Abstract
Thirty-four cefotaxime-resistant Salmonella enterica serovar Typhimurium isolates representative of the isolates that caused outbreaks of gastroenteritis in 10 hospitals in seven regions of Russia and Belarus from 1994 to 2003 were analyzed. All isolates produced the CTX-M-5-like extended-spectrum β-lactamase, which confers high-level resistance to cefotaxime and ceftriaxone and decreased susceptibility to ceftazidime. The blaCTX-M genes were located on small (7.4- to 12-kb) non-self-transferable plasmids approximately 20 bp downstream of the ISEcp1 insertion sequences. Some isolates carried additional conjugative plasmids mediating resistance to penicillin-inhibitor combinations and various non-β-lactam agents, including tetracycline, chloramphenicol, gentamicin, tobramycin, and co-trimoxazole. Despite the minor differences in susceptibility patterns, all isolates were considered clonally related on the basis of arbitrarily primed PCR and pulsed-field gel electrophoresis analysis. The similarities of the restriction profiles of the CTX-M-coding plasmids further supported the clonal origin of these isolates.
Multiple-drug resistance in salmonellae has emerged as an important problem in many countries of the world (1, 4, 11, 19, 22, 30, 32, 38, 53, 57). The development of resistance to expanded-spectrum cephalosporins is especially alarming because these drugs have been successfully used for the empirical treatment of severe salmonellosis over a relatively long time. Nevertheless, sporadic infections or nosocomial outbreaks caused by oxyimino-cephalosporin-resistant salmonellae have been reported increasingly more often over the last decade (10, 14, 15, 23, 33, 37, 56). This resistance is frequently attributed to the production of various plasmid-mediated extended-spectrum β-lactamases (ESBLs), including the TEM, SHV, PER, and CTX-M enzymes (2, 6, 8, 17, 18, 20, 25, 34, 35, 39, 41, 49-52, 55). The latter group of enzymes is one of the most commonly encountered ESBL types in Salmonella spp. The CTX-M-2 β-lactamase, initially identified in Salmonella enterica serotype Typhimurium in Buenos Aires, Argentina, in 1990 (7), has broadly disseminated among different serovars of Salmonella in Argentinean hospitals (38). Isolates of Salmonella serovar Typhimurium producing ESBLs closely related to CTX-M-2 have also been reported in eastern and southern European countries, such as Latvia (9), Greece (47), Russia (16), and Hungary (45). The last report described the spread of a single Salmonella serovar Typhimurium clone resistant to extended-spectrum cephalosporins in three European countries.
In Russia and Belarus the incidence of nosocomial infections caused by multiresistant salmonellae rose dramatically in the middle to late 1990s. The most noteworthy were the outbreaks of Salmonella serovar Typhimurium resistant to cefotaxime that occurred in some Russian hospitals in Moscow, St. Petersburg, and the Smolensk region and in many Belarussian hospitals in the Minsk, Gomel, Grodno, and Vitebsk regions from 1994 through the beginning of 2003 (3, 13). Most patients affected by these outbreaks were children younger than 1 year of age, although in one of the Moscow hospitals a large epidemic affected more than 600 adults, and in the St. Petersburg Psychiatric Institute, a resistant Salmonella serovar Typhimurium clone disseminated among elderly patients. The severity of diseases varied from mild forms of gastroenteritis to life-threatening bacteremia with high body temperatures. Some of the cases were complicated by other extraintestinal infections. In addition, asymptomatic carriage was observed among hospital personnel (3).
The resistance phenotype exhibited by representative Salmonella serovar Typhimurium isolates from multiple nosocomial outbreaks in Russia and Belarus prompted us to investigate the potential relationship between these isolates and the molecular mechanisms of their resistance to β-lactam antibiotics.
(The results of this work were presented in part at the 41st Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, Ill., 16 to 19 December 2001.)
MATERIALS AND METHODS
Bacterial isolates.
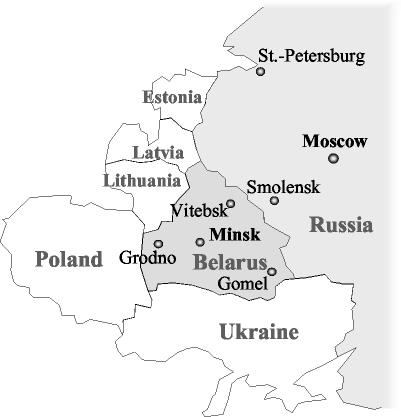
Thirty-four cefotaxime-resistant isolates (one per patient) of Salmonella serovar Typhimurium were included in this study. They were representative isolates from the outbreaks of gastroenteritis that occurred in 10 hospitals in seven regions of Russia and Belarus from 1994 to 2003 (Table 1; Fig. 1). Twenty-six strains were isolated from hospitalized children from 1 month to 14 years of age, one strain (JAR-81) was recovered from a 30-year-old woman who had apparently been infected from her child in a hospital, six strains (MOS-20 and SP-829 through SP-893) were obtained from patients older than 50 years, and one strain (JAR-137) was cultured from an environmental source (a water container used to wash hospital clothes) that was potentially implicated in the transmission of a nosocomial infection. In addition, the previously described Argentinean strain, strain CAS-5, with a resistance phenotype similar to those of the study isolates (7), and three unrelated susceptible isolates of Salmonella serovar Typhimurium were used to compare the genomic fingerprints. Isolates were identified biochemically with the API 20E system (bioMérieux, Marcy l'Etoile, France) and serotyped with respect to their cell wall (O) and flagellar (H) antigens. Salmonella serovar Typhimurium strain ATCC 14028 was used for quality control for identification and serotyping.
TABLE 1.
Characteristics of cefotaxime-resistant Salmonella serovar Typhimurium isolates
| Isolate(s) | Hospital | Location | Country | Yr of isolation |
|---|---|---|---|---|
| MI-16 | H1 | Minsk | Belarus | 1995 |
| VOL-19 | H2 | Volkovisk (Grodno region) | Belarus | 1995 |
| RET-27 | H3 | Retchitsa (Gomel region) | Belarus | 1997 |
| VTB-6570, -3078, -13526, -9837, VTB-14533 | H4 | Vitebsk | Belarus | 1999-2000 |
| VTB-1358, -1603, -14242, -700 | H5 | Vitebsk | Belarus | 2000 |
| USH-16753, USH-13205, -1845 | H6 | Ushatchy (Vitebsk region) | Belarus | 1999-2000 |
| ORS-13935 | H7 | Orsha (Vitebsk region) | Belarus | 2000 |
| SP-829, -832, -838, -891, -893 | H8 | St. Petersburg | Russia | 1996 |
| MOS-20 | H9 | Moscow | Russia | 1997 |
| JAR-637, -685, -727, -728, -735, -736, -737, -22, -79, -80, -81, 137 | H10 | Jartsevo (Smolensk region) | Russia | 2002-2003 |
FIG. 1.
