Abstract
Purpose
Azoospermia is one of the major causes of male infertility and is basically classified into obstructive (OA) and non-obstructive azoospermia (NOA). The molecular background of NOA still largely remains elusive. It has been shown that the poly(A)-binding proteins (PABPs) essentially play critical roles in stabilization and translational control of the mRNAs during spermatogenesis.
Methods
In the present study, we aim to evaluate expression levels of the PABP genes, EPAB, PABPC1, and PABPC3, in the testicular biopsy samples and in the isolated spermatocyte (SC) and round spermatid (RS) fractions obtained from men with various types of NOA including hypospermatogenesis (hyposperm), RS arrest, SC arrest, and Sertoli cell-only syndrome (SCO).
Results
In the testicular biopsy samples, both PABPC1 and PABPC3 mRNA expressions were gradually decreased from hyposperm to SCO groups (P < 0.05), whereas there was no remarkable difference for the EPAB expression among groups. The expression levels of cytoplasmically localized PABPC1 and PABPC3 proteins dramatically reduced from hyposperm to SCO groups (P < 0.05). In the isolated SC and RS fractions, the EPAB, PABPC1, and PABPC3 mRNA expressions were gradually decreased from hyposperm to SC arrest groups (P < 0.05). Similarly, both PABPC1 and PABPC3 proteins were expressed at higher levels in the SC and RS fractions from hyposperm group when compared to the SC and RS fractions from either RS arrest or SC arrest group (P < 0.05).
Conclusion
Our findings suggest that observed significant alterations in the PABPs expression may have an implication for development of different NOA forms.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-016-0654-z) contains supplementary material, which is available to authorized users.
Keywords: Male infertility, Non-obstructive azoospermia, EPAB, PABPC1, PABPC3
Introduction
Infertility is defined as failure of a couple to conceive after unprotected intercourse throughout 1 year, and it occurs in 13–18 % of couples worldwide [1, 2]. The male factor solely accounts for infertility development between 40 and 50 % of cases [1, 3], and among male factors, azoospermia is the most common factor leading to male infertility. It is briefly described as absence of sperm in the ejaculate and classified as obstructive azoospermia (OA) derived from obstruction of the genital tract, and non-obstructive azoospermia (NOA) resulted from testicular failure (reviewed in [4]). The latter one is a more severe form of azoospermia, and it can arise from congenital or developmental defects in the reproductive system, genetic anomalies, endogenous and/or exogenous testicular insults, and expressional alterations in the critically important genes during spermatogenesis [5, 6]. As a result of these defects, spermatogenic impairments such as hypospermatogenesis, spermatogenic arrest, or Sertoli cell-only syndrome (SCO) can emerge in men with NOA. While hypospermatogenesis (hyposperm) is defined as abnormally reduced and disordered sperm production, spermatogenetic activity is ceased mainly at spermatocyte (SC) or round spermatid (RS) stage in the case of spermatogenetic arrest. In the SCO, only Sertoli cells line the seminiferous epithelium, meaning that there is no spermatogenic cell type. It is important to note that there is no sperm production in the testis with either spermatogenic arrest or SCO under normal conditions. Related studies have revealed that expressional changes in the spermatogenesis-related genes may have profound adverse effects on the molecular background of azoospermia development [7–9]. However, there are a limited number of studies aimed to evaluate the molecular background of NOA development, in which irreversible spermatogenesis impairment exists to a large extent.
As what is known, spermatogenesis is a strictly regulated complex process by which a part of spermatogonia develops into mature spermatozoa, and this developmental process can be divided into three main stages: spermatogonial (spermatocytogenesis), meiosis, and spermiogenesis [10, 11]. In the spermatogonial stage, spermatogonial cells undergo a series of synchronized mitotic divisions and differentiate into spermatocytes. The spermatocytes experience meiosis in the meiotic stage to form haploid round spermatids. Then, the round spermatids undergo structural and nuclear changes during spermiogenesis stage. These consecutive processes are unique to male germ cell development and mainly rely on germ cell type-specific gene expression in a time-dependent manner. However, transcriptional activity is ceased at mid-spermiogenesis stage of spermatogenesis onward through tightly condensing chromatin structures by protamines [12, 13]. Therefore, messenger RNAs (mRNAs) needed for later period of spermatogenesis are generated and stored dormant with long poly(A) tails (∼150 nucleotides) at the early stages of spermatogenesis [14, 15]. When their translation is required during late stages of spermatogenesis, the long poly(A)-tails of target mRNAs are shortened to approximately 30 nucleotides to trigger their association with ribosomes for translation [14, 16]. In vertebrates, the post-transcriptional or translational control of these mRNAs in germ cells and early embryos are mainly performed by cognate poly(A)-binding proteins, EPAB [17–20], PABPC1 [20–23], and PABPC3 [21, 22, 24] via binding their poly(A) tails.
The embryonic poly(A)-binding protein (EPAB, also known as ePABP and PABPc1-like), is expressed in mouse [25, 26] and human [27] gonads. In the postnatal mouse testes, Epab mRNA expression is localized to only spermatogenic cell types, but it is not expressed in the somatic cells [26]. Although Epab shows specific expression patterns in mouse testis, Epab knockout (Epab−/−) male mice are found to be fertile [28]. Conversely, the Epab−/− female mice are infertile due to impairments in oocyte maturation and early embryo development processes [29]. The poly(A)-binding protein, cytoplasmic 1 (PABPC1, also known as PABP1) defined as a somatic poly(A)-binding protein is generally expressed in mouse [25, 26] and human [27, 30] gonads. On the other hand, the poly(A)-binding protein, cytoplasmic 3 (PABPC3; also known as tPABP, and PABPC2 in mouse) is synthesized only in spermatocytes and round spermatids in mouse [21, 22], but only in round spermatids in human testis [24]. The central function of the PABP proteins seems to govern translational control and stabilization of the mRNAs in the spermatogenic cells throughout spermatogenesis. Consistently, PABPC1 is enriched in polyribosomes and translationally inactive messenger ribonucleoprotein particles, and PABPC2 protein is distributed mainly in the chromatoid bodies of mouse spermatogenic cells [21].
Taken together, spatial and temporal expression patterns of the EPAB, PABPC1, and PABPC3 genes have been characterized in mouse [21, 22, 26, 28] and human [24, 27] spermatogenic cells. Based on the expressional distributions of the PABP genes, they appear to play roles in the posttranscriptional control and regulation of the translation activities of the mRNAs, being required for proper spermatogenic activity. On the other hand, the association between PABP genes and male infertility development derived from various forms of spermatogenetic impairments remained to be investigated. In the present study, we evaluate expression levels of the EPAB, PABPC1, and PABPC3 genes at mRNA and protein levels using quantitative real-time polymerase chain reaction (qRT-PCR), immunohistochemistry, RNA in situ hybridization (RNA ISH), and Western blot techniques in the testicular biopsy samples and isolated SC and RS fractions obtained from NOA groups.
Material and methods
Material collection
The testicular biopsy samples were collected from 22 infertile men (range 25–70 years), undergoing diagnostic testicular biopsy or sperm retrieval. This study was approved by the Akdeniz University ethics committee for research on human subjects, and written informed consent was obtained from each patient. The clinical examination of the infertile patients such as medical history, physical examination, and semen analyses was carried out based on the World Health Organization (WHO) guidelines [31]. Additionally, the patients not having the known risk factors associated with male infertility such as Y-chromosome microdeletions, abnormal chromosome karyotype, varicocele, orchitis, testicular torsion, and physical and endocrine abnormalities were included in this study. The testicular biopsy samples (4–6 mm3) were divided into three small pieces (1–2 mm3 each one) under a dissecting microscope (Zeiss, Oberkochen, Germany), and then each piece was used in paraffin embedding, spermatogenic cell isolation, and RNA extraction processes as explained in detail below.
Quantifying relative expression levels of genes of interest in normal spermatogenesis were not possible due to difficulty in obtaining control human testis tissues. During the study period, four testicular biopsy samples owing to tumor suspicion or orchiectomy in the presence of testicular cancer were taken to be used as fertile controls; however, detailed analysis of their hematoxylin-eosin (HE)-stained slides by a pathologist revealed that there was no successful spermatogenetic activity resulting in spermatozoa production. It is important to note that hyposperm group partially reflected normal control features since there was a moderate spermatogenetic activity, which resulted in producing a low number of sperm cells in their seminiferous tubules. Therefore, we have considered the expression levels in hyposperm group as a baseline in relative gene expression analysis.
Paraffin embedding
One piece of the testicular biopsy samples was immersed in Bouin’s solution (Sigma-Aldrich, St. Louis, MO, USA) at +4 °C for 12 h, and then it was dehydrated through a graded ethanol series. Following that the biopsy sample was cleared in xylene and subsequently embedded in paraffin. Serial cross sections at 5-μm thickness were cut from paraffin block using a rotary microtome (Leica, Nussloch, Germany) and mounted on glass slides (Menzel Gläser, Braunschweig, Germany), which were later used for HE, immunohistochemistry, and RNA in situ hybridization assays. To determine histopathologic status of the testicular biopsy samples, HE staining was performed as in our previous studies [26, 32]. The two specialists evaluated the HE-stained slides under a bright-field microscopy (Olympus, Jena, Germany) to determine their histopathological features and Johnsen scores.
