applying modern phylogenomic criterions. Apart from Stenotrophomonas maltophilia, S. africana represents another species of clinical importance. Interestingly, Pseudomonas hibsicola, P. beteli, and S. pavani that are of plant origin are closer to S. maltophilia than the majority of the environmental isolates. The genus has an open pan-genome. By providing the case study on genes encoding metallo-β-lactamase and Clustered Regularly Interspaced Short Palindrome Repeats (CRISPR) regions, we have tried to show the importance of this genomic dataset in understanding its ecology.
Keywords: Stenotrophomonas, Type Strains, phylogenomics, Average Nucleotide Identity (ANI), evolution
Background
The members of the genus Stenotrophomonas are widespread in the diverse habitats with biotechnological applications and clinical relevance (Ryan et al., 2009). According to a recent report by WHO, S. maltophilia is a leading drug-resistant pathogen in hospitals worldwide (Brooke, 2014). Currently, the genus Stenotrophomonas compromises 12 validated species in the List of Prokaryotic Names Standing in Nomenclature1 from diverse habitats. The type species of the genus S. maltophilia was initially isolated from the pleural fluid and named as Bacterium bookeri and was then reclassified as P. maltophilia (Hugh and Ryschenkow, 1961). Further, this species was transferred to genus Xanthomonas as X. maltophilia (Swings et al., 1983) and later it was designated as a distinct and new genus Stenotrophomonas (Palleroni and Bradbury, 1993). Remaining 11 species of this genus were isolated from distinct environmental sources, S. rhizophila (Wolf et al., 2002), S. pavanii (Ramos et al., 2011), S. humi, S. terrae (Heylen et al., 2007), S. ginsengisoli (Kim et al., 2010), and S. panacihumi (Yi et al., 2010), S. koreensis (Yang et al., 2006), S. nitritireducens (Finkmann et al., 2000), S. acidaminiphila (Assih et al., 2002), S. chelatiphaga (Kaparullina et al., 2009), and S. daejeonensis (Lee et al., 2011). Additionally, there are many taxonomical revisions in the genus Stenotrophomonas. S. africana was initially described as a novel species of this genus, isolated from human cerebrospinal fluid (Drancourt et al., 1997). But later based on the whole cell protein and DNA–DNA hybridization analysis S. africana was proposed as a synonym of S. maltophilia (Coenye et al., 2004). S. dokdonesis (Yoon et al., 2006) a former species of the genus Stenotrophomonas, was assigned to the new genus Pseudoxanthomonas (Lee et al., 2008). Apart from this, there are three species of genus Pseudomonas, i.e., P. beteli, P. hibiscicola, and P. geniculata which are transferred to the genus Stenotrophomonas considered as synonyms of the S. maltophilia (Van den Mooter and Swings, 1990; Anzai et al., 2000). P. pictorium, is also considered to be closer to Stenotrophomonas (Svensson-Stadler et al., 2012).
Herein we generated draft genomes of 16 type strains which include 11 currently validated species of the genus Stenotrophomonas and five genomes from the different genera which are historically associated or grouped with the Stenotrophomonas. Whole genome sequence of type strains of the S. rhizophila and P. hibisicola are available in the public database (Table 1). The complete genome of S. maltophilia type strain is available publically (Davenport et al., 2014), but we had also sequenced independently and hence included in this study. The genome sequence of the type strains will be valuable in taxonomic and evolutionary studies of genus Stenotrophomonas and its relatives.
Table 1.
Genome features of the Stenotrophomonas genomes under study.
