Abstract
Hepatitis C virus (HCV) is a serious global problem, and present therapeutics are inadequate to cure HCV infection. In the present study, various antiviral assays show that As2O3 at submicromolar concentrations is capable of inhibiting HCV replication. The 50% effective concentration (EC50) of As2O3 required to inhibit HCV replication was 0.35 μM when it was determined by a reporter-based HCV replication assay, and the EC50 was below 0.2 μM when it was determined by quantitative reverse transcription-PCR analysis. As2O3 did not cause cellular toxicity at this concentration, as revealed by an MTS [3-(4,5-dimethylthiozol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] assay. A combination of As2O3 and alpha interferon exerted synergistic effects against HCV, as revealed by a multiple linear logistic model and isobologram analysis. Furthermore, in an alternative HCV antiviral system that may recapitulate additional steps involved in HCV infection and replication, As2O3 at 0.3 μM totally abolished the HCV signal, whereas alpha interferon at a high dose (5,000 IU/ml) only partially suppressed the HCV signal. The study highlights the indications for use of a novel class of anti-HCV agent. Further elucidation of the exact antiviral mechanism of As2O3 may lead to the development of agents with potent activities against HCV or related viruses.
Approximately 170 million individuals worldwide suffer from chronic hepatitis C virus (HCV) infection (38). Although the acute phase of infection is usually associated with mild symptoms, most patients acquire a persistent infection that frequently leads to chronic liver diseases, including cirrhosis and hepatocellular carcinoma (21, 25). At present, alpha interferon (IFN-α) in combination with ribavirin or a polyethylene glycol-modified form of IFN-α in combination with ribavirin is the only recommended therapy. IFN-based therapies have shown only limited efficacy and are unsuitable for certain patient populations, although IFN-based therapies are relatively more efficacious against genotypes 2 and 3 than against genotype 1 (13, 30, 31, 34). The limited efficacy against genotype 1 and the poor tolerability of IFN-based therapies warrant the search for more effective medicines to combat HCV infection.
HCV is the only member of the genus Hepacivirus that belongs to the family Flaviviridae. HCV contains a single-stranded, positive-sense RNA genome of approximately 9.5 kb which encodes a unique polyprotein (10). The polyprotein precursor is co- and posttranslationally processed by cellular and viral proteases to yield the mature structural and nonstructural proteins, arranged in the sequence NH2-C-E1-E2-P7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH. The inability to efficiently propagate HCV in cell culture has impeded the development of drugs against this virus. This obstacle was overcome, to a degree, by the recent development of a bicistronic subgenomic HCV replicon in Huh-7 cells (5, 26). These subgenomic replicon systems have greatly facilitated studies of HCV replication. The pharmaceutical industry has also been using such replication systems to screen for anti-HCV drugs (2).
A reporter gene that can be used to monitor HCV RNA replication by simply measuring the enzymatic activity of secreted alkaline phosphatase (SEAP) in the culture medium has been developed previously (23). In this study, we focused on screening a set of marketed drugs, including drugs not approved for antiviral indications. Here, we report on the discovery that As2O3 is a potent anti-HCV agent. We also showed that As2O3 and IFN-α have synergistic antiviral effects when they are used together. Recently, the U.S. Food and Drug Administration approved As2O3 for use for the treatment of acute promyelocytic leukemia (APL) (8, 37). The possibility that As2O3 or its analogues may be used for the treatment of HCV infections is discussed.
MATERIALS AND METHODS
Compound collection.
The compounds tested were mainly from The Genesis Plus Collection (MicroSource Discovery Systems, Inc., Gaylordsville, Conn.). The 960 compounds in this library are primarily U.S. Food and Drug Administration-approved compounds. An alphabetical list of the compounds can be found at the MicroSource Discovery Systems website (http://www.msdiscovery.com/genplus3.html). In addition, several other drugs used for the treatment of HCV infections, including arsenic trioxide and IFN-α, were ordered from Sigma, St. Louis, Mo.
Cell culture.
Huh-7 parental cells and Huh-7 cells containing HCV subgenomic replicons (Ava5) were provided by Apath, LLC (St. Louis, Mo.) (5). Human embryonic kidney cells constitutively expressing the EBNA-1 protein from Epstein-Barr virus (293EBNA cells; Invitrogen, Carlsbad, Calif.) were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and 250 μg of G418 per ml. Cell viability was determined by the MTS [3-(4,5-dimethylthiozol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetra-zolium, inner salt] assay, which was performed essentially as described previously (11). Each datum point represents the mean for six repeats.
Modified HCV replicon with a reporter gene.