Immunohistochemistry
Immunoexpression of the PABPC1 and PABPC3 proteins in the testicular biopsy samples diagnosed as having hyposperm, RS arrest, SC arrest, and SCO were detected by immunohistochemistry. The paraffin sections at 5-μm thickness were deparaffinized in fresh xylene, and then rehydrated in a series of decreasing ethanol concentrations. We performed antigen retrieval as follows: sections were first boiled at 750 W in 1 mM Tris-EDTA (TE) buffer (pH 9.0), and then warmed at 250 W for 25 min in the same buffer. Afterward, the endogenous peroxidase activity was quenched by incubating the sections in 3 % hydrogen peroxide (H2O2) solution prepared in methanol for 25 min at room temperature (RT). After washing the sections in 1× phosphate-buffered saline (1× PBS), ultra V blocking (LabVision, Neueste Nachrichten, Germany) was applied at RT for 5 min. Then, we incubated the sections at +4 °C overnight with primary antibodies specific to PABPC1 (Abcam, Cambridge, UK) or PABPC3 (ProteinTech Group inc., Chicago, USA) protein (both of which diluted 1:100 in 1× PBS). Following primary antibody incubation, sections were washed in 1× PBS (3 × 15 min), and subsequently incubated with secondary goat anti-rabbit antibody conjugated with horseradish peroxidase (HRP) (Vector Labs, Peterborough, United Kingdom) (diluted 1:350 in 1× PBS) at RT for 1 h. As a negative control, a section on each slide was incubated with 1× PBS instead of primary antibody in each experiment. The immunohistochemical reaction was developed using 3, 3′-diaminobenzidine (DAB) substrate to visualize the peroxidase reaction site of the PABPC1 or PABPC3 protein. Following that, sections were washed under tap water and then counterstained with Mayer’s hematoxylin solution (Sigma-Aldrich). Finally, we dehydrated the sections in a graded ethanol solutions and mounted with Entellan (Merck) using coverslips. As a result, immunoexpression and localization of the PABPC1 and PABPC3 proteins in the NOA groups were examined with a bright-field microscopy (Zeiss). Also, we have further analyzed expression levels of the PABPC1 and PABPC3 proteins in all groups by using ImageJ software (National Institutes of Health, Bethesda, Maryland, USA), which confers us semiquantitative analysis of the immunohistochemical staining [33].
RNA in situ hybridization
To detect cellular and subcellular localization of the EPAB mRNA in the testicular biopsy samples with hyposperm (n = 3), we have used RNA ISH technique as detailed in the prior studies [26, 32]. For this purpose, we have generated complementary DNA (cDNA) probe-specific for EPAB mRNA by PCR, using cDNA synthesized from testicular samples with hyposperm. The EPAB primers used in the PCR reactions were located on exons 1 and 2. The reaction mixture contained 10× PCR buffer (Qiagen, Valencia, CA, USA), 0.125 mM of each dNTP (Roche), 0.5 μM of each primer, and two units of SuperTaq polymerase (Qiagen), and 0.02 mM Dig-UTP (digoxigenin-11-uridine-5′-triphosphate; Roche, Indianapolis, IN, USA). Amplifications were carried out with 35 cycles of PCR in which the initial 5-min denaturation at 95 °C was followed by a “touch-down” program for 10 cycles of 92 °C for 30 s, 65 °C for 30 s (−1 °C per cycle), and 72 °C for 30 s; and then 25 cycles of 92 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and final extension at 72 °C for 10 min in a volume of 50-μl reaction mixture. The amplified PCR products fractionated on a 1.5 % agarose/Tris-acetate-EDTA gels were extracted using QIAEX II gel extraction kit (Qiagen). The labeling quality and amount of the produced EPAB probe were determined by comparing probe with a labeled control DNA in a dot blot, following the manufacturer’s protocol (Roche). We stored the Dig-labeled and Dig-unlabeled EPAB probes at −20 °C until use. The RNA ISH method applied here was modified from our previous works [26, 34]. Then, we evaluated the stained sections with a bright-field microscope (Zeiss) to detect the EPAB mRNA localization in the testicular biopsy samples with hyposperm.
Spermatogenic cell isolation
Spermatocytes and round spermatids were isolated from testicular biopsy samples obtained from hyposperm, RS arrest, and SC arrest groups by using 2–4 % bovine serum albumin (BSA) (American Bioanalytical Inc, Natick, Massachusett, USA) discontinuous gradient method modified from previously described protocols [26, 35–37]. In brief, the testicular biopsy sample was transferred into 2 ml of Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 (Gibco, Grand Island, NY, USA) under sterile conditions. Then, it was digested in DMEM/F12 media containing 0.5 mg/mL collagenase (Roche) at 34 °C for 20 min so that intertubular cells were largely removed. Afterward, seminiferous tubules were washed twice with fresh media and gently pipetted with a glass Pasteur pipette during incubation at 34 °C for 20 min in medium including 0.25 mg/mL trypsin (Gibco). Thus, we recovered the mixed spermatogenic cells also involving Sertoli cells. Following centrifugation of the mixed spermatogenic cells at 1600 rpm for 6 min, the pellet suspended with fresh DMEM/F12 medium including 0.5 % BSA and 1 μg/mL DNase (Roche) was incubated at RT for 5 min. Then, mixed spermatogenic cells were centrifuged at 1600 rpm for 6 min again, and the pellet was resuspended in 0.5 % BSA, and filtered through 70- and 40-μm cell strainers (BD biosciences, California, USA), respectively.
The mixed spermatogenic cell types were separated into fractions using a discontinuous 2 to 4 % BSA gradient prepared in a 15-mL tube (BD biosciences) by consecutively adding 1 mL of 10 % BSA, and then 1 mL of 4.0, 3.8, 3.6, 3.4, 3.2, 3.0, 2.8, 2.6, 2.4, 2.2, and 2.0 % BSA. The filtered spermatogenic cells were counted using a Makler chamber (Sefi-Medical, Haifa, Israel), and 1 mL of the mixed spermatogenic cells at a concentration of 3.0 × 105 cells/mL was placed onto the gradient tube. Finally, 1 mL of 0.2 % BSA was added as a cover. Notably, we established at least four tubes in each isolation process. Then, the gradient tubes were incubated at +4 °C for 4 h with gentle rocking at a rate of 15 tilts/min to accelerate gravity sedimentation. At the end of incubation period, we have collected 1 mL fractions from each gradient tube and centrifuged at 1600 rpm for 6 min. Then, we assessed the characteristics of the fractioned spermatogenic cells under an inverted microscope (Diaphot 300, Nikon, Tokyo, Japan). The cell viability rate of both spermatocyte and round spermatid fractions was determined using a trypan blue exclusion assay (Invitrogen, Grand Island, NY, USA) at the end of each cell isolation procedure. The percentage of viable cells in each fraction was calculated as the number of viable cells in a hundred of cells randomly counted. In addition, smear slides of the SC and RS fractions prepared following each isolation experiment were stained with hematoxylin-eosin as described above. They were subsequently analyzed to determine the purity rate of the spermatocyte and round spermatid fractions. It is important to note that we identified the spermatogenic cells and somatic cells based on their typical morphological features as defined previously [35, 38].
RNA extraction, cDNA synthesis, and gene expression quantification
The total cellular RNA extraction and complementary DNA (cDNA) synthesis from testicular biopsy samples and isolated spermatogenic cells was carried out by using the RNAqueous-Micro Kit and RETROscript reverse transcription kit (Ambion) based on the manufacturer’s instructions (Ambion), respectively and kept at −80 °C until use. The qRT-PCR reaction was employed to quantitatively characterize expression levels of the EPAB, PABPC1, and PABPC3 genes in the testicular biopsy samples and isolated SC and RS fractions as detailed previously [32, 34]. The sequences and localizations of the EPAB, PABPC1, PABPC3, and β-ACTIN primers were presented in Supplementary Table 1. We used β-ACTIN as an internal control to normalize expression of the target genes. The relative expression profiles of the EPAB, PABPC1, and PABPC3 transcripts were calculated by using 2−ΔΔCt (cycle threshold) method and reported as fold changes. Of note, the specificity of qRT-PCR products was confirmed by melting curve analysis at the end of each reaction.
Western blotting
The semiquantitative expression levels of the PABPC1 and PABPC3 proteins in the isolated SC and RS fractions were also determined by Western blot. For this purpose, isolated fractions were dissolved in lysis buffer [composed of 1 % sodium dodecyl sulfate (SDS), 1.0 mM sodium ortho-vanadate, 10 mM Tris pH 7.4] supplemented with 1× protease inhibitor cocktail (Amresco, Solon, OH, USA). The total concentration of the extracted protein was measured using the Lowry method (Pierce, Rockford, IL, USA). The 20 μg of protein was separated on 10 % Tris–HCl gel and electrotransferred overnight +4 °C to polyvinylidene difluoride (PVDF) membrane (Roche). Then, the membrane was blocked with 5 % (w/v) non-fat dried skimmed milk powder prepared in TBS-T (20 mM Tris/HCl and 150 mM NaCl plus 0.05 % Tween 20 at pH 7.4) at RT for 1 h. Following that, we incubated the membrane with primary antibody specific for PABPC1, PABPC3, or β-ACTIN protein [diluted 1:500 in 5 % (w/v) non-fat dried skimmed milk powder in TBS-T for each antibody] at +4 °C overnight. Note that the primary antibodies against β-ACTIN, PABPC1, and PABPC3 proteins were purchased from Cell Signaling, Abcam, and Company ProteinTech Group Inc., respectively. After washing membrane in TBS-T three times for 15 min each, they were incubated with HRP-conjugated anti-rabbit secondary antibody (1:2000 dilution; Vector Laboratories) at RT for 1 h. The signal detection coming from target proteins were carried out using ECL Plus reagent (Pierce) via incubating the membrane in a dark place for 5 min. All Western blot data were evaluated using the ImageJ software, and expression of the PABPC1 and PABPC3 proteins were normalized to β-ACTIN expression. Finally, expression profiles of the PABPC1 and PABPC3 proteins in the SC and RS fractions obtained from NOA groups were determined.