Geographic locations of the hospitals where the cefotaxime-resistant Salmonella strains were isolated.
Susceptibility testing and phenotypic ESBL detection.
The MICs of ampicillin, amoxicillin-clavulanic acid (2:1), piperacillin, piperacillin-tazobactam (with tazobactam at a fixed concentration of 4 μg/ml), cefotaxime, ceftriaxone, ceftazidime, ceftazidime-clavulanic acid (4:1), aztreonam, and cefoxitin were determined by Etests (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar (Becton Dickinson, Sparks, Md.). Susceptibilities to non-β-lactam agents (tetracycline, chloramphenicol, gentamicin, tobramycin, trimethoprim-sulfamethoxazole, and ciprofloxacin) were determined by the disk diffusion method with commercial disks (Becton Dickinson). The results of susceptibility testing were interpreted according to the NCCLS standards (36). Escherichia coli strains ATCC 25922 and ATCC 35218 were used for quality control.
ESBL production was detected by the double-disk synergy test. Disks with cefotaxime (30 μg) and ceftazidime (30 μg) were each placed 20 and 30 mm (center to center) from a disk with amoxicillin-clavulanic acid (20/10 μg). Salmonella serovar Typhimurium strains producing the known enzymes CTX-M-2 and CTX-M-4 were used for quality control for ESBL detection.
Molecular typing.
Arbitrarily primed PCR (AP-PCR) with primers ERIC1R and ERIC2 (54) and PCR with a random primer, primer OPB-17 (26, 42), were used to type all the Salmonella isolates. In addition, the genetic relatedness of 15 isolates was assessed by the pulsed-field gel electrophoresis (PFGE) approach.
PCR typing was performed with template DNA extracted by use of the InstaGene matrix (Bio-Rad, Hercules, Calif.) from three to four colonies of each strain grown overnight on MacConkey agar. Reactions were set up in the Ready-To-Go PCR Beads format (Amersham Biosciences, Piscataway, N.J.) and contained 50 pmol of each primer and 2 μl (approximately 10 ng) of template DNA. The amplification was carried out in a PTC-200 thermocycler (MJ Research, Waltham, Mass.) under the following conditions: initial denaturation at 94°C for 2 min and 30 s, followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 47°C (in the case of the ERIC primers) or 35°C (in the case of primer OPB-17) for 1 min, and elongation at 72°C for 1 min, with the final elongation step extended to 4 min. PCR products were analyzed by agarose gel electrophoresis and ethidium bromide staining.
PFGE analysis was performed with a CHEF DRIII apparatus (Bio-Rad) as described by Struelens et al. (43). The genomic DNA of the isolates was digested with the XbaI restriction enzyme (MBI Fermentas, Vilinius, Lithuania). The results were interpreted in accordance with the criteria of Tenover et al. (46).
Isoelectric focusing (IEF) of β-lactamases.
Supernatants of bacterial sonicates (7) containing β-lactamases were examined with a PhastSystem apparatus and preformed polyacrylamide gels over the pH ranges 5 to 8 and 3 to 9 (Amersham Biosciences). β-Lactamase bands were visualized with nitrocefin (Oxoid, Basingstoke, United Kingdom). The enzymes with known pIs (TEM-1, pI 5.4; TEM-2, pI 5.6; TEM-3, pI 6.3; SHV-1, pI 7.6; and SHV-5, pI 8.2) were used as standards.
Detection and characterization of β-lactamase-encoding genes.
Detection of blaTEM genes was performed by PCR, as described earlier (29).
The blaCTX-M genes were detected with primers CTX-M/F′ (5′-TTTGCGATGTGCAGTACCAGTAA-3′) and CTX-M/R′ (5′-CGATATCGTTGGTGGTGCCATA-3′), which match conserved sequences at positions 205 to 227 and 748 to 727, respectively, relative to the blaCTX-M gene translational starting point. The 50-μl PCR mixtures contained 50 mM KCl, 10 mM Tris-HCl (pH 9), 0.1% Triton X-100, 2 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 0.5 μM each primer, 1 U of TaqBead Hot Start polymerase (Promega, Madison, Wis.), and 5 μl of template DNA prepared with Lyse-N-Go PCR reagent (Pierce, Rockford, Ill.), as recommended by the manufacturer. Amplification reactions were carried out in a PTC-200 thermocycler (MJ Research) under the following conditions: initial denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 95°C for 20 s, annealing at 51°C for 30 s, and elongation at 72°C for 30 s. The final elongation step was extended to 3 min. To verify that the PCR products corresponded to the blaCTX-M-2-related genes, we compared their restriction profiles after digestion with PstI endonuclease (Promega) with that of the amplicon of the blaCTX-M-2 gene.
Both strands of the amplified 544-bp internal fragments of blaCTX-M genes from six Salmonella strains were sequenced with primers CTX-M/F′ and CTX-M/R′ and a CEQ-2000 automated sequencer (Beckman-Coulter, Fullerton, Calif.). Sequencing was done by the Eurogene Company (Moscow, Russia).
A PCR with primers OXA-1/F (5′-ATGAAAAACACAATACATATCAAC-3′) and OXA-1/R (5′-TTTCCTGTAAGTGCGGACAC-3′) was used to detect a 755-bp internal fragment of blaOXA-1-related genes. The composition of the PCR mixtures and the amplification conditions were the same as those described above for the blaCTX-M genes, with the exception that the magnesium concentration was 1.5 mM and the denaturation and annealing temperatures were 94 and 48°C, respectively.
Detection of mobile elements upstream of the blaCTX-M genes.
The association of blaCTX-M genes with the ISEcp1 element (P. D. Stapleton, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 1457, 1999; GenBank accession number AJ242809) or the modified sul1-type integron containing open reading frame orf513 (5) (GenBank accession number AY079169) was studied by PCRs with an internal ISEcp1-specific forward primer (5′-TGTCTGGTATAATAAGAATATCATC-3′) or an internal integron-specific forward primer (5′-ATCCATCACAGAGTCGTCTC-3′) and CTX-M-specific reverse consensus primer MA3 (40). The ISEcp1 and integron primers were designed to match the 3′-end sequences of the ISEcp1 tnpA gene and orf513, respectively. These primers were used in PCRs under the same conditions described above for amplification of the blaCTX-M fragments, but with a magnesium concentration of 1.5 mM and denaturation and annealing temperatures of 94 and 43°C, respectively.
Transfer of resistance and analysis of plasmids carrying the blaCTX-M genes.
All the cefotaxime-resistant Salmonella isolates were mated in broth with E. coli AB1456 (F− Rifr). Transconjugants were selected on two types of agar plates: one containing rifampin (100 μg/ml) plus cefotaxime (10 μg/ml) and the other one containing rifampin (100 μg/ml) plus ampicillin (100 μg/ml).