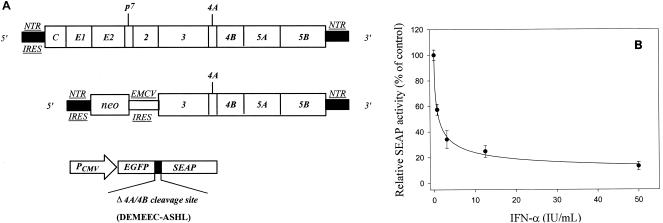
The reporter-based cell line for HCV drug screening was derived from HCV replicon cells (Ava5) (5). The subgenomic HCV RNA comprises the following elements: the 5′ end of the HCV genome, including its internal ribosomal entry site and 12 amino acid codons of the core protein, followed by the neomycin phosphotransferase (neo) gene; the internal ribosomal entry site of the encephalomyocarditis virus; the HCV NS3 to NS5B coding region; and the HCV 3′ nontranslated region (Fig. 1A, middle panel). Ava5 cells were engineered to contain a reporter gene, EG(Δ4AB)SEAP, and the stable cell line is designated Ava5-EG(Δ4AB)SEAP (22, 23). In the EG(Δ4AB)SEAP reporter gene, the enhanced green fluorescent protein (EGFP) was fused to SEAP through the NS3-NS4A protease decapeptide recognition sequence, Δ4AB, which spans the HCV NS4A-NS4B junction region (Fig. 1A, lower panel). We have previously shown that Ava5 cells that stably express the EG(Δ4AB)SEAP reporter gene can be used to reflect anti-HCV activity by measurement of the SEAP activity in the culture medium (22). In brief, Ava5-EG(Δ4AB)SEAP cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum containing 1 mg of G418 per ml and 10 μg of blasticidin per ml. Cells were seeded in 96-well plates at a density of 5 × 103 cells per well. After incubation for 1 day, the cells were treated with various concentrations of drugs. Two days after drug treatment of Ava5-EG(Δ4AB)SEAP cells, the culture medium was changed to remove the accumulated SEAP. The cultures were then replenished with fresh medium containing drugs at the same concentrations used in the initial incubation, and incubation was continued for another 24 h. The culture medium was then collected and subjected to SEAP activity assays. The SEAP activities were measured with a Phospha-Light assay kit (Tropix, Foster City, Calif.), according to the instructions of the manufacturer.
FIG. 1.
A novel reporter-based assay for anti-HCV drug screening. (A) Schematic representations of HCV full-length (upper) and subgenomic (middle) genome structures. The EGFP(Δ4AB)SEAP reporter construct is shown at the bottom. Ava5 cells contain the subgenomic replicon (5). Ava5-EGFP(Δ4AB)SEAP cells were engineered by stable integration of the EGFP(Δ4AB)SEAP reporter gene into Ava5 cells. (B) Anti-HCV activity of IFN-α in Ava5-EG(Δ4AB)SEAP cells. Ava5-EGFP(Δ4AB)SEAP cells were treated with IFN-α at various doses. The SEAP activities in the culture media were measured to reflect the extent of HCV replication inside cells. Results are expressed as the means ± standard deviations (error bars) for three replicate wells.
Preparation of total RNA and RT.
An RNeasy Mini kit (Qiagen GmbH, Hilden, Germany) or the TRIZOL reagent (Invitrogen, Carlsbad, Calif.) was used to extract total cellular RNA. One microgram of total RNA was mixed with 50 pmol of HCV-specific reverse primer (primer A9412; 5′-GATGGC CTATTG GCCTGG AGGGG-3′ ). After the secondary structure of RNA was allowed to unfold for 10 min at 65°C, reverse transcription (RT) reactions were carried out at 42°C for 1 h in a total volume of 20 μl that contained 1 mM each deoxynucleotide triphosphate, 50 U of reverse transcriptase (Expand-RT; Roche Biochemicals, Mannheim, Germany), and 20 U of RNase inhibitor (Promega, Madison, Wis.).
Q-RT-PCR.
Quantitative RT-PCR (Q-RT-PCR) with a LightCycler DNA master mixture (Roche Diagnostics GmbH, Mannheim, Germany) and SYBR Green I was used to detect the synthesis of PCR products in real time. The intensity of fluorescence versus the PCR cycle number was recorded, and the relative amount of HCV RNA was calculated from the corresponding amount on the calibration curve. The calibration curve was derived from five serial dilutions of reference DNA, which consisted of the corresponding cDNA of HCV RNA, by use of the criteria recommended by Roche Diagnostics.
The following primer sets were used to determine the relative copy numbers of HCV RNA and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Forward primer 5′-ACCAGAATACGACTTGGAGTTGATAA-3′ and reverse primer 5′-CACCCTTTTGCCAGATGCAT-3′ were used to measure the copy numbers of the positive strand of HCV. Forward primer 5′-CTCGACGTTGTCACTGA-3′ and reverse primer 5′-CCATCCGAGTACGTGC-3′ were used to measure the copy numbers of the negative strand of HCV. To measure the levels of GAPDH, we used forward primer 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse primer 5′-GAAGATGGTGATGGGATTTC-3′ in the Q-RT-PCR.
The PCR procedure was as follows: (i) denaturation at 95°C for 60 s and (ii) amplification by ramping to 95°C at 0 s, 64°C for 10 s, and 72°C for 10 s. The specificity of the amplification reaction was determined by performing a melting curve analysis with a temperature range from 65 to 95°C. Normalized HCV RNA copy numbers were obtained by dividing the HCV RNA copy numbers by the corresponding GAPDH copy numbers in the same sample. All measurements were performed in triplicate to facilitate statistical analysis.
Northern and Western blotting.
Northern blot analysis was performed as described previously (20, 23). Approximately 106 Ava5 cells were seeded onto 10-cm-diameter dishes and maintained in culture medium supplemented with 1 mg of G418 per ml. One day after the cells were seeded they were treated with the drugs tested. At various times, the cells were harvested and total cellular RNAs were isolated by use of a High Pure RNA isolation kit (Roche, Mannheim, Germany), in accordance with the instructions of the manufacturer. Three micrograms of total RNA was denatured by treatment with 2 M formaldehyde-50% formamide for 15 min at 55°C and separated by denaturing agarose gel electrophoresis. The RNA was then transferred to a positively charged nylon membrane (BrightStar-Plus; Ambion, Austin, Tex.) with the Speed Vacuum transfer apparatus (Amersham Bioscience, Buckinghamshire, England). After the RNA was dry, it was then cross-linked to the membrane by UV irradiation (Stratagene, La Jolla, Calif.). The membrane was probed separately with the NS5B gene fragment of HCV and the human GAPDH (gapdh) gene fragment labeled with [α-32P]dCTP by use of the Rediprime II random prime labeling system (Pharmacia), in accordance with the instructions of the manufacturer. Hybridization was carried out with denatured probes in Rapid-hyb hybridization buffer (Pharmacia) for 2 h at 65°C. After hybridization, the membranes were washed once in 2× SSC (1× is 0.15 M NaCl plus 0.015 M sodium citrate)-0.2% sodium dodecyl sulfate (SDS) for 20 min at 60°C, once in 1× SSC-0.2% SDS for 20 min at 60°C, and twice in 0.1× SSC-0.2% SDS for 15 min at 65°C. The results of hybridization were visualized by autoradiography. A standard procedure was used for Western blotting (17). Anti-NS3 and antiactin antibodies were obtained from LTK Biolaboratories (Taipei, Taiwan) and CHEMICON International, Inc. (Temecula, Calif.), respectively. Signals were revealed with an ECL Western blotting system (Amersham Pharmacia Biotech United Kingdom, Amersham PLC, Little Chalfont, England).