Statistical analysis
The one-way analysis of variance (one-way ANOVA) followed by Dunnett’s T3 post hoc test was used for analysis of the qRT-PCR data of testicular biopsy samples and isolated spermatogenic cells. We evaluated the immunohistochemistry results of the PABPC1 and PABPC3 proteins by using one-way ANOVA followed by Scheffe and Dunnett’s T3 post hoc tests, respectively. The Dunnett’s T3 post hoc test was also conducted to analyze the statistical significance of the Western blot findings of both PABPC1 and PABPC3 proteins in the SC fraction. On the other hand, the Student’s t test was used for analyzing Western blot data of the PABPC1 and PABPC3 proteins in the RS fraction. The statistical calculations were carried out using Statistical Package for the Social Sciences (SPSS) (IBM SPSS Statistics version 20; IBM Corporation, NY, USA). In all tests, P < 0.05 was considered as statistically significant.
Results
In the current study, expression levels of the EPAB, PABPC1, and PABPC3 genes were first characterized in the testicular biopsy samples and isolated SC and RS fractions obtained from men with various types of NOA.
Histopathology of the testicular biopsy samples
The hematoxylin and eosin-stained testicular biopsy sections were evaluated under the bright-field microscopy (Olympus) to characterize their histopathologic features and Johnsen scores (Supplementary Fig. 1a–d and Table 2). Based on their spermatogenic activity and germinal epithelial cell content, testicular biopsies were classified into four groups: hyposperm, RS arrest, SC arrest, and SCO. In the hyposperm group, spermatogenic activity had partially taken place, and all germinal epithelial cells (spermatogonia, primary/secondary spermatocytes, round spermatids, elongating/elongated spermatids, spermatozoa, and Sertoli cells) and intertubular cells were observed (Supplementary Fig. 1a). However, spermatogenic activity is ceased at the RS or primary/secondary SC stages in the RS arrest or SC arrest group, respectively (Supplementary Fig. 1b, c). In the SCO group, only Sertoli cells were present, and there was no germline cell as expected (Supplementary Fig. 1d). It is noteworthy that some groups had histopathologic changes such as Leydig cell hyperplasia, atrophic tubules, basement membrane thickening, fibrin clot formation, and hyperplasia in the intertubular area. The histopathological evaluation and Johnsen score results of all NOA groups were presented in the Supplementary Table 2.
Expression analysis of the EPAB, PABPC1, and PABPC3 genes in the NOA groups
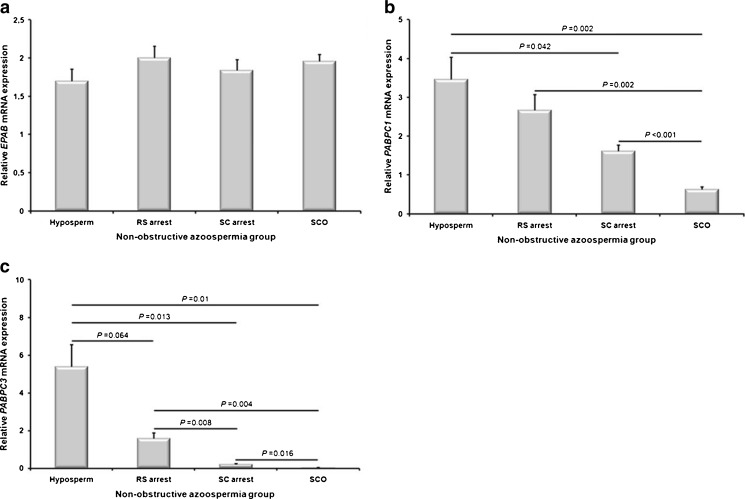
EPAB, PABPC1, and PABPC3 mRNA expression levels have been analyzed for the first time in the hyposperm, RS arrest, SC arrest, and SCO groups using qRT-PCR technique. Although EPAB was transcribed in all NOA groups, there was no statically significant difference between groups (P > 0.05; Fig. 1a).
Fig. 1.
Relative expression levels of the EPAB, PABPC1, and PABPC3 mRNA in the NOA groups including hyposperm, RS arrest, SC arrest, and SCO. The quantitative real-time PCR (qRT-PCR) technique was used to determine expression levels of the PABP genes in the testicular biopsy samples obtained from NOA groups. a EPAB gene expression was detected in all groups, and there was no statistical difference between groups (P > 0.05). b The PABPC1 expression was gradually decreased from hyposperm to SCO groups (P < 0.05). c Similarly, we found that PABPC3 expression was gradually decreased from hyposperm to SC arrest groups, and there was no expression detected in the SCO group. Hyposperm hypospermatogenesis, RS arrest round spermatid arrest, SC arrest spermatocyte arrest, SCO Sertoli cell-only syndrome. The statistical significance among groups was analyzed by using one-way analysis of variance (one-way ANOVA) followed by Dunnett’s T3 post hoc test, and their P values were given on columns using line charts. Note that P < 0.05 was considered as statistically significant. Data are presented as mean ± standard error of mean (SEM)
Similar to EPAB, all NOA groups expressed the PABPC1 mRNA, but in different levels (Fig. 1b). Intriguingly, the PABPC1 expression was gradually decreased from hyposperm to SCO groups, and the hyposperm, RS arrest, and SC arrest groups had significantly higher PABPC1 levels than that of the SCO group (P < 0.05; Fig. 1b). Additionally, PABPC1 expression in the hyposperm group was predominantly higher than in the SC arrest group (P < 0.05; Fig. 1b). Similarly, PABPC3 transcript levels were found to be progressively decreased from hyposperm to SCO groups (Fig. 1c). The hyposperm group possessed the highest PABPC3 expression when compared to the remaining groups, and the RS arrest group had higher expression levels than that of the SC arrest and SCO groups (P < 0.05; Fig. 1c). Moreover, PABPC3 was transcribed at higher levels in the SC arrest group in comparison to the SCO group (P < 0.05; Fig. 1c).
Immunoexpression of the PABPC1 and PABPC3 proteins in the NOA groups
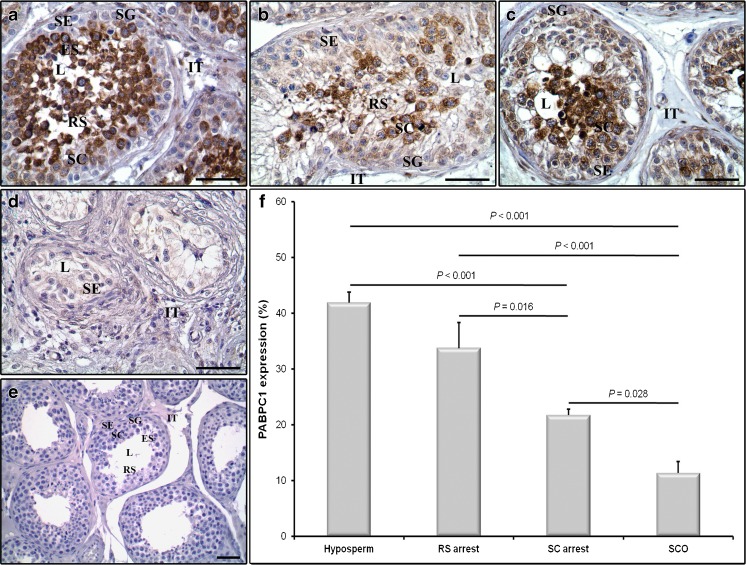
In the present work, immunoexpression of the PABPC1 and PABPC3 proteins in the hyposperm, RS arrest, SC arrest, and SCO groups were characterized by using immunohistochemistry. The immunohistochemical staining exhibited that all groups expressed PABPC1 in both germinal epithelial cells and intertubular cells (Fig. 2a–d). In the germinal epithelial cells, PABPC1 was strongly expressed in the cytoplasm of spermatocytes and round spermatids and in relatively low levels in the spermatogonial cells and Sertoli cells resided at the edge of seminiferous tubules in the corresponding groups. Likewise, we observed weak PABPC1 expression in the cytoplasm of the intertubular cells (Fig. 2a–d). Notably, there was no immunopositivity in the negative control section as expected (Fig. 2e).
Fig. 2.