In addition, plasmids purified from 18 isolates with a Wizard Plus SV Minipreps kit (Promega) were used to transform E. coli TOP10 (Invitrogen Corp., Carlsbad, Calif.) competent cells. Transformants were selected on agar containing cefotaxime (10 μg/ml). Native plasmids isolated from the transformants and restriction fragments obtained after digestion of these plasmids with the PstI and PvuII endonucleases (Promega) were analyzed by agarose gel electrophoresis.
RESULTS AND DISCUSSION
Susceptibility.
The resistance phenotypes of nosocomial Salmonella isolates are shown in Table 2. The most distinctive feature of these isolates was their high levels of resistance to cefotaxime, ceftriaxone, and aztreonam (MICs, ≥256, ≥256, and 64 μg/ml, respectively). Most of the isolates remained susceptible to ceftazidime at the breakpoints advocated by NCCLS (16 to 32 μg/ml), although the MICs of this drug for the strains were markedly elevated (3 to 24 μg/ml) compared to those for the naturally susceptible strains (0.06 to 0.25 μg/ml). A synergy between the oxyimino-β-lactams and clavulanic acid indicated the production of ESBLs. The higher levels of resistance to cefotaxime and ceftriaxone than to ceftazidime corresponded well to the resistance phenotype conferred by the CTX-M-type ESBLs in Salmonella strains described previously (7, 9, 16, 38, 45). Apparently, the resistance was not related to impermeability or the production of class C β-lactamases, since all the isolates retained susceptibility to cefoxitin (31). Of the 34 isolates studied, 27 (79%) demonstrated high-level resistance to all penicillin-inhibitor combinations, whereas the remaining 7 isolates were susceptible to piperacillin-tazobactam.
TABLE 2.
Susceptibilities and β-lactamase patterns of Salmonella serovar Typhimurium isolates, transconjugants and transformants
| Strain | Etest MICs (mg/liter)a
|
Coresistance markersb | PCR result for bla genes
|
β-Lactamase pI(s) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AM | XL | PP | PTc | CT | TX | TZ | TZL | FX | AT | TEM | OXA-1 | CTX-M | |||
| MI-16 | ≥256 | 96 | ≥256 | ≥256 | ≥256 | ≥256 | 12 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| VOL-19 | ≥256 | 96 | ≥256 | 128 | ≥256 | ≥256 | 12 | 1 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| RET-27 | ≥256 | 32 | ≥256 | 2 | ≥256 | ≥256 | 12 | 0.75 | 2 | 64 | Gm, Tb | − | − | + | ≥8.4 |
| VTB-3078 | ≥256 | 192 | ≥256 | 128 | ≥256 | ≥256 | 4 | 1 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| VTB-6570 | ≥256 | 96 | ≥256 | 128 | ≥256 | ≥256 | 8 | 1 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| VTB-9837 | ≥256 | 8 | ≥256 | 3 | ≥256 | ≥256 | 3 | 0.75 | 2 | 64 | − | − | + | ≥8.4 | |
| VTB-13526 | ≥256 | 96 | ≥256 | ≥256 | ≥256 | ≥256 | 12 | 0.5 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| VTB-14533 | ≥256 | 192 | ≥256 | ≥256 | ≥256 | ≥256 | 12 | 0.75 | 2 | 64 | Tet, Chl | − | + | + | 7.5, ≥8.4 |
| VTB-700 | ≥256 | 12 | ≥256 | 3 | ≥256 | ≥256 | 12 | 0.5 | 2 | 64 | Tet, Chl, Gm, Tb | − | − | + | ≥8.4 |
| VTB-1358 | ≥256 | 32 | ≥256 | 2 | ≥256 | ≥256 | 12 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb | − | − | + | ≥8.4 |
| VTB-1603 | ≥256 | 16 | ≥256 | 2 | ≥256 | ≥256 | 16 | 1 | 2 | 64 | Tet, Chl, Gm, Tb | − | − | + | ≥8.4 |
| VTB-14242 | ≥256 | 96 | ≥256 | ≥256 | ≥256 | ≥256 | 16 | 1 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| USH-16753 | ≥256 | 96 | ≥256 | ≥256 | ≥256 | ≥256 | 16 | 1 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| USH-1845 | ≥256 | 192 | ≥256 | ≥256 | ≥256 | ≥256 | 24 | 1 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| USH-13205 | ≥256 | 64 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 0.38 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| ORS-13935 | ≥256 | 96 | ≥256 | ≥256 | ≥256 | ≥256 | 12 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| SP-829 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | 4 | 0.75 | 2 | 64 | Tet, Chl | − | + | + | 7.5, ≥8.4 |
| SP-832 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | 6 | 0.75 | 2 | 64 | Tet, Chl | − | + | + | 7.5, ≥8.4 |
| SP-838 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 1 | 2 | 64 | Tet, Chl, Gm, Tb | + | + | + | 5.4, 7.5, ≥8.4 |
| SP-891 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | 12 | 1.5 | 2 | 64 | Tet, Chl, Gm, Tb | + | + | + | 5.4, 7.5, ≥8.4 |
| SP-893 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | 4 | 0.75 | 2 | 64 | Tet, Chl | − | + | + | 7.5, ≥8.4 |
| MOS-20 | ≥256 | 12 | ≥256 | 8 | ≥256 | ≥256 | 8 | 1.5 | 2 | 64 | Tet, Chl, Gm, Tb | − | − | + | ≥8.4 |
| JAR-637 | ≥256 | 32 | ≥256 | ≥256 | ≥256 | ≥256 | 4 | 0.75 | 2 | 64 | Tet, Chl, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-685 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 1 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-727 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 1 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-728 | ≥256 | 128 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-735 | ≥256 | 128 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-736 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-737 | ≥256 | 64 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-22 | ≥256 | 128 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-79 | ≥256 | 128 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 1 | 2 | 64 | Tet, Chl, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-80 | ≥256 | 24 | ≥256 | 2 | ≥256 | ≥256 | 8 | 0.75 | 2 | 64 | − | − | + | ≥8.4 | |
| JAR-81 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| JAR-137 | ≥256 | 128 | ≥256 | ≥256 | ≥256 | ≥256 | 8 | 0.75 | 2 | 64 | Tet, Chl, Gm, Tb, Sxt | − | + | + | 7.5, ≥8.4 |
| TCs type 1c | ≥256 | ≥256 | ≥256 | 64 | 0.5 | 0.125 | 0.5 | 0.5 | 4 | 0.064 | Tet, Chl, ±Gm, ±Tb, ±Sxt | − | + | − | 7.5 |
| TCs type 2c | ≥256 | 64 | ≥256 | 2 | ≥256 | ≥256 | 16 | 1 | 4 | 64 | − | − | + | ≥8.4 | |
| TFs type 1d | ≥256 | 128 | ≥256 | 2 | ≥256 | ≥256 | 16 | 1.5 | 8 | 64 | − | − | + | ≥8.4 | |
| TFs type 2d | ≥256 | 126 | ≥256 | 2 | ≥256 | ≥256 | 16 | 1.5 | 8 | 64 | + | − | + | 5.4, ≥8.4 | |
AM, ampicillin; XL, amoxicillin-clavulanic acid; PP, piperacillin; PTc, piperacillin-tazobactam; CT, cefotaxime; TX, ceftriaxone; TZ, ceftazidime; TZL, ceftazidime-clavulanic acid; FX, cefoxitin; AT, aztreonam. Boldface data indicate resistance.