Assay of HCV infection of plasma.
Yeh and colleagues (39) have discovered a crucial gene, SiP-L, that enables an otherwise nonpermissive cell line to support HCV replication after cells are infected by HCV virions from HCV-positive plasma. The human liver cDNA clone Sip-L was inserted into a vector, pDR2, downstream of a Rous sarcoma virus long terminal repeat promoter (39). The resulting plasmid, pDR2-Sip-L, was transfected into 293EBNA cells by the standard calcium phosphate precipitation method. Transformants were selected by the addition of 150 μg of hygromycin B per ml into the culture medium. The resulting cells, 293EBNA-Sip-L cells, were capable of supporting HCV replication or maintaining HCV RNA when cells were infected by HCV-positive plasma (39).
HCV-positive plasma samples with different HCV genotypes (genotypes 1a, 1b, 2a, and 2b, as determined by a Inno-LIPA HCV II assay [Innogenetics, Zwijndrecht, Belgium]), were tested for their abilities to infect 293EBNA-Sip-L cells. These samples all contained 107 to 108 copies of HCV RNA per ml, as measured by a branched DNA method (Quantiplex HCV RNA assay, version 2.0; Chiron, Emeryville, Calif.). Infectivity by serum samples infected with genotypes 1a, 1b, and 2a was observed. In this study, HCV genotype 1b-infected plasma containing 107 copies of HCV RNA per ml, as measured by the branched DNA method, was used in the HCV virion infection assay. Cells in a 60-mm-diameter culture dish were incubated in culture medium containing 50 μl of HCV-positive plasma for 12 h. The cells were then incubated in fresh medium without HCV-positive serum, and the medium was changed everyday. To detect HCV RNA, infected cells were trypsinized and washed twice with fresh medium by centrifugation. The primers used for nested PCR were C1 (5′-CGGCAACAGGTAAACTCCAC-3′; antisense; nucleotides 114 to 95), C2 (5′-CCCTGTGAGGAACTACTGTC-3′; sense; nucleotides −299 to −280), C3 (5′-ACGATCTGACCACCGCCCGG-3′; antisense; nucleotides 92 to 73), and C4 (5′-TTCACGCAGAAAGCGTCTAG-3′; sense; nucleotides −279 to −260). A digoxigenin-labeled probe was used for the subsequent Southern blotting analysis.
Analysis of synergism.
The effects of drug combinations were examined by statistical analyses. A multiple linear logistic regression model (14) was used to quantify the dose-response relationship, which accounts for the interaction effect between As2O3 and IFN-α. Since isobolograms are frequently used to demonstrate the effects of multiple drugs for their additivity, synergism, or antagonism, an isobologram analysis (14, 16, 29) was also performed. The resulting isoeffect curve of 50% inhibition was given to depict the synergistic effect of the drug-drug interaction.
RESULTS
Development of Ava5-EG(Δ4AB)SEAP as drug screening system.
We have previously shown that Ava5 cells, which contain HCV subgenomic replicon RNA, could be engineered to express a reporter gene so that the level of HCV replication in Ava5 cells could be determined by simply measuring the SEAP enzyme activity in the cell culture medium (23). Thus, a reporter-based cell line, Ava5-EG(Δ4AB)SEAP, was created to constitutively express the EG(Δ4AB)SEAP reporter gene in Ava5 cells (22). A schematic diagram of the reporter gene is shown in Fig. 1A (bottom panel). In this reporter system for HCV replication, cleavage of the EGFP-SEAP reporter is dependent on viral NS3-NS4A protease cleavage of the reporter protein in trans and the fact that the presence of protease is only as a result of viral replication and translation. The Ava5-EG(Δ4AB)SEAP cells were tested with IFN-α to establish that anti-HCV activity can indeed be reflected by the levels of SEAP activity in the culture medium (Fig. 1B). Ava5-EG(Δ4AB)SEAP cells were treated with IFN-α at various doses up to 50 IU/ml. After 48 h, the culture medium were replaced with fresh medium containing the same concentration of drug used in the initial incubation, and the cells were incubated for another 24 h. Subsequently, the culture medium was harvested for analysis of SEAP activities. There was a clear dose-dependent decline in SEAP activity upon IFN-α treatment (Fig. 1B). The 50% effective concentration (EC50) of IFN-α required to inhibit HCV replication was approximately 2 IU/ml when the EC50 was measured by using Ava5-EG(Δ4AB)SEAP cells.
Effects of As2O3 on HCV replication.