| S. No | Species | Genome size (Mbp) | No. of contigs | Coverage fold | N50 (bp) | GC Content (%) | No. of CDS | Isolation source | Accession no. | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | S. maltophilia ATCC 13637T | 4.98921 | 1 | 417 | 4989212 | 66.1 | 4645 | Blood | CP008838 | Davenport et al., 2014 |
| 2 | S. maltophilia MTCC 434T | 4.88156 | 306 | 52.5 | 41743 | 66.20 | 4327 | Blood | JALV00000000 | This study |
| 3 | S. africana LMG22072T | 4.51217 | 173 | 116 | 47895 | 66.30 | 3991 | Cerebrospinal fluid | LLXW00000000 | This study |
| 4 | P. hibsicola ATCC 19867T | 4.42403 | 20 | NA | 411451 | 66.40 | 3928 | Plant | ARNB01000000 | DOE-Joint Genome Institute∗ |
| 5 | P. beteli LMG00978T | 4.48462 | 109 | 184 | 83392 | 66.80 | 3907 | Piper betle | LLXV00000000 | This study |
| 6 | S. pavanii DSM 25135T | 4.3138 | 129 | 115 | 79654 | 67.40 | 3783 | Stem of sugarcane | LDJN00000000 | This study |
| 7 | P. geneculata JCM13324T | 4.80979 | 170 | 154 | 55144 | 66.20 | 4339 | Tap water | LLXT00000000 | This study |
| 8 | S. chelatiphaga DSM 21508T | 3.96773 | 148 | 115 | 51682 | 66.90 | 3366 | Municipal sewage | LDJK00000000 | This study |
| 9 | S. rhizophilia DSM 14405T | 4.64898 | 8 | 38 | 736911 | 67.30 | 3928 | Rhizosphere soil | CP007597 | Alavi et al., 2014 |
| 10 | S. panacihumi JCM 16536T | 3.92315 | 141 | 203 | 56105 | 68.80 | 3403 | Soil | LLXU00000000 | This study |
| 11 | S. koreensis DSM 17805T | 3.0299 | 58 | 185 | 188329 | 66.10 | 2662 | Compost | LDJH00000000 | This study |
| 12 | S. ginsengisoli DSM24757T | 3.37411 | 99 | 157 | 85121 | 65.90 | 2928 | Field Soil | LDJM00000000 | This study |
| 13 | S. acidaminiphila JCM13310T | 3.94252 | 126 | 116 | 71728 | 68.80 | 3405 | Sewage bioreactor | LDJO00000000 | This study |
| 14 | S. daeonesis JCM 16244T | 3.28486 | 124 | 154 | 46611 | 68.60 | 2816 | Sewage | LDJP00000000 | This study |
| 15 | P. pictorium JCM 9942T | 3.508292 | 84 | 193 | 89339 | 66.00 | 3099 | Soil | LLXS00000000 | This study |
| 16 | S. humi DSM 18929T | 4.12205 | 92 | 143 | 171493 | 64.00 | 3549 | Soil | LDJI00000000 | This study |
| 17 | S. nitritireducens DSM 12575T | 3.98349 | 95 | 140 | 167790 | 68.30 | 3387 | Bio filters | LDJG00000000 | This study |
| 18 | S. terrae DSM 18941T | 4.41032 | 143 | 131 | 96150 | 63.90 | 3670 | Soil | LDJJ00000000 | This study |
| 19 | S. dokdonesis DSM 21858T | 3.55366 | 34 | 169 | 325708 | 64.50 | 3063 | Soil | LDJL00000000 | This study |
Methods
Bacterial Strains and Culture Conditions
Type strains of genus Stenotrophomonas and related species (Table 1) were procured from different culture collection centers, Microbial Type Culture Collection (MTCC), Belgian Coordinated Collections of Microorganisms/LMG (BCCM/LMG) and The Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures GmbH (DSM). The high molecular weight genomic DNA of S. africana LMG 22072 was procured from BCCM/LMG for whole genome sequencing. All isolates were grown as per the media and conditions recommended by the respective culture collection centers.
Genome Sequencing, Assembly, and Annotation
Genomic DNA was extracted by using ZR Fungal/Bacterial DNA MiniPrep Kit (Zymo Research Corporation, Irvine, CA, USA) and quantified using Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Illumina sequencing library of genomic DNA was prepared using Nextera XT sample preparation kit (Illumina, Inc., San Diego, CA, USA) with dual indexing adapters. Illumina sequencing library was sequenced using in-house Illumina Miseq (Illumina, Inc., San Diego, CA, USA) platform using paired-end sequencing kits. The Illumina adapters were trimmed by the internal software during the base calling. In addition, to that adapter contamination identified by NCBI during the submission was removed by manual trimming. Raw reads were assembled using CLC Genomics Workbench v7.5 (CLC Bio-Qiagen, Aarhus, Denmark) and annotation was using NCBI Prokaryotic Genome Annotation Pipeline through NCBI2. CRISPR was identified using the CRISPR recognition tool (Bland et al., 2007).
Genome Similarity Assessment
For genome similarity assessment we used BLAST-based average nucleotide identity (ANIb) and Genome to Genome Distance calculator or digital DNA-DNA hybridization (dDDH) values. Pairwise ANI was calculated using JSpecies (Richter and Rosselló-Móra, 2009) and digital DDH (Auch et al., 2010) was calculated using web tool GGDC 2.03. Xanthomonas campestris pv. campestries ATCC 33913 and P. aeruginosa DSM 50071 were included as outgroups.