Immunoexpression of the PABPC1 protein in the NOA groups. Cellular and subcellular localization of the PABPC1 protein in the hyposperm (a), RS arrest (b), SC arrest (c), and SCO (d) groups were evaluated using immunohistochemistry. e Concurrently, the negative control sections not involving the primary antibody were used in each immunohistochemistry application to detect presence of any non-specific staining. We found that PABPC1 protein was expressed in cytoplasm of all germinal epithelial cells as well as in the intertubular cells, which is consistent with being a somatic PABP protein. Furthermore, to compare semiquantitative expression levels of the PABPC1 protein in these groups, we have used ImageJ software (f). The PABPC1 expression was found to be progressively reduced from hyposperm to SCO groups (P < 0.05) (f). SG spermatogonia, SC spermatocyte, RS round spermatid, ES elongated spermatid, SE Sertoli cell, IT intertubular area, L lumen. The statistical significance between groups was evaluated by using one-way ANOVA followed by Scheffe post hoc test, and their P values were given on columns using line charts. Note that P < 0.05 was considered as statistically significant. Scale bar: 50 μm. Data are presented as mean ± SEM
The relative expression of the PABPC1 protein in the NOA groups was further analyzed by using the ImageJ software. This analysis revealed that PABPC1 expression gradually decreased from hyposperm to SCO groups (Fig. 2f), coinciding with its mRNA expression profiles in the same groups (Fig. 1b). The hyposperm group had significantly higher PABPC1 expression when compared to the SC arrest and SCO groups, and the RS arrest group had predominantly higher PABPC1 expression levels than the SC arrest and SCO groups (P < 0.05; Fig. 2f). Furthermore, PABPC1 was produced at higher profile in the SC arrest than in the SCO group (P < 0.05; Fig. 2f).
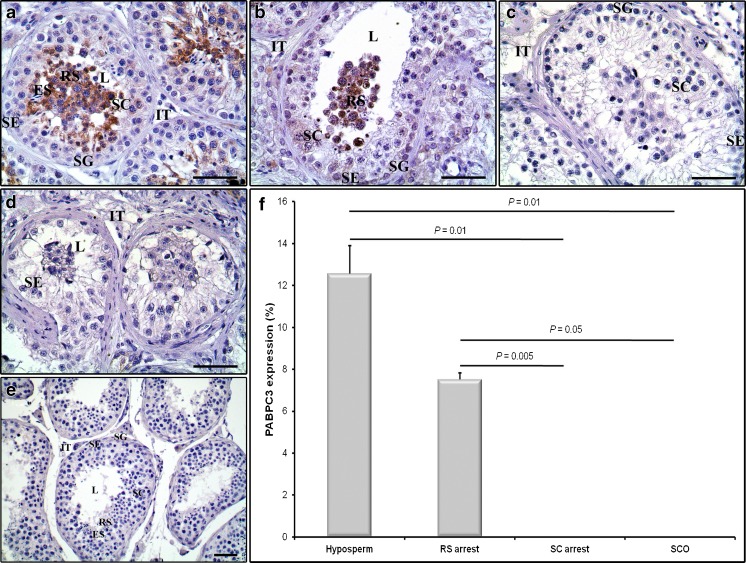
The cellular localization and relative expression levels of the PABPC3 protein were also determined in the NOA groups. The PABPC3 expression was only detected in the hyposperm and RS arrest groups, and there was no expression detected in the SC arrest and SCO groups (Fig. 3a–d). In both hyposperm and RS arrest groups, PABPC3 was cytoplasmically expressed solely in the spermatocytes and round spermatids, in which round spermatids seemed to have slightly stronger PABPC3 immunopositivity with respect to spermatocytes (Fig. 3a, b). It is important to note that we did not observe any PABPC3 immunoexpression in the spermatogonia, elongated spermatids, and Sertoli cells as well as in the intertubular cells in all studied groups (Fig. 3a–d). Note that we have not detected any PABPC3 staining in the negative control section (Fig. 3e).
Fig. 3.
Immunoexpression of the PABPC3 protein in the NOA groups. Cellular and subcellular localization of the PABPC3 protein in the hyposperm (a), RS arrest (b), SC arrest (c), and SCO (d) groups were detected using immunohistochemistry. e The negative control section not including primary antibody was used in each immunohistochemistry application to detect presence of any non-specific staining. We found that PABPC3 protein was expressed only in the cytoplasm of spermatocytes and round spermatids in the hyposperm and RS arrest groups. Furthermore, to compare semiquantitative expression levels of the PABPC3 protein in these groups, we have used ImageJ software (f). This analysis revealed that hyposperm group had the highest PABPC3 expression compared to the remaining groups, and its expression gradually decreased to the RS arrest group (P < 0.05). There was no PABPC3 expression determined in the SC arrest and SCO groups (f). SG spermatogonia, SC spermatocyte, RS round spermatid, ES elongated spermatid, SE Sertoli cell, IT intertubular area, L lumen. The statistical significance among groups was analyzed by using one-way ANOVA followed by Dunnett’s T3 post hoc test, and their P values were presented on columns using line charts. Note that P < 0.05 was considered as statistically significant. Scale bar: 50 μm. Data are presented as mean ± SEM
When PABPC3 immunoexpression was additionally analyzed by using ImageJ software, we revealed that its expression gradually decreased from hyposperm to RS arrest groups, and there was no staining observed in the SC arrest and SCO groups (Fig. 3f). Both hyposperm and RS arrest groups exhibited statistically significant higher PABPC1 expression than either the SC arrest or SCO group (P < 0.05; Fig. 3f). Also, the findings were found to be largely comparable with the PABPC3 expression at mRNA levels in the same groups (Fig. 1c).
Localization of the EPAB mRNA
To determine localization of the EPAB mRNA in the testicular sample with hyposperm, we have applied RNA in situ hybridization technique. We found that both intertubular and germinal epithelial cells cytoplasmically expressed the EPAB mRNA (Supplementary Fig. 2a). In the germinal epithelial cells, all spermatogenic cell types and Sertoli cells expressed the EPAB transcript in different levels. Also, spermatocytes seemed to have slightly higher EPAB mRNA expression than that of the other spermatogenic cell types (Supplementary Fig. 2A). The presence of EPAB expression in both germinal epithelial and intertubular cells suggested that EPAB is generated in both somatic and germline cells, meaning that it is not a germline-specific PABP gene. Notably, there was no EPAB expression in the negative control section, in which unlabeled EPAB probe had been used (Supplementary Fig. 2b).
Particular features of the isolated SC and RS fractions
The SC and RS fractions isolated by using 2–4 % discontinuous BSA gradient were first checked under an inverted microscope (Olympus), and their representative photomicrographs were presented in Supplementary Fig. 3a, b. As can be seen in the relative photomicrographs, spermatocytes are large cells with a diameter of 12–15 μm and include fine spots (Supplementary Fig. 3a), whereas round spermatids are in a diameter of 6–7 μm and have a typical nucleolus in their nuclei (Supplementary Fig. 3b).
The percentage cell viability of the SC and RS fractions was found to be 96.4 and 98 %, respectively. Also, the purity rates of the SC and RS fractions were determined via evaluating the smear slides, stained with hematoxylin and eosin (Supplementary Fig. 3c, d). The purity rates of the SC and RS fractions collected from hyposperm, RS arrest, and SC arrest groups were shown in Supplementary Fig. 3e. In the hyposperm group, SC and RS fractions were in the purities of ∼94 and ∼78 %, respectively. However, in the RS arrest group, although SC fraction was quite high (∼95 %), the RS purity was found to be ∼24 % (Supplementary Fig. 3e).
Expression of the EPAB, PABPC1, and PABPC3 genes in the SC and RS fractions
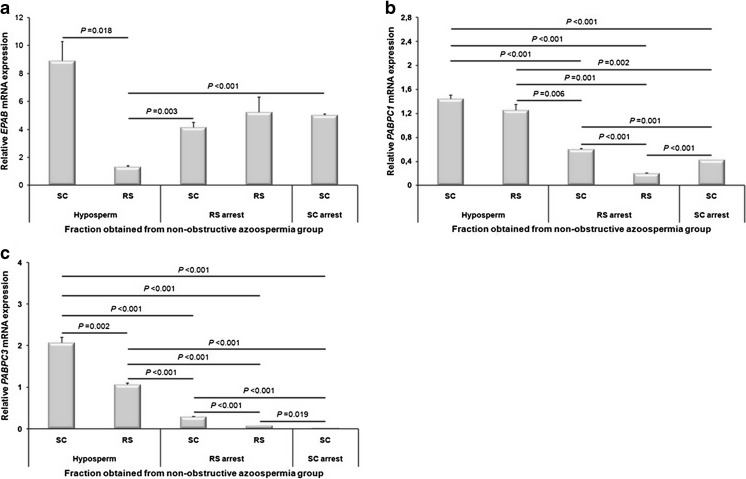
Expression levels of the EPAB, PABPC1, and PABPC3 genes in the SC and RS fractions obtained from hyposperm, RS arrest, and SC arrest groups were characterized by using qRT-PCR method (Fig. 4a–c). The EPAB mRNA expression was found at the highest and lowest levels in the SC and RS fractions, respectively, both of which had been obtained from hyposperm group when compared with the SC and RS fractions from the remaining groups (Fig. 4a). In the hyposperm group, the SC fraction exhibited strongly higher EPAB mRNA expression than that of the RS fraction from the same group (P < 0.05, Fig. 4a). However, the RS fraction from the hyposperm group had remarkably lower EPAB expression when compared to the SC fraction either from RS arrest or SC arrest group (P < 0.05, Fig. 4a).
Fig. 4.