Tet, tetracycline; Chl, chloramphenicol; Gm, gentamicin; Tb, tobramycin; Sxt, co-trimoxazole.
TCs, transconjugants. Type 1 transconjugants were obtained from all the Salmonella isolates producing the OXA-1-type β-lactamase. Type 2 transconjugants were obtained from strains VTB-6570 and VTB-1358.
TFs, transformants. Type 1 transformants were obtained from 17 Salmonella isolates producing the CTX-M-type ESBL. Type 2 transformants were obtained from strain SP-891.
The profiles of resistance to non-β-lactams differed among the isolates. Nineteen of these isolates were simultaneously resistant to tetracycline, chloramphenicol, gentamicin, tobramycin, and co-trimoxazole; six were simultaneously resistant to tetracycline, chloramphenicol, gentamicin, and tobramycin; two were simultaneously resistant to tetracycline, chloramphenicol, and co-trimoxazole; one was simultaneously resistant to aminoglycosides only; and the remaining two isolates were susceptible to all drugs mentioned above. None of the isolates were resistant to ciprofloxacin.
β-Lactamase characterization.
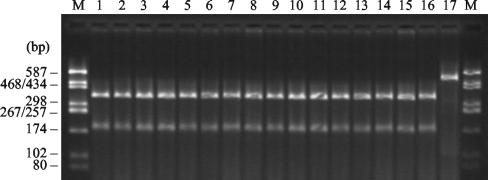
IEF revealed the production of β-lactamases with pIs ≥8.4 in all the isolates, which indicated further that enzymes of the CTX-M family could be responsible for their ESBL phenotypes. The presence of CTX-M-encoding genes was confirmed by PCR with blaCTX-M gene-specific consensus primers. By restriction analysis, all PCR products showed the same PstI banding pattern, which is characteristic of blaCTX-M-2 and closely related genes (Fig. 2). Direct sequencing of the PCR products was carried out for five isolates: VTB-1358, VTB-6570, USH-1845, VTB-14533, and MOS-20. The amplified regions of their blaCTX-M genes were identical to those of the previously described blaCTX-M-5 gene found in a Latvian Salmonella serovar Typhimurium strain (9). The sequence of the blaCTX-M gene from St. Petersburg strain SP-893 (R-893), included in this study, was previously determined by Tassios and colleagues (45). The amino acid sequence of the CTX-M-4 β-lactamase encoded by this gene is 96.5% homologous to that of CTX-M-5. Thus, at least two related CTX-M variants conferring similar resistance phenotypes were present among the epidemic nosocomial Salmonella serovar Typhimurium strains isolated in Russia and Belarus from 1994 to 2003.
FIG. 2.
PstI-digested PCR products of blaCTX-M genes of representative cefotaxime-resistant isolates. Lanes 1 to 10, Belarussian isolates MI-16, VOL-19, RET-27, VTB-6570, VTB-3078, VTB-13526, VTB-14533, VTB-1358, USH-1845, and ORS-13935, respectively; lanes 11 to 15, Russian isolates MOS-20, SP-891, SP-893, JAR-637, and JAR-81, respectively; lane 16, Salmonella serovar Typhimurium CAS-5 (CTX-M-2); lane 17, undigested PCR product from strain CAS-5; lanes M, molecular size markers (pUC18-HaeIII restriction fragments).
In addition to the CTX-M enzyme, 27 isolates from different hospitals produced a second β-lactamase that focused at pH 7.5 in IEF experiments. All these isolates were resistant to piperacillin-tazobactam, whereas the isolates lacking a pI 7.5 β-lactamase were fully susceptible to this drug. The presence of these secondary β-lactamases and resistance to penicillin-inhibitor combinations correlated well with the positive results of PCR with blaOXA-1-specific primers (Table 2). Plasmid-mediated penicillinases of the OXA-1 group are weakly inhibited by clavulanic acid and tazobactam (28) and are frequently found in salmonellae (24, 27, 48). Therefore, the production of OXA-1-like β-lactamases was most likely responsible for the additional resistance to penicillin-inhibitor combinations displayed by some of the Salmonella isolates studied.
Two isolates from St. Petersburg expressed a third β-lactamase with a pI of 5.4 and gave positive results by PCR with blaTEM-specific primers. Production of this enzyme (most likely TEM-1) was masked phenotypically by the CTX-M- and OXA-type β-lactamases, which a broader range of resistance to β-lactam antibiotics.
Transfer of resistance.
Two different types of transconjugants were obtained by mating isolates resistant to cefotaxime and piperacillin-tazobactam. Transconjugants of the first group were selected at high frequencies (10−3 to 10−4) on plates containing rifampin and ampicillin. All these transconjugants produced only β-lactamases of the OXA-1 type (pI 7.5) and exhibited similar levels of resistance to penicillins and penicillin-inhibitor combinations, with no more than 1-dilution difference in the MICs for the isolates. They were also resistant to chloramphenicol and tetracycline but differed in their resistance to aminoglycosides and co-trimoxazole (Table 2).
Transconjugants of the second group were obtained on plates containing rifampin and cefotaxime. Despite multiple attempts, only two isolates (VTB-1358 and VTB-6570) transferred resistance to cefotaxime in the conjugation experiments. In both cases the frequency of transfer was extremely low (∼10−7 to 10−8), suggesting that the plasmids conferring resistance to cefotaxime were probably non-self-transmissible but were mobilized by coexisting conjugative plasmids. The respective transconjugants produced only CTX-M-type β-lactamases (pI, ∼8.4) conferring the ESBL phenotype but lacked resistance to piperacillin-tazobactam and non-β-lactam antimicrobials.
Likewise, all cefotaxime-resistant transformants carrying the wild-type plasmids of 18 arbitrarily selected Salmonella isolates produced the CTX-M- but not the OXA-type β-lactamase and appeared to be susceptible to piperacillin-tazobactam, chloramphenicol, tetracycline, aminoglycosides, and co-trimoxazole. Notably, a clone obtained after transformation with plasmid DNA of strain SP-891 produced TEM-1, in addition to the CTX-M β-lactamase. The data presented above suggested that the blaCTX-M gene-carrying plasmids of all but one of the Salmonella isolates contained no additional resistance determinants.
Analysis of plasmids carrying the blaCTX-M genes and associated mobile elements.