Pan-Genome Analysis
Pan-genome analysis of representatives of the genus Stenotrophomonas and related species under study was carried out by using the PGAP pipeline version 1.12 (Zhao et al., 2012). The MultiParanoid (MP) method was used for Pan-genome analysis with minimum score value 40 and e-value e1 × 10-10 used as cut off for BLAST. Pan-genome was visualized by using PanGP (Zhao et al., 2014). The flower pot diagram to represent the core and unique genes was drawn by using python script of Matplotlib (Hunter, 2007).
Data Deposition
The genome sequence data of the 16 type strains sequenced under this study has been deposited in NCBI GenBank and their accession numbers are mentioned in Table 1. Single point access to download genomes in FASTA format is also available at Figshare4.
Interpretation of Data Set
Genome Sequences and Phylogenic Inference
The general features of the newly sequenced genomes of type strains and their assembly statistics are summarized in Table 1. The ANI and dDDH values of representatives of the genus Stenotrophomonas are summarized in Supplementary Table S1. In microbial taxonomy for species delineation, 94 and 70% cut-off is used for ANI values and dDDH values, respectively, (Richter and Rosselló-Móra, 2009; Auch et al., 2010). No two strains have >94% ANI and >70% dDDH values suggesting that all the 18 members belong to distinct species.
Stenotrophomonas africana was initially described as a novel species but later reclassified as a synonym of S. maltophilia. Interestingly ANI and dDDH values of S. africana with the type strain of S. maltophilia are 90 and 49%, respectively, suggesting that the S. africana is a separate species.
The taxonomic status of the misclassified species P. genicualata, P. hibsicola, and P. betele is unclear and they are considered as synonyms of the S. maltophilia. But based on ANI and dDDH values with the type strain of the S. maltophilia, these species P. genicualata, P. hibsicola, and P. betele do not belong to S. maltophilia and represent separate species.
Pseudomonas pictorium is also considered as a misclassified Pseudomonas and is closely related to Stenotrophomonas sp. It shows <94% ANI and <70% dDDH with type strains of all Stenotrophomonas species and should be reclassified as a distinct species of the genus Stenotrophomonas.
Stenotrophomonas dokdonensis which was transferred to a new genus Pseudoxanthomonas exhibits ANI in the range of 73–75% and dDDH around 20% with the type strain of the species of Stenotrophomonas. Hence, there is need to re-examine its classification into a separate genus.
Pangenome Analysis of the Genus Stenotrophomonas
Genome sequences of the 18 Stenotrophomonas species under study were used to analyze the pan-genome and core genome. A total of 11052 genes were identified and among them, 1328 (12%) genes make the core genome of genus Stenotrophomonas. The pan and core genome sizes were plotted against the number of genomes under study. The pan-genome curve shows that the power trend line has not attended the plateau (Figure 1) suggesting that Stenotrophomonas displays an open pan-genome and that the number of genomes analyzed here is not sufficient to describe the complete gene repertoire and it requires more sequencing in order to describe all genes of the genus. The remaining 9724 gene clusters make the accessory genome which also includes the species-specific unique genes that range from 146 to 501 genes (Figure 1).
FIGURE 1.