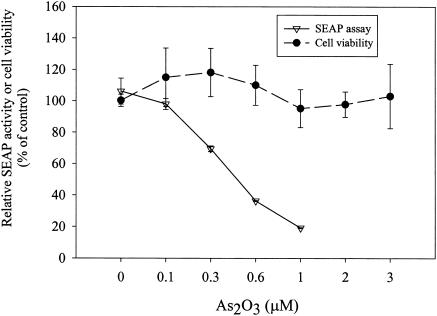
Using Ava5-EG(Δ4AB)SEAP cells, we screened a few hundred marketed drugs for their anti-HCV activities. In this study, we showed that As2O3 at a low concentration (less than 1 μM) can strongly inhibit HCV replication in cell culture. As2O3 decreased the SEAP activity in Ava5-EG(Δ4AB)SEAP cells in a dose-dependent manner (Fig. 2). When cells were treated with 1 μM As2O3, 7% SEAP activity remained compared to that in untreated (control) cells, while treatment with 50 IU of IFN-α per ml reduced SEAP activity to 18% of that for the control cells (Fig. 1B). No toxicity was observed for cells treated with As2O3 over this concentration range, as revealed by the MTS assay (Fig. 2), and the concentration required to inhibit cell viability by 50% (TD50) was measured to be 6.3 μM (data not shown).
FIG. 2.
Inhibition of HCV replication by As2O3 in reporter-based cells. By using Ava5-EGFP(Δ4AB)SEAP cells, As2O3 was found to possess potent anti-HCV activity at a concentration that did not cause cellular toxicity. The cellular toxicity was evaluated by the MTS assay. Results are expressed as the means ± standard deviations (error bars) for three replicate wells.
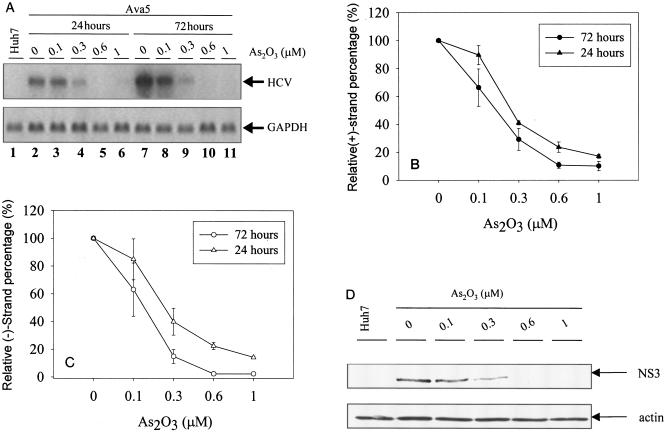
Various assays such as Q-RT-PCR, Northern blotting, and Western blotting were used to ascertain the effect of As2O3 on the inhibition of HCV replication. Ava5 cells were treated with As2O3, and the cells were harvested at 24 and 72 h after drug treatment. Subsequently, total cellular RNAs were extracted and analyzed by Northern blotting and Q-RT-PCR assays and the cell lysates were analyzed by Western blotting. As shown in Fig. 3A, there was a clear dose-dependent decline in HCV subgenomic RNA levels upon As2O3 treatment (lanes 2 to 6 and lanes 7 to 11), while the mRNA level of a housekeeping gene, the gene for GAPDH, remained unchanged. No signal corresponded to HCV subgenomic RNA in the parental Huh-7 cell line (lane 1).
FIG. 3.
Inhibition of HCV replication in Ava5 cells by As2O3. (A) Northern blot analysis of HCV replicon RNAs from Ava5 cells after treatment with 0, 0.1, 0.3, 0.6, or 1 μM As2O3 (lanes 2 to 6 and lanes 7 to 11, respectively). RNAs were extracted at 1 day (lanes 2 to 6) or 3 days (lanes 7 to 11) after drug treatment. As a control, RNA was extracted from parental cell line Huh-7 (lane 1). The amount of GAPDH transcript was used as a control for mRNA expression in cells treated with As2O3. Arrows indicate the HCV replicon RNA and GAPDH RNA. (B and C) Total cellular RNAs were also analyzed by Q-RT-PCR to measure the levels of positive-sense (B) and negative-sense (C) HCV RNA. Viral RNA levels were normalized with GAPDH levels, and levels obtained for samples not treated with drug were designated 100%. (D) Western blotting analysis of cell lysates. Ava5 cells were exposed to different concentrations of As2O3 for 72 h. Arrows indicate HCV NS3 and actin proteins. The results in panels B and C are expressed as the means ± standard deviations of three experiments.
The extracted cellular RNA samples were also analyzed by Q-RT-PCR to determine HCV RNA levels. The positive and negative strands of HCV RNA were measured and normalized according to the GAPDH RNA levels in each samples. In Fig. 3B and C, the relative copy numbers of the HCV positive and negative strands reflect the percentages of the remaining HCV RNA copy numbers after drug treatment compared to the copy numbers for the control, which received no drug treatment. The EC50s of As2O3 for HCV positive-sense RNA were determined to be 0.26 and 0.18 μM after 24 and 72 h of drug treatment, respectively. By using negative-sense viral RNA as the antiviral indicator, the EC50s of As2O3 were 0.28 and 0.15 μM after 24 and 72 h of drug treatment, respectively. These results indicate that As2O3 inhibits the generation of both positive- and negative-sense RNA in a similar kinetic profile. The TD50s of As2O3 after 1 and 3 days of treatment were 6.3 and 12.3 μM, respectively. Thus, when positive-sense viral RNA levels, as measured by the Q-RT-PCR used in this study, were used to reflect antiviral activity, the TD50/EC50 ratios were 47.3 (12.3 μM /0.26 μM) and 35.0 (6.3 μM/0.18 μM) for 1 day and 3 days of drug treatment, respectively, whereas by the SEAP reporter assay the TD50/EC50 ratio was 18.0 (6.3 μM /0.35 μM).