Relative expression levels of the EPAB, PABPC1, and PABPC3 genes in the isolated SC and RS fractions. a The SC fraction obtained from hyposperm group had the highest EPAB expression, and the SC and RS fractions from other NOA groups exhibited different expression profiles. b Both SC and RS fractions from hyposperm group highly expressed the PABPC1 mRNA when compared to the SC and RS fractions from remaining NOA groups (P < 0.05). c Similarly, PABPC3 was transcribed at higher amounts in the SC and RS fractions from hyposperm group in comparison to SC and RS fractions from other NOA groups (P < 0.05). The statistical significance among groups was analyzed by using one-way ANOVA followed by Dunnett’s T3 post hoc test, and their P values were depicted on columns using line charts. Note that P < 0.05 was considered as statistically significant. Data are presented as mean ± SEM
When we explored the PABPC1 mRNA expression in the SC and RS fractions, we revealed that both SC and RS fractions from hyposperm group had the highest expression levels in comparison to the SC and RS fractions from other groups (Fig. 4b). Both SC and RS fractions from hyposperm group expressed the higher levels of PABPC1 transcript than that of the SC and RS fractions from RS arrest, and of the SC fraction from SC arrest group (P < 0.001; Fig. 4b). Similarly, the SC fraction from RS arrest group had higher PABPC1 expression in comparison to RS fraction from RS arrest and SC fraction from SC arrest groups (P < 0.05; Fig. 4b). On the other hand, PABPC1 was transcribed at lower levels in the RS fraction from RS arrest group than in the SC fraction from SC arrest group (P < 0.001; Fig. 4b).
Similar to the PABPC1 expression, the SC and RS fractions from hyposperm group possessed the highest PABPC3 mRNA expression when compared to the SC and RS fractions collected from the remaining NOA groups (Fig. 4c). The SC fraction from SC arrest group had the lowest PABPC3 mRNA expression level (Fig. 4c). Also, PABPC3 expressed significantly higher levels in the SC and RS fractions from hyposperm group than that of the SC fractions from RS arrest or SC arrest group, and of the RS fraction from RS arrest group (P < 0.001; Fig. 4c). Notably, the SC fraction from hyposperm group had higher PABPC3 expression when compared to the RS fraction from the same group (P = 0.002; Fig. 4c). Moreover, the SC fraction from RS arrest group possessed significantly higher PABPC3 transcript levels than that of the RS fraction from RS arrest group and SC fraction from SC arrest group (P < 0.001; Fig. 4c). On the other hand, the RS fraction from RS arrest group exhibited prominently higher PABPC3 mRNA expression than that of the SC fraction from SC arrest group (P = 0.019; Fig. 4c). It is important to note that both PABPC1 and PABPC3 genes showed almost similar mRNA expression patterns in the SC and RS fractions obtained from NOA groups.
Expression of the PABPC1 and PABPC3 proteins in the SC and RS fractions
In addition to analyzing mRNA levels of the PABPC1 and PABPC3 genes, we have also characterized their protein expression profiles in the SC and RS fractions using Western blot. Since there is no commercially available EPAB-specific antibody, its protein expression could not be evaluated in the current work.
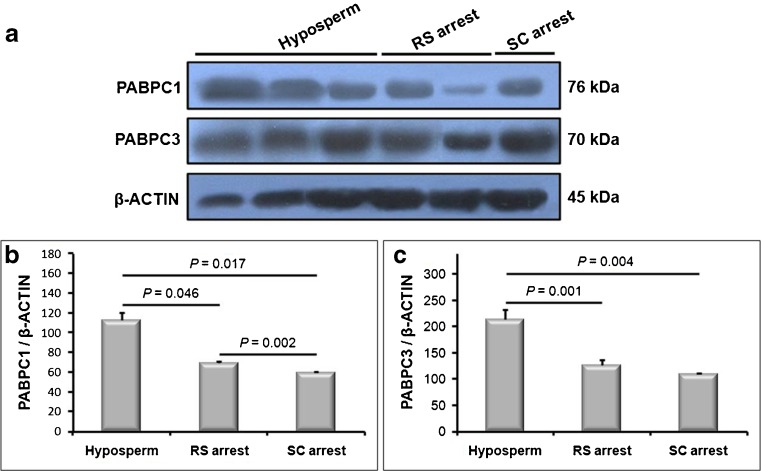
Both PABPC1 and PABPC3 proteins were expressed in significantly higher levels in the SC fraction from hyposperm group when compared to the SC fraction either from RS or SC arrest group (P < 0.05; Fig. 5a–c), similar to their mRNA expression patterns in the same groups. Additionally, the PABPC1 protein was predominantly produced in the SC fraction from RS arrest group than that of the SC fraction obtained from SC arrest group (P < 0.05; Fig. 5b). However, there was no statistically significant difference for the PABPC3 expression in the SC fractions between RS arrest and SC arrest groups (Supplementary Fig. c).
Fig. 5.
Relative expression levels of PABPC1 and PABPC3 proteins in the SC fraction. We used Western blot technique to determine relative expression profiles of these proteins. a Western blot results of the PABPC1 and PABPC3 proteins in the SC and RS fractions were given. Both PABPC1 and PABPC3 proteins had higher expression levels in the SC fraction obtained from hyposperm group than that of the SC fraction isolated from either RS arrest or SC arrest group. b ImageJ analysis was performed to further evaluate the Western blot results of the PABPC1 protein. We found that the SC fraction isolated from hyposperm group possessed significantly higher PABPC1 protein expression when compared to the SC fractions from RS arrest and SC arrest groups (P < 0.05). c Relative expression of PABPC3 protein in the SC fraction was also analyzed by using ImageJ software. We revealed that the SC fraction from hyposperm group expressed the PABPC3 at higher levels in the hyposperm group in comparison to the SC fractions from remaining NOA groups (P < 0.05). The statistical significance among groups was analyzed by using one-way ANOVA Dunnett’s T3 post hoc test, and their P values were presented on columns using line charts. Note that P < 0.05 was considered as statistically significant. Data are presented as mean ± SEM
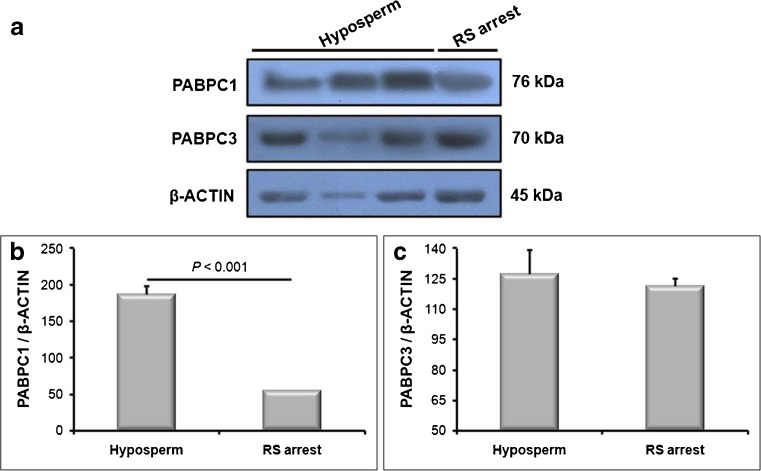
When expression of the PABPC1 and PABPC3 proteins have been evaluated in the RS fractions collected from hyposperm and RS arrest groups, both proteins were found to be highly expressed in the RS fraction from hyposperm group (Fig. 6a–c). Although there was statistically significant difference for PABPC1 expression in the RS fractions between hyposperm and RS arrest groups (P < 0.001; Fig. 6b), we have not observed any remarkable difference for PABPC3 expression profiles between hyposperm and RS groups (Fig. 6c).
Fig. 6.
PABPC1 and PABPC3 protein expression in the RS fraction. The relative expression levels of the PABPC1 and PABPC3 proteins in the RS fractions isolated from hyposperm and RS arrest groups were characterized using Western blot, and Western blot results were further analyzed using ImageJ software. a Western blot results of the PABPC1 and PABPC3 proteins in the RS fractions were presented. The RS fraction from hyposperm group had higher PABPC1 and PABPC3 expression levels compared to the RS fraction isolated from RS arrest group. b The imageJ analysis revealed that PABPC1 expression in the RS fraction from hyposperm group was remarkably higher than that of the RS fraction from RS arrest group (P < 0.05). c Similarly, RS fraction from hyposperm group exhibited higher PABPC3 expression when compared to the RS fraction from RS arrest group, but it was not found as significant (P > 0.05). The statistical significance among groups was analyzed by using the student’s t-test, and the P value was demonstrated on column using a line chart. Note that P < 0.05 was considered as statistically significant. Data are presented as mean ± SEM
Discussion
In the current study, expression levels of the EPAB, PABPC1, and PABPC3 genes were characterized for the first time in infertile men with various forms of NOA. We found here that expression of the PABPs at mRNA and protein levels significantly decreased in the testicular biopsy samples from hyposperm to SCO group and in the isolated spermatogenic cells. This suggested that the altered expression of the PABP genes may adversely affect spermatogenic process and subsequently may result in development of infertility in the NOA groups.