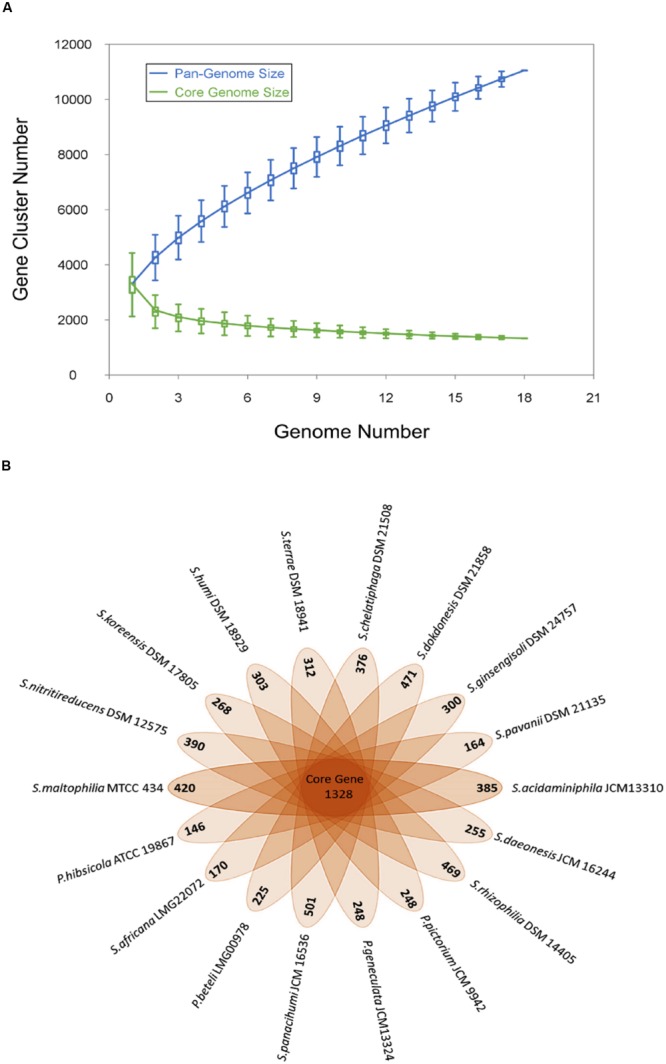
Pan and core genome of the genus Stenotrophomonas. (A) The number of gene clusters in pan-genome and core-genome are plotted against number of Stenotrophomonas genomes sequenced. (B) Flower plot diagram showing numbers of unique genes in each Stenotrophomonas species in the petals and Stenotrophomonas core orthologous gene number in the center.
Distribution of Chromosomal Metallo-β-Lactamase in Genus Stenotrophomonas
Type species of the genus Stenotrophomonas, i.e., S. maltophila has two chromosomally encoded β-lactamases, L1 and L2 which are characteristics of S. maltophilia and gives resistance to almost all β-lactam group of antibiotics (Denton and Kerr, 1998). L1 is a metallo-β-lactamase (Walsh et al., 1994; Ullah et al., 1998) and L2 is a clavulanic acid sensitive serine β-lactamases (Walsh et al., 1997). Here in we accessed the distribution of the L1 metallo-β-lactamase in the representative of the species of the genus. L1 metallo-β-lactamase is exclusively present only in the S. africana, S. pavani, P. genicualata, P. hibsicola, and P. betele along with the S. maltophilia and absent in the other species of the genus. The G+C content of the L1 metallo-β-lactamase is same as that of the genomic GC content, i.e., 67% suggesting that it is not acquired through the lateral gene transfer.
CRISPR-cas System in Stenotrophomonas
CRISPR-cas system is important to the bacteria for the adaptive immunity against the invasive elements in bacteria (Barrangou et al., 2007). Among 18 genomes analyzed representing 12 validated species and newly identified species of genus Stenotrophomonas, CRISPR loci are absent in the S. humi, S. koreensis, S. chelatiphaga, S. dokdonesis, S. daeonesis, P. pictorium, S. panacihumi, P. hibsicola and P. geniculata. Interestingly S. terrae, and S. acidaminiphila have multiple distinct CRISPR locus. The distribution of CRISPR locus across Stenotrophomonas genus along with repeat sequences and a number of repeats is mentioned in the Supplementary Table S2. It is important to note that the CRISPR loci are not widespread in this genus as 9 out of 18 genomes of the representative of the genus Stenotrophomonas are not having any CRISPR repeats.