The anti-HCV effect of As2O3 was also evaluated by Western blotting. Ava5 cells were treated with As2O3 at various concentrations for 72 h. Cell lysates were harvested and analyzed by Western blotting with anti-NS3 antiserum. As shown in Fig. 3D, the level of NS3 protein declined upon As2O3 treatment in a dose-dependent manner, while the level of actin remained unchanged. No HCV NS3 antigen could be detected in cells after treatment with As2O3 at 0.6 or 1.0 μM. Huh-7 cells do not contain viral NS3 protein. Clearly, As2O3 has potent activity against HCV. We also examined the activities of As2O3 against other viruses and found that As2O3 did not inhibit unrelated viruses such as enterovirus 71 (EV71), human cytomegalovirus (HCMV), or porcine transmissible gastroenteritis coronavirus (TGEV) (S. R. Shih, L. J. Ruan, and C. M. Chen, respectively, personal communications).
Synergistic anti-HCV effects by use of As2O3 and IFN-α in combination.
The anti-HCV effects of As2O3 and IFN-α in combination were evaluated by using the Ava5-EG(Δ4AB)SEAP reporter-based cells. Ava5-EG(Δ4AB)SEAP cells were treated with these two drugs in combination in a checkerboard titration manner. After 48 h, the culture medium was changed and drug treatments were continued for another 24 h. The SEAP activities in the culture media were then measured. The dose-response inhibition of HCV RNA replication by various As2O3 concentrations (0, 0.1, 0.3, 0.6, and 1.0 μM) was evaluated in the presence of various doses of IFN-α (0, 1.25, 2.5, 5, and 10 IU/ml) (Table 1).
TABLE 1.
Effects of combination of As2O3 and IFN-α on HCV replication
| IFN-α concn (IU/ml) | Relative SEAP activity with the following As2O3 concn (μM)a:
|
|||
|---|---|---|---|---|
| 0 | 0.1 | 0.3 | 0.6 | |
| 0 | 100 ± 9.2 | 87 ± 3.9 | 58 ± 2.0 | 23 ± 0.4 |
| 1.25 | 79 ± 8.8 | 48 ± 1.9 | 39 ± 11.3 | 13 ± 5.4 |
| 2.5 | 58 ± 6.5 | 30 ± 4.3 | 14 ± 3.0 | 5 ± 3.5 |
| 5 | 45 ± 8.2 | 26 ± 5.2 | 14 ± 7.6 | 7 ± 4.4 |
| 10 | 32 ± 8.5 | 18 ± 4.0 | 5 ± 3.5 | 4 ± 3.8 |
The values represent the percentage of the control activity, and the results are expressed as the means ± standard deviations of three experiments.
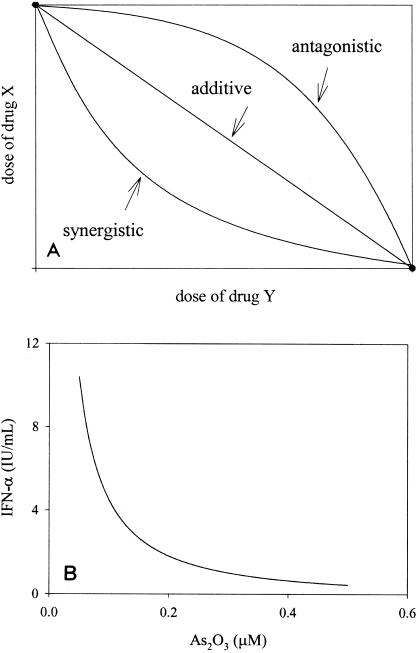
Multiple linear logistic models (14) and isobologram analysis (29) were used to determine whether the combination of As2O3 and IFN-α exerts a synergistic, additive, or antagonistic effect. In the multiple linear logistic regression model, the interaction effect between As2O3 and IFN-α was investigated by adding an interaction term. The estimated regression coefficient of this interaction term is positive, which indicates a synergistic effect, and an F test for the interaction term in a nested fashion reveals that it is statistically significant (P value < 0.05). In addition, isobologram analysis was performed to investigate the strength of the synergism. A general representation of an isobologram used to measure the drug-drug interaction is shown in Fig. 4A. It has been proposed that synergism, additivity, and antagonism are represented by concave, linear, and convex isoeffective curves (isoboles), respectively. We used equation 4 of Machado and Robinson (29) as the isobole-based model and used the corresponding nonadditivity parameter in the model to estimate the joint action ratio, R, an isobole-based measure for the joint potency of As2O3 and IFN-α, to quantify the degree of synergism (16). The resulting estimate of R is 1.2311 ± 0.0249, which is significantly different from the additivity hypothesis of R equal to 1 (P < 0.05). This indicates that the combination of As2O3 and IFN-α exerts a synergistic effect on the inhibition of HCV replication. The concave shape of the resulting 50% isobole for inhibition of HCV replication is displayed in Fig. 4B.
FIG. 4.
Anti-HCV effects of As2O3 and IFN-α in combination. (A) Example of the use of isobolograms to represent drug interactions. The synergy, additivity, and antagonism are represented by concave, linear, and convex isoeffective curves (isoboles), respectively. (B) Isobole of 50% inhibition of HCV replication by combinations of IFN-α and As2O3 at various doses. The concave shape of the resulting isobole of 50% inhibition is displayed.
Anti-HCV activity of As2O3 in an alternative viral infection system.