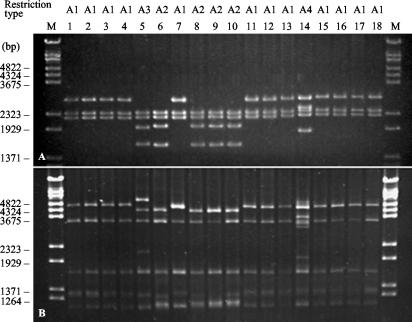
A single low-molecular-weight plasmid was detected in each of the CTX-M-β-lactamase-producing transformants. Therefore, genes encoding the CTX-M β-lactamases resided on small plasmids (7.4 to 12 kb). However, when the plasmids of different Salmonella isolates were digested with the PstI or PvuII restriction endonuclease, the patterns obtained were very similar (Fig. 3). CTX-M-encoding plasmids from 12 isolates from four Belarussian and three Russian hospitals shared the same restriction profile (profile A1). The plasmids from the other five isolates (profiles A2 and A3) were approximately 750 bp longer and contained an additional PstI restriction site. The larger size (12 kb) of the plasmid derived from isolate SP-891, the restriction profile of the plasmid (profile A4), and the presence of the blaTEM gene on this plasmid indicated that it may have evolved by insertion of a TnA-type transposon.
FIG. 3.
Restriction profiles of CTX-M-coding plasmids digested with endonucleases PstI (A) and PvuII (B). Lanes 1 to 12, Belarussian isolates RET-27, VTB-6570, VTB-3078, VTB-13526, VTB-1358, VTB-1603, VTB-14533, VTB-700, VTB-16753, USH-1845, USH-13205, and ORS-13935, respectively; lanes 13 to 18, Russian isolates MOS-20, SP-891, SP-893, JAR-637, JAR-727, and JAR-81, respectively; lanes M, molecular size markers (bacteriophage λ-BstEII restriction fragments).
It is worth mentioning that the blaCTX-M-5-carrying plasmid found in the epidemic Salmonella serovar Typhimurium strain from Latvia was also small (10 kb) and non-self-transferable (9). In contrast, plasmid pMVP-4 of Argentinean Salmonella strain CAS-5 carrying the blaCTX-M-2 gene, which is most closely related to blaCTX-M-5, was large (142 kb) and self-transferable (7). Moreover, the CTX-M-2-encoding plasmids of the Salmonella isolates from Argentina were reported to contain a modified sul1-type integron that includes orf513 upstream of the blaCTX-M-2 gene (12, 38), whereas the blaCTX-M-5 gene found on the plasmid of the Latvian strain was shown to be associated with another mobile element, ISEcp1 (21).
In order to characterize the genetic environment of the blaCTX-M genes in Salmonella serovar Typhimurium isolates from Russia and Belarus, we performed PCRs with forward primers specific for the ISEcp1 element or orf513 and the reverse blaCTX-M-specific primer. All the isolates studied produced a single amplicon of approximately 470 bp upon amplification with the ISEcp1-related primer. The size of this fragment perfectly matched the distance between the primer binding sites identified in the sequence of the CTX-M-5-encoding plasmid of the Latvian strain (GenBank accession number AF286192). The fact that ISEcp1 was detected at the same distance upstream from the blaCTX-M gene in the isolates from Russia, Belarus, and Latvia provided further evidence of the similarities of their plasmids. PCR with the orf513-related primer yielded a product only in the case of Argentinean strain CAS-5, which, on the other hand, gave no product in the experiment with the ISEcp1-specific primer. Consequently, our data confirmed that the blaCTX-M genes identified in the eastern European and South American strains of Salmonella serovar Typhimurium were located in different genetic structures.
Molecular typing.
AP-PCR with primers highly discriminative for Salmonella serovar Typhimurium (26, 42) and PFGE analysis of XbaI macrorestriction fragments (44, 45) were used to verify whether the strains isolated in different hospitals across western Russia and Belarus were related at the chromosomal level. Argentinean strain CAS-5 and three susceptible epidemiologically unrelated isolates of Salmonella serovar Typhimurium were included in the analysis for comparative purposes.
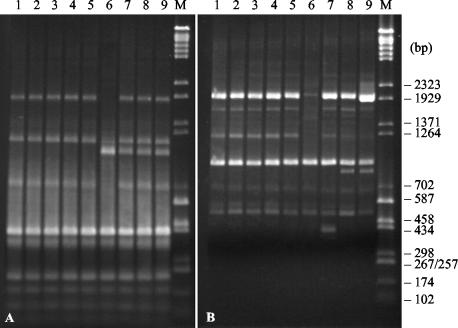
All the cefotaxime-resistant isolates from Russia and Belarus exhibited identical AP-PCR patterns; hence, only the AP-PCR patterns of five representative strains are shown in Fig. 4, along with the clearly distinguishable fingerprints of CTX-M-2-producing strain CAS-5 from Argentina and susceptible isolates from Russia and Belarus.
FIG. 4.
AP-PCR profiles of representative cefotaxime-resistant and -susceptible Salmonella serovar Typhimurium isolates amplified with primers ERIC1R and ERIC2 (A) and OPB-17 (B). Lanes 1 to 5, cefotaxime-resistant isolates RET-27, VTB-6570, MOS-20, SP-891, and JAR-637, respectively; lane 6, Salmonella serovar Typhimurium CAS-5 (CTX-M-2); lanes 7 to 9, epidemiologically unrelated susceptible isolates 21, 1745, and 4112, respectively; lanes M, molecular size markers (a mixture of pUC18-HaeIII and bacteriophage λ-BstEII restriction fragments).
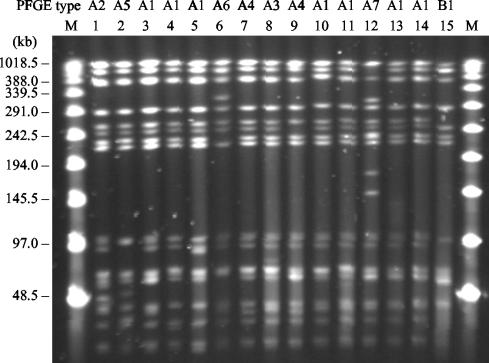
PFGE analysis further confirmed the genetic relationships among Salmonella isolates from multiple outbreaks (Fig. 5). The macrorestriction patterns (pattern A1) of the seven isolates from Vitebsk, Moscow, and St. Petersburg were indistinguishable from each other; and those (patterns A2 to A7) of the other seven isolates tested differed by three bands at most, identifying them as closely related, according to the criteria suggested by Tenover et al. (46). The four-band difference observed between the patterns of strain CAS-5 (pattern B1) and the patterns of some of the eastern European isolates were more difficult to interpret, considering the extreme distance between their geographic sources. However, on the basis of the results of AP-PCR typing and plasmid analysis, the Argentinean strain should be considered unrelated to the outbreak isolates described here.
FIG. 5.
PFGE profiles of representative cefotaxime-resistant Salmonella serovar Typhimurium isolates. Lanes 1 to 12, Belarussian isolates VTB-1358, RET-27, VTB-1603, VTB-6570, VTB-3078, USH-13205, ORS-13935, USH-1845, VTB-14533, VTB-13526, VTB-700, and VTB-16753, respectively; lanes 13 and 14, Russian isolates SP-891 and MOS-20, respectively; lane 15, Salmonella serovar Typhimurium CAS-5 (CTX-M-2); lanes M, molecular size markers (bacteriophage λ ladder PFGE marker; New England BioLabs, Beverly, Mass.).