Author Contributions
PP and SM carried out whole genome sequencing and prepared sequin files for submissions. PP and SK carried out the genome analysis. PP drafted the manuscript. PBP conceived the study and participated in its design and coordination. All authors have read and approved the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
PP is supported by the fellowship from UGC and SM is supported by the fellowship from CSIR. We acknowledge the funding from CSIR Network projects (BSC-402H and BSC-119E/HUM) and OLP-0062.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00309
References
- Alavi P., Starcher M. R., Thallinger G. G., Zachow C., Müller H., Berg G. (2014). Stenotrophomonas comparative genomics reveals genes and functions that differentiate beneficial and pathogenic bacteria. BMC Genomics 15:482 10.1186/1471-2164-15-482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzai Y., Kim H., Park J.-Y., Wakabayashi H., Oyaizu H. (2000). Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50 1563–1589. 10.1099/00207713-50-4-1563 [DOI] [PubMed] [Google Scholar]
- Assih E. A., Ouattara A. S., Thierry S., Cayol J.-L., Labat M., Macarie H. (2002). Stenotrophomonas acidaminiphila sp. nov., a strictly aerobic bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int. J. Syst. Evol. Microbiol. 52 559–568. 10.1099/00207713-52-2-559 [DOI] [PubMed] [Google Scholar]
- Auch A. F., Von Jan M., Klenk H.-P., Göker M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2:117 10.4056/sigs.531120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 1709–1712. 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- Bland C., Ramsey T. L., Sabree F., Lowe M., Brown K., Kyrpides N. C., et al. (2007). CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 8:209 10.1186/1471-2105-8-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke J. S. (2014). New strategies against Stenotrophomonas maltophilia: a serious worldwide intrinsically drug-resistant opportunistic pathogen. Expert Rev. Anti-Infect. Ther. 12 1–4. 10.1586/14787210.2014.864553 [DOI] [PubMed] [Google Scholar]
- Coenye T., Vanlaere E., Falsen E., Vandamme P. (2004). Stenotrophomonas africana Drancourt et al 1997 is a later synonym of Stenotrophomonas maltophilia (Hugh 1981) Palleroni and Bradbury 1993. Int. J. Syst. Evol. Microbiol. 54 1235–1237. 10.1099/ijs.0.63093-0 [DOI] [PubMed] [Google Scholar]
- Davenport K., Daligault H., Minogue T., Broomall S., Bruce D., Chain P., et al. (2014). Complete genome sequence of Stenotrophomonas maltophilia type strain 810-2 (ATCC 13637). Genome Announc. 2:e974 10.1128/genomeA.00974-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton M., Kerr K. G. (1998). Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drancourt M., Bollet C., Raoult D. (1997). Stenotrophomonas africana sp. nov., an opportunistic human pathogen in Africa. Int. J. Syst. Bacteriol. 47 160–163. 10.1099/00207713-47-2-606 [DOI] [PubMed] [Google Scholar]
- Finkmann W., Altendorf K., Stackebrandt E., Lipski A. (2000). Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 50 273–282. 10.1099/00207713-50-1-273 [DOI] [PubMed] [Google Scholar]
- Heylen K., Vanparys B., Peirsegaele F., Lebbe L., De Vos P. (2007). Stenotrophomonas terrae sp. nov. and Stenotrophomonas humi sp. nov., two nitrate-reducing bacteria isolated from soil. Int. J. Syst. Evol. Microbiol. 57 2056–2061. 10.1099/ijs.0.65044-0 [DOI] [PubMed] [Google Scholar]
- Hugh R., Ryschenkow E. (1961). Pseudomonas maltophilia, an Alcaligenes-like species. J. Gen. Microbiol. 26 123–132. 10.1099/00221287-26-1-123 [DOI] [PubMed] [Google Scholar]
- Hunter J. D. (2007). Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9 90–95. 10.1109/MCSE.2007.55 [DOI] [Google Scholar]
- Kaparullina E., Doronina N., Chistyakova T., Trotsenko Y. (2009). Stenotrophomonas chelatiphaga sp. nov., a new aerobic EDTA-degrading bacterium. Syst. Appl. Microbiol. 32 157–162. 10.1016/j.syapm.2008.12.003 [DOI] [PubMed] [Google Scholar]
- Kim H.-B., Srinivasan S., Sathiyaraj G., Quan L.-H., Kim S.-H., Bui T. P. N., et al. (2010). Stenotrophomonas ginsengisoli sp. nov., isolated from a ginseng field. Int. J. Syst. Evol. Microbiol. 60 1522–1526. 10.1099/ijs.0.014662-0 [DOI] [PubMed] [Google Scholar]