In the literature, there are a limited number of studies aimed at elucidating the molecular background of NOA development. One of them by Malcher et al. (2013) revealed that there are significantly downregulated (AKAP4, UBQLN3, CAPN11, GGN, SPACA4, SPATA3, and FAM71F1) and upregulated (WBSCR28, ADCY10, TMEM225, SPATS1, FSCN3, GTSF1L, and GSG1) genes in the testicular tissues of infertile men with various types of NOA when compared to control counterparts using microarray [7]. The researchers concluded that the set of genes differently expressed in the NOA groups can be used as biomarkers to understand the degree of spermatogenic impairment in the idiopathic NOA groups. In that study, the men with NOA had been classified into four groups based on their histopathologic features as follows: Postmeiotic arrest, meiotic arrest, premeiotic arrest, and Sertoli cell-only syndrome. In our study, we have classified the NOA groups according to their histopathologic description into hyposperm, RS arrest, SC arrest, and SCO. It is important to note that the classified groups in both studies largely overlap; except for there is no premeiotic group in the current work. The two studies documented that expression levels of the genes functioning in different cellular activities during spermatogenesis are significantly altered in the NOA groups from hyposperm or postmeiotic arrest to SCO groups.
In another study, Lin et al. (2006) found significant downregulation of 300 genes in the testicular samples from infertile men with maturation arrest or SCO compared to controls with normal spermatogenesis [8]. Among 300 genes, they identified sterility-related and testis-specific 10 novel genes, which encode proteins with functional domains related to spermatogenesis and/or spermiogenesis or non-functional domains [8]. Similarly, significant changes in global gene expression between infertile and fertile human testis tissues have also been determined in the previous investigations by using microarrays [9, 39]. Although the studies briefly summarized above revealed that global gene expression is widely changed in the azoospermic testicular tissues as is in the PABP genes, further studies are required to understand the impact of upregulation or downregulation of the specific genes during spermatogenesis. This would help providing more accurate diagnosis and treatment of male infertility due to azoospermia.
Although there are certain limitations on studying human testis materials, the present and previous studies bring new insights into illuminating the molecular background of NOA development. In all studies, the patients with NOA were classified based on their histopathologic characteristics. However, histopathologic data could be obtained from microscopic examination of a small biopsy taken from only one part of the testis. Therefore, this analysis may not have reflected histopathologic status of the whole testis of the infertile men. Consistently, it has been shown that multiple biopsy samples from different parts of the testis resulted in sperm retrieval in patients with azoospermic testis and even in the patients with SCO in certain cases [40, 41]. As a result, since obtaining testicular biopsy materials from one part of human testis cannot represent histopathologic status of the whole testis, it may affect the accuracy of findings. As a result, majority of the studies investigated the testis transcriptome showing differential expression of certain genes in the NOA groups when compared to controls. In the testicular biopsy analysis, this change may derive from testicular sampling differences and/or gradually reducing the spermatogenic cell types from control or hyposperm to SCO groups. While control or hyposperm group includes all spermatogenic cell types, the RS arrest group does not have elongating/elongated spermatids and spermatozoa, and there were no round spermatids onward in the SC arrest group. However, the SCO group does not possess any spermatogenic cell types. The gradual decrease of the spermatogenic cells from control or hyposperm to SCO groups may potentially influence expression levels of the genes which are known to be generated in the spermatogenic cells. In addition, we monitored certain histopathologic features including Leydig cell hyperplasia, atrophic tubules, and hyperplasia in the intertubular area in certain NOA groups in the current study. Because expression of the target genes at mRNA and protein levels were determined after normalizing to β-ACTIN expression, the abnormally increased somatic cell numbers in the testicular tissues may be potential in changing the expression levels of the target genes indirectly. To counteract this challenge, we have further analyzed the expression of the PABP genes in the isolated spermatogenic cells.
In the present study, we also revealed for the first time that EPAB was expressed in the somatic cells as well as germline cells in human testis. This finding is compatible with the work by Guzeloglu-Kayisli et al. (2008), in which they detected EPAB mRNA expression in the human somatic tissues as well as in the ovary and testis tissues [27]. Notably, in a recently published study, the EPAB mRNA being transcribed in the human somatic cells and 8-cell and blastocyst stage early embryos includes a premature stop codon in the exon 8 [30]. In marked contrast, mouse Epab is found to be transcribed only in the ovary and testis tissues, but not in the somatic tissues [25]. Further analysis on the postnatal mouse testes and isolated spermatogenic cells showed that it is expressed solely in the spermatogenic cells, and exhibits prominent expressional differences during testicular development [26], and in the postnatal mouse ovaries [32]. Collectively, the findings indicated that the spatial expression of the EPAB gene represents remarkable difference between mouse and human testes.
Apart from our study, PABPC1 and PABPC3 expression in human testis had been analyzed in a previous study by Feral et al. (2001) [24]. In that work, they revealed that while PABPC1 mRNA is expressed in both gonadal and somatic tissues, PABPC3 is synthesized only in the testis tissue. Furthermore, RNA in situ hybridization localized the PABPC1 mRNA to both spermatocytes and round spermatids, but PABPC3 was observed only in the round spermatids [24]. The findings for PABPC1 expression are to a large extent compatible with our results: we analogously detected PABPC1 mRNA expression in almost similar levels in the spermatocyte and round spermatid fractions isolated from hyposperm and RS arrest groups. However, our results related to PABPC3 gene expression in the SC and RS fractions does not completely overlap with the former study [24]. Although they did not observe PABPC3 mRNA expression in the spermatocytes by RNA in situ hybridization, we have observed it in the SC fraction as well as in the RS fraction. The difference between former and present studies may arise from existing genetic differences among men used in these studies and employing distinct technical methods. The qRT-PCR technique used in the present study is accepted as a reliable and robust method in detecting PABPC3 mRNA in quite low levels than that of the RNA in situ hybridization, which is capable of sensing PABPC3 mRNA expression above a certain amount. Moreover, it is noteworthy that we have analyzed here testicular samples including various types of pathology (Supplementary Table 2). Hence, use of pathologic testicular samples may lead to changes in spatial and temporal expression patterns of the PABPC1 and PABPC3 genes; therefore, new studies are required for these genes on testicular samples obtained from healthy and fertile men. Notably, the expression levels of the Pabpc1 and Pabpc2 (known as PABPC3 in human) genes in the mouse spermatogenic cells were found to be compatible with our findings [22].
To further examine the PABP expression in each cell type, we have isolated SC and RS fractions by using 2–4 % BSA gradient method. The mean purity percent of the SC fractions was higher than that of the RS fractions. The different purity rates between the two fractions most likely results from the presence of more cell types having almost similar cell density with round spermatids than spermatocytes. On the other hand, there was a sharp decline of the RS proportion in the RS arrest group that may derive from decreasing number of round spermatids in this group since spermatogenesis was arrested at the round spermatid stages.
Based on our literature analysis, there is no study endeavored to ascertain expression levels of both PABPC1 and PABPC3 proteins in the testicular tissues and isolated spermatogenic cells from distinct NOA groups. We found that the RS and SC fractions obtained from hyposperm group possessed significantly higher PABPC1 and PABPC3 protein expression than that of the RS and SC fractions collected from either RS or SC arrest group. The finding suggests that both PABPC1 and PABPC3 proteins seem to be important PABP proteins to achieve spermatogenesis successfully, which is partially achieved in the hyposperm group. The accuracy of this hypothesis should be tested in the spermatogenic cells obtained from larger NOA groups via comparing them with controls. In contrast, immunolocalization of the two proteins in the hyposperm group is partially correlated with the findings in a prior study, in which Kimura et al. (2009) characterized expression of the PABPC1 and PABPC2 proteins in mouse testis by using immunofluorescence technique [21]. Although PABPC1 is observed in all germinal epithelial cells as well as in the intertubular cells, PABPC2 is specifically generated in spermatocytes and round spermatids in mouse [21], but we detected PABPC3 only in round spermatids.
In the current work, we have observed significant alterations in the PABP genes in the NOA groups; it should be kept in mind that these changes may also originate from the following possible reasons: (i) impaired global gene expression profiles in the spermatogenic cells of the NOA groups. Global gene expression changes in the spermatogenic cells may occur due to male infertility development. (ii) Impairment in the processes such as epigenetic mechanism and PABP-interacting proteins (PAIP1 and PAIP2) controlling the expression of the PABP genes. For example, changes in the epigenetic regulation mechanism such as DNA methylation may lead to alterations in the PABPs expression. (iii) Possible changes in the transcriptional and translational activities and posttranslational mechanisms in the spermatogenic cell types in the infertile men may result in changed PABPs expression levels. These possibilities should be evaluated in detail in future investigations.
In conclusion, we here characterized expression levels of the EPAB, PABPC1, and PABPC3 genes in the testicular biopsy samples and isolated spermatogenic cells obtained from various types of NOA groups including hyposperm, RS arrest, SC arrest, and SCO. Our findings indicated that the decreased expression levels of the PABP genes may have potential in affecting spermatogenetic activity adversely. It is important to note that the major challenge in our work was the lack of control human testis tissues with preserved spermatogenesis. Absence of control human testis tissue prevented us from comparing NOA groups with control counterparts. Therefore, our findings need to be confirmed in a large number of infertile samples via comparing controls to precisely reveal the relationship between PABP expression and male infertility development. On the other hand, since there is not enough evidence related to functional importance of the PABP genes in the spermatogenic cells during spermatogenesis, the findings in the current work may not reflect the examined phenomena. Further studies are needed to understand biological importance of the PABP genes in the spermatogenic cells during spermatogenesis. These functional studies may provide establishing a new and different relationship with male infertility development. Also, it should be kept in mind that possible alteration in the upstream genes controlling the PABPs expression may have an effect on the expression changes in the PABP genes observed in the testicular tissues. This issue should be analyzed in future studies. Besides, investigating the effect of altered PABPs expression on the testis transcriptome and proteome would help identify the biological pathways being regulated by the PABPs. Further studies that will be designed in the light of our results would provide better understanding of the molecular background of the hyposperm, RS arrest, SC arrest, and SCO development.