Epidemiologies of the CTX-M-producing Salmonella isolates from Russia and Belarus.
The genetic similarity among cefotaxime-resistant Salmonella isolates from nosocomial outbreaks that occurred in Russia and Belarus suggested the possible transmission of a single clone between different hospitals. Unfortunately, the epidemiological data that could be used to establish the relationship between the early outbreaks in Minsk, Grodno, Gomel, and St. Petersburg were lacking. However, the data concerning the more recent cases clearly support the possibility of clonal transmission mediated by the transfer of patients from one hospital to another. For example, nosocomial infections due to cefotaxime-resistant Salmonella serovar Typhimurium were registered in hospitals H5, H6, and H7, located in Vitebsk and smaller settlements within the region, soon after the transfer of children patients from hospital H4, where the outbreak was ongoing at the time.
In one case documented by Akimkin and Pokrovsky (3), a resistant strain was apparently transferred from one hospital to another by a 65-year-old man who visited and nursed his grandson, who presented with symptoms of an acute gastrointestinal infection that developed in a children's clinic. Shortly after visiting the child, the man was admitted to hospital H10 due to suspected appendicitis, but after examination he was found to be infected with cefotaxime-resistant Salmonella serovar Typhimurium. New cases of infection with the same strain were reported as soon as 1 week after he had been admitted. The subsequent epidemic in that hospital affected 584 patients and 54 health care workers from 1994 through 1998.
It is noteworthy that, unlike many outbreaks due to multiresistant S. enterica strains of animal origin, most of the cases described here were obviously associated with the person-to-person transfer of a resistant clone in a hospital environment. Several reports have pointed out the long-term postinfection carriage by children and asymptomatic carriage by adults (including patients and health care workers) as an important factor facilitating the transmission of this clone (3, 13).
On the basis of the analysis of PFGE patterns and plasmid and β-lactamase contents, Tassios et al. (45) have established that a clonal relationship exists between the outbreak isolates of Salmonella serovar Typhimurium from St. Petersburg that were also included in our study and strains isolated from sporadic cases of gastroenteritis in Hungary and Greece. In one of those cases the epidemiological data indicated transfer of the cefotaxime-resistant strain by an immigrant from southern Russia (47). Interestingly, Zirnstein et al. (G. W. Zirnstein, B. Swaminathan, F. Angulo, F. Tenover, and J. Rasheed, 2nd Int. Conf. Emerg. Infect. Dis., 2000) have reported the first isolation in the United States of a CTX-M-5-producing Salmonella serovar Typhimurium strain from an infant adopted from southern Russia. The blaCTX-M-5-carrying plasmid found in the respective isolate was small (approximately 9 kb), similar to those carried by epidemic strains from Latvia (9), Russia, and Belarus. The Russian strains reported here were isolated in hospitals in the west-central (Moscow) and western (Smolensk and St. Petersburg) parts of the country, on the border with Belarus and Latvia, but until now it was impossible to obtain information on the presence of cefotaxime-resistant Salmonella serovar Typhimurium isolates in the southern parts of Russia. After evaluation of the epidemiological and molecular analysis data, however, it seems likely that the isolates found in Greece and the United States belong to the same clone which may have spread widely in Russia.
Conclusions.
The results of our study suggest that the multiple nosocomial outbreaks which occurred in the three regions of Russia and in various parts of Belarus between 1994 and 2003 were probably caused by the same clone of Salmonella serovar Typhimurium. Its resistance to expanded-spectrum cephalosporins was attributed to the production of CTX-M-4- and CTX-M-5-like β-lactamases. Some isolates of this clone were also resistant to penicillin-inhibitor combinations and various non-β-lactam agents due to the presence of additional conjugative plasmids carrying the genes for OXA-1-type penicillinase and other resistance determinants. The present situation with the international spread of Salmonella strains resistant to most of the clinically important β-lactams is of particular concern. It urges the need for consistent epidemiological monitoring and effective prevention of infections due to such strains by stringent hospital hygiene and control of personnel for intestinal colonization.
Acknowledgments
We thank Adolf Bauernfeind (Munich, Germany) for providing the reference strain of Salmonella serovar Typhimurium producing the CTX-M-2 β-lactamase and Marek Gniadkowski (Warsaw, Poland) for reviewing the manuscript of this paper.
REFERENCES
- 1.Ackers, M. L., N. D. Puhr, R. V. Tauxe, and E. D. Mintz. 2000. Laboratory-based surveillance of Salmonella serotype Typhi infections in the United States: antimicrobial resistance on the rise. JAMA 283:2668-2673. [DOI] [PubMed] [Google Scholar]
- 2.AitMhand, R., A. Soukri, N. Moustaoui, H. Amarouch, N. ElMdaghri, D. Sirot, and M. Benbachir. 2002. Plasmid-mediated TEM-3 extended-spectrum beta-lactamase production in Salmonella typhimurium in Casablanca. J. Antimicrob. Chemother. 49:169-172. [DOI] [PubMed] [Google Scholar]
- 3.Akimkin, V. G., and V. I. Pokrovsky. 2002. Epidemiology of nosocomial salmonellosis, p. 35-72. In V. G. Akimkin and V. I. Pokrovsky (ed.), Nosocomial salmonellosis in adults. Russian Academy of Medical Sciences, Moscow, Russia.
- 4.Alvseike, O., T. Leegaard, P. Aavitsland, and J. Lassen. 2002. Trend of multiple drug resistant Salmonella Typhimurium in Norway. Euro. Surveill. 7:5-7. [DOI] [PubMed] [Google Scholar]
- 5.Arduino, S. M., P. H. Roy, G. A. Jacoby, B. E. Orman, S. A. Pineiro, and D. Centron. 2002. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes Orf513. Antimicrob. Agents Chemother. 46:2303-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baraniak, A., E. Sadowy, W. Hryniewicz, and M. Gniadkowski. 2002. Two different extended-spectrum beta-lactamases (ESBLs) in one of the first ESBL-producing salmonella isolates in Poland. J. Clin. Microbiol. 40:1095-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauernfeind, A., J. M. Casellas, M. Goldberg, M. Holley, R. Jungwirth, P. Mangold, T. Rohnisch, S. Schweighart, and R. Wilhelm. 1992. A new plasmidic cefotaximase from patients infected with Salmonella typhimurium. Infection 20:158-163. [DOI] [PubMed] [Google Scholar]
- 8.Blahova, J., M. Lesicka-Hupkova, K. Kralikova, V. Krcmery, Sr., T. Krcmeryova, and K. Kubonova. 1998. Further occurrence of extended-spectrum beta-lactamase-producing Salmonella enteritidis. J. Chemother. 10:291-294. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, P. A., Y. Yang, D. Sahm, I. Grope, D. Gardovska, and G. Storch. 1998. CTX-M-5, a novel cefotaxime-hydrolyzing β-lactamase from an outbreak of Salmonella typhimurium in Latvia. Antimicrob. Agents Chemother. 42:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carattoli, A., F. Tosini, W. P. Giles, M. E. Rupp, S. H. Hinrichs, F. J. Angulo, T. J. Barrett, and P. D. Fey. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casin, I., J. Breuil, A. Brisabois, F. Moury, F. Grimont, and E. Collatz. 1999. Multidrug-resistant human and animal Salmonella typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded beta-lactamase PSE-1. J. Infect. Dis. 179:1173-1182. [DOI] [PubMed] [Google Scholar]
- 12.Di Conza, J., J. A. Ayala, P. Power, M. Mollerach, and G. Gutkind. 2002. Novel class 1 integron (InS21) carrying blaCTX-M-2 in Salmonella enterica serovar Infantis. Antimicrob. Agents Chemother. 46:2257-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dmitrachenko, T. I. 2002. Salmonellosis, shigellosis: clinical, epidemiological and bacteriological criteria of the rational antibacterial therapy. Ph.D. thesis. Belarusian State Medical University, Minsk, Belarus.