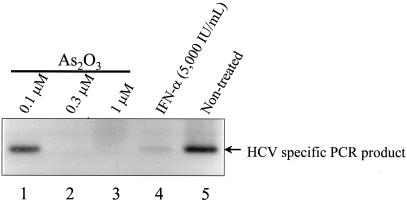
Although Ava5 and Ava5-EG(Δ4AB)SEAP cells containing the HCV subgenomic replicon were used to study HCV replication in this study, these subgenomic replicon systems do not sustain all steps of the HCV replication cycle. Thus, we intended to examine whether As2O3 has anti-HCV activity using an alternative HCV infection and replication assay that might constitute all steps involved in the full cycle of HCV infection and replication. For this purpose, we used an engineered cell line that could be infected by type 1b HCV-positive plasma (39). Yeh and colleagues (39) have discovered a crucial gene, SiP-L, that enables an otherwise nonpermissive cell line to support HCV replication after the cells are infected by HCV virions from HCV-positive plasma. Figure 5 shows that As2O3 at a concentration of 0.3 μM or greater was able to abolish the HCV signal in the 293EBNA-Sip-L cell HCV infection and replication system (Fig. 5, lanes 2 and 3). In this assay, a high dose of IFN-α (5,000 IU/ml) only partially suppressed the HCV signal, as can be seen from the faint HCV band following IFN-α treatment cells (Fig. 5, lane 4). These results indicate that 0.3 μM As2O3 is more potent than the high dose of IFN-α in suppressing the HCV signal in the whole virion infection ‘nd replication assay. Thus, As2O3 might inhibit HCV replication or interfere with the persistence of the genomes placed into HCV-infected 293EBNA-Sip-L cells.
FIG. 5.
Effect of As2O3 on HCV replication in 293EBNA-Sip-L cells determined by Southern blotting analysis of the RT-PCR products derived from HCV RNAs. 293EBNA-Sip-L cells were infected with HCV-positive serum containing 107 copies of HCV RNA per ml, and infected cells were treated with As2O3 (lanes 1 to 3). As controls, cells that were treated with 5,000 U of IFN-α per ml (lane 4) and cells that received no treatment (lane 5) were also tested. All drugs were added on day 4 after the cells were infected with HCV-positive serum. The source of the RNA for the HCV RNA assays came from cells that were harvested on the day 7 postinfection.
DISCUSSION
Because IFN-α-based therapies are not adequate for the treatment of HCV and IFN-α is poorly tolerated, expensive, and suitable only for certain patient populations, it is critical to identify new alternative therapeutics that are more effective and economical than the present therapies. We describe herein that As2O3 can effectively inhibit the replication of HCV in cells with HCV subgenomic replicons. In the reporter-based subgenomic replication system (Ava5-EG[Δ4AB]SEAP cells), the EC50 of As2O3 for inhibition of HCV replication, as determined by measurement of the SEAP levels in the culture medium, was approximately 0.35 μM. The anti-HCV activity of As2O3 has been confirmed by several other assays with Ava5 cells containing the HCV subgenomic RNA replicon (Fig. 3). Since it is important to consider strategies with drug combinations for the treatment of HCV infections, we also showed that As2O3 at low doses was able to enhance the anti-HCV efficacy of IFN-α in a synergistic manner (Fig. 4B). We further showed that As2O3 at 0.3 μM is able to totally abolish the HCV signal in an alternative HCV infection and replication system that might constitute all steps involved in the complete HCV infection and replication cycle (39) (Fig. 5). Our results indicate that a low dose of As2O3 would facilitate IFN-α-mediated antiviral activity.
It is interesting that IFN-α at 5,000 IU/ml could only partially inhibit HCV replication in the infection and replication assay with the whole HCV virion. In contrast to this result, the EC50 of IFN-α was approximately 2 IU/ml, as measured with Ava5-EG(Δ4AB)SEAP cells (Fig. 1B). Thus, IFN-α was much less potent in inhibiting the replication of the whole HCV genome than in inhibiting the replication of subgenomic HCV RNA. This is presumably due to the presence of viral structural proteins, C, E1, and E2, in the replication system with the whole HCV virion but not in the system with subgenomic HCV RNA. Viral protein E2 is known to inhibit PKR activities, which, in turn, might result in the attenuated antiviral effects of IFN-α (36). Furthermore, Keskinen et al. (19) showed that expression of HCV structural proteins impaired the antiviral activities of IFNs. Thus, expression of HCV structural proteins might have compromised the anti-HCV activity of IFN-α. In contrast, this study showed that As2O3 is a potent anti-HCV agent in two alternative antiviral assays.
The underlying mechanism of inhibition of HCV by As2O3 is not clear. As2O3 did not inhibit NS3-NS4A protease activity (results not shown). Arsenicals are known to inhibit many enzymes by reacting with biological ligands that possess available thiol groups in adjacent positions (3, 12, 18). In particular, it has been shown that arsenic can interact with thiol groups among the Cys113, Cys172, and Cys422 residues that are in spatial proximity in the metalloactivation domain of the ArsA ATPase (4). Similarly, by structural analysis based on molecular mechanics optimizations, we have observed that Cys303, Cys311, and Cys324 on three separate secondary structures of viral RNA polymerase (NS5B) are clustered in proximity (1, 6, 7, 24). This cluster of thiol groups might be able to form a stable arsenic adduct with reasonable bond distances and angles (unpublished observation). However, further investigations are needed to elucidate whether As2O3 or its metabolites inhibit NS5B viral polymerase. It is also tempting to speculate that As2O3 might exert its anti-HCV activity by interfering with cellular factors necessary for HCV replication. It has been shown that HCV replication is tightly coupled with cell growth (35), suggesting that the availability of cellular factors necessary for HCV replication is cell cycle dependent. Hartwig et al. (15) reported that very low concentrations of arsenite suppress poly(ADP-ribosyl)ation in mammalian cells. Nasr et al. (33) showed that As2O3 and IFN synergistically induce massive down-regulation of cell cycle-related genes and cause cell cycle arrest, leading to apoptosis. In view of all these findings, further investigations are needed to elucidate the exact antiviral mechanism of As2O3. Meanwhile, selection and examination of an HCV subgenomic RNA replicon resistant to As2O3 are also in progress. Elucidation of the antiviral mechanism by which As2O3 inhibits HCV replication may lead to the development of agents with potent activities against HCV or related viruses.