Electronic supplementary material
Representative image of hematoxylin and eosin (HE)-stained sections of the NOA groups. The HE-stained sections of each group were analyzed under a bright-field microscopy, and thereby their histopathological characteristics and Johnsen scores were determined. Classification of the testicular biopsy samples into different non-obstructive azoospermia (NOA) groups were performed based on their spermatogenic activity and germinal epithelial cell content, and they were accordingly separated into groups as follows: a hypospermatogenesis, b round spermatid arrest, c spermatocyte arrest, and d Sertoli cell-only syndrome. SG spermatogonia, SC spermatocyte, RS round spermatid, ES elongated spermatid, SE Sertoli cell, IT intertubular area, L lumen. Scale bar: 50 μm. (JPG 1.39 MB)
Localization of the EPAB transcript in the human testis tissue. We have detected EPAB mRNA localization in the testicular biopsy samples obtained from men with hyposperm. For this purpose, EPAB probe specific to exons 1 and 2 was produced as the following processes: RNA isolation, cDNA synthesis, and PCR amplification. a A representative photomicrograph of the RNA in situ hybridization on the testis section from hyposperm group. We observed that EPAB mRNA was cytoplasmically expressed in the spermatogenic cells as well as in the intertubular somatic cells. b A representative photomicrograph of the negative control section. As expected, there was no reaction for the EPAB unlabeled probe in the negative control section. SG spermatogonia, SC spermatocyte, RS round spermatid, ES elongated spermatid, SE Sertoli cell, IT intertubular area, L lumen. Scale bar: 50 μm. (JPG 1.49 MB)
Particular features of the isolated SC and RS fractions from hyposperm, RS arrest, and SC arrest groups using 2–4 % bovine serum albumin (BSA) gradient. Representative photomicrographs of the SC (a) and RS (b) fractions were captured by an inverted microscopy (Olymus) at original magnification of ×400. Scale bar: 50 μm. To determine purity percent of the isolated spermatocyte and round spermatid fractions, their smear slides were air-dried, and then stained with hematoxylin-eosin. Representative photomicrographs of hematoxylin-eosin-stained SC (c) and RS (d) fractions were taken at original magnification of ×200; inserts were at ×1000 magnification. Scale bar: 50 μm. e The HE-stained slides were examined with the bright-field microscope (Zeiss) to determine purity percent of the isolated SC and RS fractions, which were identified based on their morphological characteristics. In general, the purity rates of isolated spermatocyte and round spermatid fractions were in usable levels (>77 %); however, the RS fraction obtained from RS arrest group was found to be in low levels (∼24 %). SG spermatogonia, SC spermatocyte, RS round spermatid, SE Sertoli cell, OC other cell, Hyposperm hypospermatogenesis, RS arrest round spermatid arrest, SC arrest spermatocyte arrest. Data are presented as mean ± standard deviation (SD). (JPG 1.76 MB)
Sequences and localizations of primers for EPAB, PABPC1, and PABPC3 genes used in the current study. F, Forward; R, Reverse; bp, base pair. (JPG 690 KB)
Histopathologic features of the testicular biopsy samples obtained from various types of NOA groups. J.S., Johnsen score. (JPG 2.30 MB)
Acknowledgments
The authors thank Deniz Ozel-Erkan, M.Sc. for her contribution to statistical analysis of data, and thank Asli Uyar, Ph.D. for critically reading the article.
Authors’ roles
S.O. and N.D. designed the study. S.O., B.S., and F.U. created all data, and S.O. wrote the article. I.C.B. performed pathological analysis of the testicular biopsies. M.F.U. provided the testicular biopsy samples. N.D. and G.A. critically read the article.
Compliance with ethical standards
Funding
This study was supported by TUBITAK (Grant No. 112S169).
Conflict of interest
The authors declare that there is no conflict of interest.
Footnotes
Capsule The EPAB, PABPC1 and PABPC3 proteins seem to be important RNA-binding proteins in regulating translation control of the mRNAs in spermatogenic cells during spermatogenesis.
References
- 1.Dube E, Hermo L, Chan PT, Cyr DG. Alterations in gene expression in the caput epididymides of nonobstructive azoospermic men. Biol Reprod. 2008;78:342–51. doi: 10.1095/biolreprod.107.062760. [DOI] [PubMed] [Google Scholar]
- 2.Gnoth C, Godehardt E, Frank-Herrmann P, Friol K, Tigges J, Freundl G. Definition and prevalence of subfertility and infertility. Hum Reprod. 2005;20:1144–7. doi: 10.1093/humrep/deh870. [DOI] [PubMed] [Google Scholar]
- 3.Hamada A, Esteves SC, Nizza M, Agarwal A. Unexplained male infertility: diagnosis and management. Int Braz J Urol. 2012;38:576–94. doi: 10.1590/S1677-55382012000500002. [DOI] [PubMed] [Google Scholar]
- 4.Wosnitzer M, Goldstein M, Hardy MP. Review of azoospermia. Spermatogenesis. 2014;4:e28218. doi: 10.4161/spmg.28218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hargreave TB. Genetic basis of male fertility. Br Med Bull. 2000;56:650–71. doi: 10.1258/0007142001903454. [DOI] [PubMed] [Google Scholar]
- 6.Safran A, Reubinoff BE, Porat-Katz A, Schenker JG Lewin A. Assisted reproduction for the treatment of azoospermia. Hum Reprod. 1998;13(Suppl 4):47–60. doi: 10.1093/humrep/13.suppl_4.47. [DOI] [PubMed] [Google Scholar]
- 7.Malcher A, Rozwadowska N, Stokowy T, Kolanowski T, Jedrzejczak P, Zietkowiak W, et al. Potential biomarkers of nonobstructive azoospermia identified in microarray gene expression analysis. Fertil Steril. 2013;100:1686–94e1–7. doi: 10.1016/j.fertnstert.2013.07.1999. [DOI] [PubMed] [Google Scholar]
- 8.Lin YH, Lin YM, Teng YN, Hsieh TY, Lin YS Kuo PL. Identification of ten novel genes involved in human spermatogenesis by microarray analysis of testicular tissue. Fertil Steril. 2006;86:1650–8. doi: 10.1016/j.fertnstert.2006.04.039. [DOI] [PubMed] [Google Scholar]
- 9.Fox MS, Ares VX, Turek PJ, Haqq C, Reijo Pera RA. Feasibility of global gene expression analysis in testicular biopsies from infertile men. Mol Reprod Dev. 2003;66:403–21. doi: 10.1002/mrd.10364. [DOI] [PubMed] [Google Scholar]
- 10.Nayernia K, Adham I, Kremling H, Reim K, Schlicker M, Schluter G, et al. Stage and developmental specific gene expression during mammalian spermatogenesis. Int J Dev Biol. 1996;40:379–83. [PubMed] [Google Scholar]
- 11.Holstein AF, Schulze W, Davidoff M. Understanding spermatogenesis is a prerequisite for treatment. Reprod Biol Endocrinol. 2003;1:107. doi: 10.1186/1477-7827-1-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hecht NB. Molecular mechanisms of male germ cell differentiation. Bioessays. 1998;20:555–61. doi: 10.1002/(SICI)1521-1878(199807)20:7<555::AID-BIES6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 13.Steger K. Transcriptional and translational regulation of gene expression in haploid spermatids. Anat Embryol. 1999;199:471–87. doi: 10.1007/s004290050245. [DOI] [PubMed] [Google Scholar]
- 14.Kleene KC. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermiogenesis in the mouse. Development. 1989;106:367–73. doi: 10.1242/dev.106.2.367. [DOI] [PubMed] [Google Scholar]
- 15.Elliott D. Pathways of post-transcriptional gene regulation in mammalian germ cell development. Cytogenet Genome Res. 2003;103:210–6. doi: 10.1159/000076806. [DOI] [PubMed] [Google Scholar]
- 16.Kleene KC, Distel RJ, Hecht NB. Translational regulation and deadenylation of a protamine mRNA during spermiogenesis in the mouse. Dev Biol. 1984;105:71–9. doi: 10.1016/0012-1606(84)90262-8. [DOI] [PubMed] [Google Scholar]
- 17.Wilkie GS, Gautier P, Lawson D, Gray NK. Embryonic poly(A)-binding protein stimulates translation in germ cells. Mol Cell Biol. 2005;25:2060–71. doi: 10.1128/MCB.25.5.2060-2071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voeltz GK, Ongkasuwan J, Standart N, Steitz JA. A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev. 2001;15:774–88. doi: 10.1101/gad.872201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Friend K, Brook M, Bezirci FB, Sheets MD, Gray NK, Seli E. Embryonic poly(A)-binding protein (ePAB) phosphorylation is required for Xenopus oocyte maturation. Biochem J. 2012;445:93–100. doi: 10.1042/BJ20120304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorgoni B, Richardson WA, Burgess HM, Anderson RC, Wilkie GS, Gautier P, et al. Poly(A)-binding proteins are functionally distinct and have essential roles during vertebrate development. Proc Natl Acad Sci U S A. 2011;108:7844–9. doi: 10.1073/pnas.1017664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura M, Ishida K, Kashiwabara S, Baba T. Characterization of two cytoplasmic poly(A)-binding proteins, PABPC1 and PABPC2, in mouse spermatogenic cells. Biol Reprod. 2009;80:545–54. doi: 10.1095/biolreprod.108.072553. [DOI] [PubMed] [Google Scholar]