- 14.Dunne, E. F., P. D. Fey, P. Kludt, R. Reporter, F. Mostashari, P. Shillam, J. Wicklund, C. Miller, B. Holland, K. Stamey, T. J. Barrett, J. K. Rasheed, F. C. Tenover, E. M. Ribot, and F. J. Angulo. 2000. Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC beta-lactamase. JAMA 284:3151-3156. [DOI] [PubMed] [Google Scholar]
- 15.Gaillot, O., C. Clement, M. Simonet, and A. Philippon. 1997. Novel transferable beta-lactam resistance with cephalosporinase characteristics in Salmonella enteritidis. J. Antimicrob. Chemother. 39:85-87. [DOI] [PubMed] [Google Scholar]
- 16.Gazouli, M., S. V. Sidorenko, E. Tzelepi, N. S. Kozlova, D. P. Gladin, and L. S. Tzouvelekis. 1998. A plasmid-mediated beta-lactamase conferring resistance to cefotaxime in a Salmonella typhimurium clone found in St Petersburg, Russia. J. Antimicrob. Chemother. 41:119-121. [DOI] [PubMed] [Google Scholar]
- 17.Gazouli, M., E. Tzelepi, A. Markogiannakis, N. J. Legakis, and L. S. Tzouvelekis. 1998. Two novel plasmid-mediated cefotaxime-hydrolyzing beta-lactamases (CTX-M-5 and CTX-M-6) from Salmonella typhimurium. FEMS Microbiol. Lett. 165:289-293. [DOI] [PubMed] [Google Scholar]
- 18.Gierczynski, R., J. Szych, A. Cieslik, W. Rastawicki, and M. Jagielski. 2003. The occurrence of the first two CTX-M-3 and TEM-1 producing isolates of Salmonella enterica serovar Oranienburg in Poland. Int. J. Antimicrob. Agents 21:497-499. [DOI] [PubMed] [Google Scholar]
- 19.Graham, S. M. 2002. Salmonellosis in children in developing and developed countries and populations. Curr. Opin. Infect. Dis. 15:507-512. [DOI] [PubMed] [Google Scholar]
- 20.Hanson, N. D., E. S. Moland, A. Hossain, S. A. Neville, I. B. Gosbell, and K. S. Thomson. 2002. Unusual Salmonella enterica serotype Typhimurium isolate producing CMY-7, SHV-9 and OXA-30 beta-lactamases. J. Antimicrob. Chemother. 49:1011-1014. [DOI] [PubMed] [Google Scholar]
- 21.Humeniuk, C., G. Arlet, V. Gautier, P. Grimont, R. Labia, and A. Philippon. 2002. β-Lactamases of Kluyvera ascorbata, probable progenitors of some plasmid-encoded CTX-M types. Antimicrob. Agents Chemother. 46:3045-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izumiya, H., J. Terajima, S. Matsushita, K. Tamura, and H. Watanabe. 2001. Characterization of multidrug-resistant Salmonella enterica serovar Typhimurium isolated in Japan. J. Clin. Microbiol. 39:2700-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, R. N., P. R. Rhomberg, D. J. Varnam, and D. Mathai. 2002. A comparison of the antimicrobial activity of meropenem and selected broad-spectrum antimicrobials tested against multi-drug resistant gram-negative bacilli including bacteraemic Salmonella spp.: initial studies for the MYSTIC programme in India. Int. J. Antimicrob. Agents 20:426-431. [DOI] [PubMed] [Google Scholar]
- 24.Kariuki, S., C. Gilks, J. Corkill, J. Kimari, A. Benea, P. Waiyaki, and C. A. Hart. 1996. Multi-drug resistant non-typhi salmonellae in Kenya. J. Antimicrob. Chemother. 38:425-434. [DOI] [PubMed] [Google Scholar]
- 25.Lee, K., D. Yong, J. H. Yum, H. H. Kim, and Y. Chong. 2003. Diversity of TEM-52 extended-spectrum β-lactamase-producing non-typhoidal Salmonella isolates in Korea. J. Antimicrob. Chemother. 52:493-496. [DOI] [PubMed] [Google Scholar]
- 26.Lin, A. W., M. A. Usera, T. J. Barrett, and R. A. Goldsby. 1996. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella enteritidis. J. Clin. Microbiol. 34:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling, J. M., G. M. Zhou, T. H. Woo, and G. L. French. 1991. Antimicrobial susceptibilities and beta-lactamase production of Hong Kong isolates of gastroenteric salmonellae and Salmonella typhi. J. Antimicrob. Chemother. 28:877-885. [DOI] [PubMed] [Google Scholar]
- 28.Livermore, D. M. 1995. β-Lactamases in laboratory and clinical resistance. Clin. Microbiol. Rev. 8:557-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mabilat, C., and S. Goussard. 1993. PCR detection and identification of genes for extended-spectrum β-lactamases, p. 553-562. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 30.Mammina, C., L. Cannova, S. Massa, E. Goffredo, and A. Nastasi. 2002. Drug resistances in salmonella isolates from animal foods, Italy 1998-2000. Epidemiol. Infect. 129:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Martinez, L., M. C. Conejo, A. Pascual, S. Hernandez-Alles, S. Ballesta, D. A.-R. Ramirez, V. J. Benedi, and E. J. Perea. 2000. Activities of imipenem and cephalosporins against clonally related strains of Escherichia coli hyperproducing chromosomal β-lactamase and showing altered porin profiles. Antimicrob. Agents Chemother. 44:2534-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metzer, E., V. Agmon, N. Andoren, and D. Cohen. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium phage-type DT104 among salmonellae causing enteritis in Israel. Epidemiol. Infect. 121:555-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miriagou, V., R. Filip, G. Coman, and L. S. Tzouvelekis. 2002. Expanded-spectrum cephalosporin-resistant salmonella strains in Romania. J. Clin. Microbiol. 40:4334-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morosini, M. I., R. Canton, J. Martinez-Beltran, M. C. Negri, J. C. Perez-Diaz, F. Baquero, and J. Blazquez. 1995. New extended-spectrum TEM-type β-lactamase from Salmonella enterica subsp. enterica isolated in a nosocomial outbreak. Antimicrob. Agents Chemother. 39:458-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulvey, M. R., G. Soule, D. Boyd, W. Demczuk, and R. Ahmed. 2003. Characterization of the first extended-spectrum β-lactamase-producing Salmonella isolate identified in Canada. J. Clin. Microbiol. 41:460-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing: 13th informational supplement. M100-S13 (M7). National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 37.Navarro, F., E. Perez-Trallero, J. M. Marimon, R. Aliaga, M. Gomariz, and B. Mirelis. 2001. CMY-2-producing Salmonella enterica, Klebsiella pneumoniae, Klebsiella oxytoca, Proteus mirabilis and Escherichia coli strains isolated in Spain (October 1999-December 2000). J. Antimicrob. Chemother. 48:383-389. [DOI] [PubMed] [Google Scholar]