As2O3 is effective in treating APL and other types of cancers (9, 41). It has been suggested that the mechanism of action is through, at least in part, the degradation of the aberrant promyelocytic leukemia-retinoic acid receptor α fusion protein (32, 40). In this study, we demonstrate that As2O3 is a potent inhibitor of HCV replication. Thus, the development of As2O3 or its analogues as new anti-HCV therapeutics may be promising. In this regard, it is important to consider the pharmaceutical aspects of arsenicals or the use of drug delivery systems that will reduce the notorious toxicities of arsenical compounds. Realgar and orpiment have been used as traditional Chinese medicines that contain As4S4 and As2S3 as major ingredients, respectively (28). Interestingly, Lu et al. (27) have pointed out that As4S4 and As2S3 are far less toxic than As2O3. When As4S4 was taken orally, uptake was shown to be relatively nontoxic compared with the toxicity of As2O3 when it was administered by the intravenous route for the treatment of APL (27). Clinical pharmacokinetic studies demonstrated that arsenic levels in blood close to 1 μM could be achieved with As4S4 by a very safe dosing regimen. In summary, in view of its potent antiviral activity, As2O3 or its analogues deserve further evaluation for their anti-HCV mechanisms and potentials for the treatment of HCV infections.
Acknowledgments
We thank Yu-Sheng Chao, Chungming Chang, and Hsin-Pang Hsieh at the National Health Research Institutes (NHRI) of Taiwan, Yung-Chi Cheng at Yale University, and Monto Ho at the University of Pittsburgh for suggestions on this study. We also thank Charles Rice at Rockefeller University and Apath, LLC, for providing Huh-7 and Ava5 cells. We also thank Shin-Ru Shih at Chang-Gung University, Li-Jung Juan at NHRI, and Chimin Chen at the Animal Technology Institute of Taiwan for anti-EV71, anti-HCMV, and anti-TGEV testing.
This work was supported by funds and facilities of NHRI of Taiwan, provided to John Hsu.
REFERENCES
- 1.Ago, H., T. Adachi, A. Yoshida, M. Yamamoto, N. Habuka, K. Yatsunami, and M. Miyano. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Structure Fold Des. 7:1417-1426. [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager, R. 2002. Hepatitis C virus replicons: potential role for drug development. Nat. Rev. Drug Discov. 1:911-916. [DOI] [PubMed] [Google Scholar]
- 3.Berleth, E. S., and C. M. Pickart. 1996. Mechanism of ubiquitin conjugating enzyme E2-230K: catalysis involving a thiol relay? Biochemistry 35:1664-1671. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharjee, H., and B. P. Rosen. 1996. Spatial proximity of Cys113, Cys172, and Cys422 in the metalloactivation domain of the ArsA ATPase. J. Biol. Chem. 271:24465-24470. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 6.Bressanelli, S., L. Tomei, F. A. Rey, and R. De Francesco. 2002. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76:3482-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bressanelli, S., L. Tomei, A. Roussel, I. Incitti, R. L. Vitale, M. Mathieu, R. De Francesco, and F. A. Rey. 1999. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc. Natl. Acad. Sci. USA 96:13034-13039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, G. Q., X. G. Shi, W. Tang, S. M. Xiong, J. Zhu, X. Cai, Z. G. Han, J. H. Ni, G. Y. Shi, P. M. Jia, M. M. Liu, K. L. He, C. Niu, J. Ma, P. Zhang, T. D. Zhang, P. Paul, T. Naoe, K. Kitamura, W. Miller, S. Waxman, Z. Y. Wang, H. de The, S. J. Chen, and Z. Chen. 1997. Use of arsenic trioxide (As2O3) in the treatment of acute promyelocytic leukemia (APL). I. exerts As2O3 dose-dependent dual effects on APL cells. Blood 89:3345-3353. [PubMed] [Google Scholar]
- 9.Chen, Z., G. Q. Chen, Z. X. Shen, G. L. Sun, J. H. Tong, Z. Y. Wang, and S. J. Chen. 2002. Expanding the use of arsenic trioxide: leukemias and beyond. Semin. Hematol. 39:22-26. [DOI] [PubMed] [Google Scholar]
- 10.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 11.Cory, A. H., T. C. Owen, J. A. Barltrop, and J. G. Cory. 1991. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 3:207-212. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham, M. L., M. J. Zvelebil, and A. H. Fairlamb. 1994. Mechanism of inhibition of trypanothione reductase and glutathione reductase by trivalent organic arsenicals. Eur. J. Biochem. 221:285-295. [DOI] [PubMed] [Google Scholar]
- 13.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 14.Greco, W. R., G. Bravo, and J. C. Parsons. 1995. The search for synergy: a critical review from a response surface perspective. Pharmacol. Rev. 47:331-385. [PubMed] [Google Scholar]
- 15.Hartwig, A., A. Pelzer, M. Asmuss, and A. Burkle. 2003. Very low concentrations of arsenite suppress poly(ADP-ribosyl)ation in mammalian cells. Int. J. Cancer 104:1-6. [DOI] [PubMed] [Google Scholar]
- 16.Hewlett, P. S. 1969. Measurement of the potencies of drug mixtures. Biometrics 25:477-487. [PubMed] [Google Scholar]