- Lee D. S., Ryu S. H., Hwang H. W., Kim Y.-J., Park M., Lee J. R., et al. (2008). Pseudoxanthomonas sacheonensis sp. nov., isolated from BTEX-contaminated soil in Korea, transfer of Stenotrophomonas dokdonensis Yoon et al. 2006 to the genus Pseudoxanthomonas as Pseudoxanthomonas dokdonensis comb. nov. and emended description of the genus Pseudoxanthomonas. Int. J. Syst. Evol. Microbiol. 58 2235–2240. 10.1099/ijs.0.65678-0 [DOI] [PubMed] [Google Scholar]
- Lee M., Woo S.-G., Chae M., Shin M.-C., Jung H.-M., Ten L. N. (2011). Stenotrophomonas daejeonensis sp. nov., isolated from sewage. Int. J. Syst. Evol. Microbiol. 61 598–604. 10.1099/ijs.0.017780-0 [DOI] [PubMed] [Google Scholar]
- Palleroni N. J., Bradbury J. F. (1993). Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int. J. Syst. Bacteriol. 43 606–609. 10.1099/00207713-43-3-606 [DOI] [PubMed] [Google Scholar]
- Ramos P. L., Van Trappen S., Thompson F. L., Rocha R. C., Barbosa H. R., De Vos P., et al. (2011). Screening for endophytic nitrogen-fixing bacteria in Brazilian sugar cane varieties used in organic farming and description of Stenotrophomonas pavanii sp. nov. Int. J. Syst. Evol. Microbiol. 61 926–931. 10.1099/ijs.0.019372-0 [DOI] [PubMed] [Google Scholar]
- Richter M., Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106 19126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P., Monchy S., Cardinale M., Taghavi S., Crossman L., Avison M. B., et al. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7 514–525. 10.1038/nrmicro2163 [DOI] [PubMed] [Google Scholar]
- Svensson-Stadler L. A., Mihaylova S. A., Moore E. R. (2012). Stenotrophomonas interspecies differentiation and identification by gyrB sequence analysis. FEMS Microbiol. Lett. 327 15–24. 10.1111/j.1574-6968.2011.02452.x [DOI] [PubMed] [Google Scholar]
- Swings J., De Vos P., Den Mooter M. V., De Ley J. (1983). Transfer of Pseudomonas maltophilia Hugh 1981 to the Genus Xanthomonas as Xanthomonas maltophilia (Hugh 1981) comb. nov. Int. J. Syst. Bacteriol. 33 409–413. 10.1099/00207713-33-2-409 [DOI] [Google Scholar]
- Ullah J., Walsh T., Taylor I., Emery D., Verma C., Gamblin S., et al. (1998). The crystal structure of the L1 metallo-β-lactamase from Stenotrophomonas maltophilia at 1.7 Å resolution. J. Mol. Biol. 284 125–136. 10.1006/jmbi.1998.2148 [DOI] [PubMed] [Google Scholar]
- Van den Mooter M., Swings J. (1990). Numerical analysis of 295 phenotypic features of 266 Xanthomonas strains and related strains and an improved taxonomy of the genus. Int. J. Syst. Bacteriol. 40 348–369. 10.1099/00207713-40-4-348 [DOI] [PubMed] [Google Scholar]
- Walsh T., Macgowan A., Bennett P. (1997). Sequence analysis and enzyme kinetics of the L2 serine beta-lactamase from Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 41 1460–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T. R., Hall L., Assinder S. J., Nichols W. W., Cartwright S. J., Macgowan A. P., et al. (1994). Sequence analysis of the L1 metallo-β-lactamase from Xanthomonas maltophilia. Biochim. Biophys. Acta 1218 199–201. 10.1016/0167-4781(94)90011-6 [DOI] [PubMed] [Google Scholar]
- Wolf A., Fritze A., Hagemann M., Berg G. (2002). Stenotrophomonas rhizophila sp. nov., a novel plant-associated bacterium with antifungal properties. Int. J. Syst. Evol. Microbiol. 52 1937–1944. 10.1099/00207713-52-6-1937 [DOI] [PubMed] [Google Scholar]
- Yang H.-C., Im W.-T., Kang M. S., Shin D.-Y., Lee S.-T. (2006). Stenotrophomonas koreensis sp. nov., isolated from compost in South Korea. Int. J. Syst. Evol. Microbiol. 56 81–84. 10.1099/ijs.0.63826-0 [DOI] [PubMed] [Google Scholar]
- Yi H., Srinivasan S., Kim M. K. (2010). Stenotrophomonas panacihumi sp. nov., isolated from soil of a ginseng field. J. Microbiol. 48 30–35. 10.1007/s12275-010-0006-0 [DOI] [PubMed] [Google Scholar]
- Yoon J.-H., Kang S.-J., Oh H. W., Oh T.-K. (2006). Stenotrophomonas dokdonensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 56 1363–1367. 10.1099/ijs.0.64070-0 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Jia X., Yang J., Ling Y., Zhang Z., Yu J., et al. (2014). PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics 30 1297–1299. 10.1093/bioinformatics/btu017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Wu J., Yang J., Sun S., Xiao J., Yu J. (2012). PGAP: pan-genomes analysis pipeline. Bioinformatics 28 416–418. 10.1093/bioinformatics/btr655 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