- 22.Kleene KC, Wang MY, Cutler M, Hall C, Shih D. Developmental expression of poly(A) binding protein mRNAs during spermatogenesis in the mouse. Mol Reprod Dev. 1994;39:355–64. doi: 10.1002/mrd.1080390403. [DOI] [PubMed] [Google Scholar]
- 23.Wormington M, Searfoss AM, Hurney CA. Overexpression of poly(A) binding protein prevents maturation-specific deadenylation and translational inactivation in Xenopus oocytes. EMBO J. 1996;15:900–9. [PMC free article] [PubMed] [Google Scholar]
- 24.Feral C, Guellaen G, Pawlak A. Human testis expresses a specific poly(A)-binding protein. Nucleic Acids Res. 2001;29:1872–83. doi: 10.1093/nar/29.9.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seli E, Lalioti MD, Flaherty SM, Sakkas D, Terzi N, Steitz JA. An embryonic poly(A)-binding protein (ePAB) is expressed in mouse oocytes and early preimplantation embryos. Proc Natl Acad Sci U S A. 2005;102:367–72. doi: 10.1073/pnas.0408378102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ozturk S, Guzeloglu-Kayisli O, Demir N, Sozen B, Ilbay O, Lalioti MD, et al. Epab and Pabpc1 are differentially expressed during male germ cell development. Reprod Sci. 2012;19:911–22. doi: 10.1177/1933719112446086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzeloglu-Kayisli O, Pauli S, Demir H, Lalioti MD, Sakkas D, Seli E. Identification and characterization of human embryonic poly(A) binding protein (EPAB) Mol Hum Reprod. 2008;14:581–8. doi: 10.1093/molehr/gan047. [DOI] [PubMed] [Google Scholar]
- 28.Ozturk S, Guzeloglu-Kayisli O, Lowther KM, Lalioti MD, Sakkas D, Seli E. Epab is dispensable for mouse spermatogenesis and male fertility. Mol Reprod Dev. 2014;81:390. doi: 10.1002/mrd.22319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guzeloglu-Kayisli O, Lalioti MD, Aydiner F, Sasson I, Ilbay O, Sakkas D, et al. Embryonic poly(A)-binding protein (EPAB) is required for oocyte maturation and female fertility in mice. Biochem J. 2012;446:47–58. doi: 10.1042/BJ20120467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guzeloglu-Kayisli O, Lalioti MD, Babayev E, Torrealday S, Karakaya C, Seli E. Human embryonic poly(A)-binding protein (EPAB) alternative splicing is differentially regulated in human oocytes and embryos. Mol Hum Reprod. 2013;20:59–65. doi: 10.1093/molehr/gat061. [DOI] [PubMed] [Google Scholar]
- 31.WHO. Laboratory manual for the examination of human semen and sperm-cervical mucus interaction 1999.
- 32.Ozturk S, Sozen B Demir N. Epab and Pabpc1 are differentially expressed in the postnatal mouse ovaries JARG. 2014. [DOI] [PMC free article] [PubMed]
- 33.Chatterjee S, Malhotra R, Varghese F, Bukhari AB, Patil A, Budrukkar A, et al. Quantitative immunohistochemical analysis reveals association between sodium iodide symporter and estrogen receptor expression in breast cancer. PLoS One. 2013;8:e54055. doi: 10.1371/journal.pone.0054055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ozturk S, Yaba-Ucar A, Sozen B, Mutlu D, Demir N. Superovulation alters embryonic poly(A)-binding protein (epab) and poly(A)-binding protein, cytoplasmic 1 (Pabpc1) gene expression in mouse oocytes and early embryos. Reprod Fertil Dev. 2014 doi: 10.1071/RD14106. [DOI] [PubMed] [Google Scholar]
- 35.Kotaja N, Kimmins S, Brancorsini S, Hentsch D, Vonesch JL, Davidson I, et al. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat Methods. 2004;1:249–54. doi: 10.1038/nmeth1204-249. [DOI] [PubMed] [Google Scholar]
- 36.Bellve AR. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 1993;225:84–113. doi: 10.1016/0076-6879(93)25009-Q. [DOI] [PubMed] [Google Scholar]
- 37.Romrell LJ, Bellve AR, Fawcett DW. Separation of mouse spermatogenic cells by sedimentation velocity. A morphological characterization. Dev Biol. 1976;49:119–31. doi: 10.1016/0012-1606(76)90262-1. [DOI] [PubMed] [Google Scholar]
- 38.Vasco C, Zuccotti M, Redi CA, Garagna S. Identification, isolation, and RT-PCR analysis of single stage-specific spermatogenetic cells obtained from portions of seminiferous tubules classified by transillumination microscopy. Mol Reprod Dev. 2009;76:1173–7. doi: 10.1002/mrd.21086. [DOI] [PubMed] [Google Scholar]
- 39.Rockett JC, Patrizio P, Schmid JE, Hecht NB, Dix DJ. Gene expression patterns associated with infertility in humans and rodent models. Mutat Res. 2004;549:225–40. doi: 10.1016/j.mrfmmm.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 40.Ghalayini IF, Al-Ghazo MA, Hani OB, Al-Azab R, Bani-Hani I, Zayed F, et al. Clinical comparison of conventional testicular sperm extraction and microdissection techniques for non-obstructive azoospermia. J Clin Med Res. 2011;3:124–31. doi: 10.4021/jocmr542w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turek PJ, Ljung BM, Cha I, Conaghan J. Diagnostic findings from testis fine needle aspiration mapping in obstructed and nonobstructed azoospermic men. J Urol. 2000;163:1709–16. doi: 10.1016/S0022-5347(05)67526-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Representative image of hematoxylin and eosin (HE)-stained sections of the NOA groups. The HE-stained sections of each group were analyzed under a bright-field microscopy, and thereby their histopathological characteristics and Johnsen scores were determined. Classification of the testicular biopsy samples into different non-obstructive azoospermia (NOA) groups were performed based on their spermatogenic activity and germinal epithelial cell content, and they were accordingly separated into groups as follows: a hypospermatogenesis, b round spermatid arrest, c spermatocyte arrest, and d Sertoli cell-only syndrome. SG spermatogonia, SC spermatocyte, RS round spermatid, ES elongated spermatid, SE Sertoli cell, IT intertubular area, L lumen. Scale bar: 50 μm. (JPG 1.39 MB)
Localization of the EPAB transcript in the human testis tissue. We have detected EPAB mRNA localization in the testicular biopsy samples obtained from men with hyposperm. For this purpose, EPAB probe specific to exons 1 and 2 was produced as the following processes: RNA isolation, cDNA synthesis, and PCR amplification. a A representative photomicrograph of the RNA in situ hybridization on the testis section from hyposperm group. We observed that EPAB mRNA was cytoplasmically expressed in the spermatogenic cells as well as in the intertubular somatic cells. b A representative photomicrograph of the negative control section. As expected, there was no reaction for the EPAB unlabeled probe in the negative control section. SG spermatogonia, SC spermatocyte, RS round spermatid, ES elongated spermatid, SE Sertoli cell, IT intertubular area, L lumen. Scale bar: 50 μm. (JPG 1.49 MB)
Particular features of the isolated SC and RS fractions from hyposperm, RS arrest, and SC arrest groups using 2–4 % bovine serum albumin (BSA) gradient. Representative photomicrographs of the SC (a) and RS (b) fractions were captured by an inverted microscopy (Olymus) at original magnification of ×400. Scale bar: 50 μm. To determine purity percent of the isolated spermatocyte and round spermatid fractions, their smear slides were air-dried, and then stained with hematoxylin-eosin. Representative photomicrographs of hematoxylin-eosin-stained SC (c) and RS (d) fractions were taken at original magnification of ×200; inserts were at ×1000 magnification. Scale bar: 50 μm. e The HE-stained slides were examined with the bright-field microscope (Zeiss) to determine purity percent of the isolated SC and RS fractions, which were identified based on their morphological characteristics. In general, the purity rates of isolated spermatocyte and round spermatid fractions were in usable levels (>77 %); however, the RS fraction obtained from RS arrest group was found to be in low levels (∼24 %). SG spermatogonia, SC spermatocyte, RS round spermatid, SE Sertoli cell, OC other cell, Hyposperm hypospermatogenesis, RS arrest round spermatid arrest, SC arrest spermatocyte arrest. Data are presented as mean ± standard deviation (SD). (JPG 1.76 MB)
Sequences and localizations of primers for EPAB, PABPC1, and PABPC3 genes used in the current study. F, Forward; R, Reverse; bp, base pair. (JPG 690 KB)
Histopathologic features of the testicular biopsy samples obtained from various types of NOA groups. J.S., Johnsen score. (JPG 2.30 MB)