- 38.Orman, B. E., S. A. Pineiro, S. Arduino, M. Galas, R. Melano, M. I. Caffer, D. O. Sordelli, and D. Centron. 2002. Evolution of multiresistance in nontyphoid salmonella serovars from 1984 to 1998 in Argentina. Antimicrob. Agents Chemother. 46:3963-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson, D. L. 2001. Extended-spectrum β-lactamases: the European experience. Curr. Opin. Infect. Dis. 14:697-701. [DOI] [PubMed] [Google Scholar]
- 40.Saladin, M., V. T. Cao, T. Lambert, J. L. Donay, J. L. Herrmann, Z. Ould-Hocine, C. Verdet, F. Delisle, A. Philippon, and G. Arlet. 2002. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol. Lett. 209:161-168. [DOI] [PubMed] [Google Scholar]
- 41.Simarro, E., F. Navarro, J. Ruiz, E. Miro, J. Gomez, and B. Mirelis. 2000. Salmonella enterica serovar Virchow with CTX-M-like β-lactamase in Spain. J. Clin. Microbiol. 38:4676-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soto, S. M., B. Guerra, M. A. Gonzalez-Hevia, and M. C. Mendoza. 1999. Potential of three-way randomly amplified polymorphic DNA analysis as a typing method for twelve Salmonella serotypes. Appl. Environ. Microbiol. 65:4830-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Struelens, M. J., F. Rost, A. Deplano, A. Maas, V. Schwam, E. Serruys, and M. Cremer. 1993. Pseudomonas aeruginosa and Enterobacteriaceae bacteremia after biliary endoscopy: an outbreak investigation using DNA macrorestriction analysis. Am. J. Med. 95:489-498. [DOI] [PubMed] [Google Scholar]
- 44.Tamada, Y., Y. Nakaoka, K. Nishimori, A. Doi, T. Kumaki, N. Uemura, K. Tanaka, S. I. Makino, T. Sameshima, M. Akiba, M. Nakazawa, and I. Uchida. 2001. Molecular typing and epidemiological study of Salmonella enterica serotype Typhimurium isolates from cattle by fluorescent amplified-fragment length polymorphism fingerprinting and pulsed-field gel electrophoresis. J. Clin. Microbiol. 39:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tassios, P. T., M. Gazouli, E. Tzelepi, H. Milch, N. Kozlova, S. Sidorenko, N. J. Legakis, and L. S. Tzouvelekis. 1999. Spread of a Salmonella typhimurium clone resistant to expanded-spectrum cephalosporins in three European countries. J. Clin. Microbiol. 37:3774-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tzouvelekis, L. S., M. Gazouli, A. Markogiannakis, E. Paraskaki, N. J. Legakis, and E. Tzelepi. 1998. Emergence of resistance to third-generation cephalosporins amongst Salmonella typhimurium isolates in Greece: report of the first three cases. J. Antimicrob. Chemother. 42:273-275. [DOI] [PubMed] [Google Scholar]
- 48.Tzouvelekis, L. S., V. Lukova, P. T. Tassios, A. C. Fluit, R. N. Jones, and N. J. Legakis. 2003. Resistance to beta-lactams among blood isolates of Salmonella spp. in European hospitals: results from the SENTRY Antimicrobial Surveillance Program 1997-98. Clin. Microbiol. Infect. 9:149-152. [DOI] [PubMed] [Google Scholar]
- 49.Tzouvelekis, L. S., E. Tzelepi, P. T. Tassios, and N. J. Legakis. 2000. CTX-M-type beta-lactamases: an emerging group of extended-spectrum enzymes. Int. J. Antimicrob. Agents 14:137-142. [DOI] [PubMed] [Google Scholar]
- 50.Vahaboglu, H., S. Dodanli, C. Eroglu, R. Ozturk, G. Soyletir, I. Yildirim, and V. Avkan. 1996. Characterization of multiple-antibiotic-resistant Salmonella typhimurium stains: molecular epidemiology of PER-1-producing isolates and evidence for nosocomial plasmid exchange by a clone. J. Clin. Microbiol. 34:2942-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vahaboglu, H., M. Fuzi, S. Cetin, S. Gundes, E. Ujhelyi, F. Coskunkan, and O. Tansel. 2001. Characterization of extended-spectrum β-lactamase (TEM-52)-producing strains of Salmonella enterica serovar Typhimurium with diverse resistance phenotypes. J. Clin. Microbiol. 39:791-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vahaboglu, H., L. M. Hall, L. Mulazimoglu, S. Dodanli, I. Yildirim, and D. M. Livermore. 1995. Resistance to extended-spectrum cephalosporins, caused by PER-1 beta-lactamase, in Salmonella typhimurium from Istanbul, Turkey. J. Med. Microbiol. 43:294-299. [DOI] [PubMed] [Google Scholar]
- 53.van Duijkeren, E., W. J. Wannet, D. J. Houwers, and W. van Pelt. 2003. Antimicrobial susceptibilities of salmonella strains isolated from humans, cattle, pigs, and chickens in The Netherlands from 1984 to 2001. J. Clin. Microbiol. 41:3574-3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Villa, L., C. Pezzella, F. Tosini, P. Visca, A. Petrucca, and A. Carattoli. 2000. Multiple-antibiotic resistance mediated by structurally related IncL/M plasmids carrying an extended-spectrum β-lactamase gene and a class 1 integron. Antimicrob. Agents Chemother. 44:2911-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC β-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang, Y. J., C. C. Liu, S. M. Wang, J. J. Wu, A. H. Huang, and C. P. Cheng. 1998. High rates of antimicrobial resistance among clinical isolates of nontyphoidal Salmonella in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 17:880-883. [DOI] [PubMed] [Google Scholar]