- 17.Hsu, T. A., J. J. Eiden, P. Bourgarel, T. Meo, and M. J. Betenbaugh. 1994. Effects of co-expressing chaperone BiP on functional antibody production in the baculovirus system. Protein Expr. Purif. 5:595-603. [DOI] [PubMed] [Google Scholar]
- 18.Kapahi, P., T. Takahashi, G. Natoli, S. R. Adams, Y. Chen, R. Y. Tsien, and M. Karin. 2000. Inhibition of NF-kappa B activation by arsenite through reaction with a critical cysteine in the activation loop of Iκ B kinase. J. Biol. Chem. 275:36062-36066. [DOI] [PubMed] [Google Scholar]
- 19.Keskinen, P., K. Melen, and I. Julkunen. 2002. Expression of HCV structural proteins impairs IFN-mediated antiviral response. Virology 299:164. [DOI] [PubMed] [Google Scholar]
- 20.Kevil, C. G., L. Walsh, F. S. Laroux, T. Kalogeris, M. B. Grisham, and J. S. Alexander. 1997. An improved, rapid Northern protocol. Biochem. Biophys. Res. Commun. 238:277-279. [DOI] [PubMed] [Google Scholar]
- 21.Lauer, G. M., and B. D. Walker. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41-52. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J.-C., C.-F. Chang, Y.-H. Chi, D.-R. Hwang, and J. T.-A. Hsu. 2004. A reporter-based assay for identifying hepatitis C virus inhibitors based on subgenomic replicon cells. J. Virol. Methods 116:27-33. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J. C., Y. F. Shih, S. P. Hsu, T. Y. Chang, L. H. Chen, and J. T. Hsu. 2003. Development of a cell-based assay for monitoring specific hepatitis C virus NS3/4A protease activity in mammalian cells. Anal. Biochem. 316:162-170. [DOI] [PubMed] [Google Scholar]
- 24.Lesburg, C. A., M. B. Cable, E. Ferrari, Z. Hong, A. F. Mannarino, and P. C. Weber. 1999. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat. Struct. Biol. 6:937-943. [DOI] [PubMed] [Google Scholar]
- 25.Liang, T. J., B. Rehermann, L. B. Seeff, and J. H. Hoofnagle. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann. Intern. Med. 132:296-305. [DOI] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Lu, D.-P., J.-Y. Qiu, B. Jiang, Q. Wang, K.-Y. Liu, Y.-R. Liu, and S.-S. Chen. 2002. Tetra-arsenic tetra-sulfide for the treatment of acute promyelocytic leukemia: a pilot report. Blood 99:3136-3143. [DOI] [PubMed] [Google Scholar]
- 28.Lu, D. P., and Q. Wang. 2002. Current study of APL treatment in China. Int. J. Hematol. 76(Suppl. 1):316-318. [DOI] [PubMed] [Google Scholar]
- 29.Machado, S. G., and G. A. Robinson. 1994. A direct, general approach based on isobolograms for assessing the joint action of drugs in pre-clinical experiments. Stat. Med. 13:2289-2309. [DOI] [PubMed] [Google Scholar]
- 30.McHutchison, J. G., and M. W. Fried. 2003. Current therapy for hepatitis C: pegylated interferon and ribavirin. Clin. Liver Dis. 7:149-161. [DOI] [PubMed] [Google Scholar]
- 31.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 32.Miller, W. H., Jr., H. M. Schipper, J. S. Lee, J. Singer, and S. Waxman. 2002. Mechanisms of action of arsenic trioxide. Cancer Res. 62:3893-3903. [PubMed] [Google Scholar]
- 33.Nasr, R., A. Rosenwald, M. E. El-Sabban, B. Arnulf, P. Zalloua, Y. Lepelletier, F. Bex, O. Hermine, L. Staudt, H. de The, and A. Bazarbachi. 2003. Arsenic/interferon specifically reverses 2 distinct gene networks critical for the survival of HTLV-1-infected leukemic cells. Blood 101:4576-4582. [DOI] [PubMed] [Google Scholar]
- 34.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, J. Albrecht, et al. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 35.Stuyver, L. J., T. R. McBrayer, P. M. Tharnish, A. E. Hassan, C. K. Chu, K. W. Pankiewicz, K. A. Watanabe, R. F. Schinazi, and M. J. Otto. 2003. Dynamics of subgenomic hepatitis C virus replicon RNA levels in Huh-7 cells after exposure to nucleoside antimetabolites. J. Virol. 77:10689-10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor, D. R., S. T. Shi, and M. M. Lai. 2000. Hepatitis C virus and interferon resistance. Microbes Infect. 2:1743-1756. [DOI] [PubMed] [Google Scholar]
- 37.Waxman, S., and K. C. Anderson. 2001. History of the development of arsenic derivatives in cancer therapy. Oncologist 6(Suppl. 2):3-10. [DOI] [PubMed] [Google Scholar]
- 38.World Health Organization. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341-344. [PubMed] [Google Scholar]
- 39.Yeh, C. T., H. Y. Lai, T. C. Chen, C. M. Chu, and Y. F. Liaw. 2001. Identification of a hepatic factor capable of supporting hepatitis C virus replication in a nonpermissive cell line. J. Virol. 75:11017-11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu, J., Z. Chen, V. Lallemand-Breitenbach, and H. De The. 2002. Timeline: how acute promyelocytic leukaemia revived arsenic. Nat. Rev. Cancer 2:705-714. [DOI] [PubMed] [Google Scholar]
- 41.Zhu, Q., J. W. Zhang, H. Q. Zhu, Y. L. Shen, M. Flexor, P. M. Jia, Y. Yu, X. Cai, S. Waxman, M. Lanotte, S. J. Chen, Z. Chen, and J. H. Tong. 2002. Synergic effects of arsenic trioxide and cAMP during acute promyelocytic leukemia cell maturation subtends a novel signaling cross-talk. Blood 99:1014-1022. [PubMed] [Google Scholar]