Abstract
We have generated a database of expression profiles carrying the transcriptional responses of the model organism Bacillus subtilis following treatment with 37 well-characterized antibacterial compounds of different classes. The database was used to build a predictor for the assignment of the mechanisms of action (MoAs) of antibacterial compounds by the use of support vector machines. This predictor was able to correctly classify the MoA class for most compounds tested. Furthermore, we provide evidence that the in vivo MoA of hexachlorophene does not match the MoA predicted from in vitro data, a situation frequently faced in drug discovery. A database of this kind may facilitate the prioritization of novel antibacterial entities in drug discovery programs. Potential applications and limitations are discussed.
Understanding the in vivo mechanism of action (MoA) of biologically active compounds is an elementary requirement for drug discovery and development. Until recently, in vivo MoA analysis was mostly limited to cases in which in-depth knowledge of the respective biological pathway was available. With the advancement of genomics, however, new technologies with the potential to address this question on a genomic scale have emerged. Recently, Bandow et al. (2) presented the results of a proteomics study in which they analyzed the MoAs of antibacterial compounds. Earlier, Hughes et al. (15) used DNA microarray analysis to correlate the gene expression profiles derived from defined yeast mutants with those derived from compound-treated yeast cells. In that paper the high degree of correlation of an expression profile of a drug (dyclonine)-treated culture with the expression profile obtained from a mutant (erg2) elucidated the molecular target of an antifungal compound. Despite its elegance, this approach faces two practical problems: (i) it is limited to systems with well-developed genetics, and (ii) it assumes that a single compound-target interaction is responsible for a given MoA.
We have explored the potential of using gene expression profiling for in vivo analysis of the MoAs of antibacterial compounds. For this purpose we analyzed the transcriptional response of Bacillus subtilis 168 (19) following treatment with 37 antibacterial agents with known MoAs using whole-genome arrays. Since it is known that various technological pitfalls are associated with expression profiling projects of this scale (5, 23, 24), we standardized and automated the experimental steps wherever possible.
We demonstrate that such a data set will facilitate classification of the MoAs of antibacterial compounds. We also present evidence that the in vivo MoA of the antibacterial compound hexachlorophene (HCP) is not as expected from in vitro biochemical data. This finding emphasizes the practical value of the expression profile database described herein. However, we also describe potential limitations of the approach.
MATERIALS AND METHODS
Bacterial strains and growth of cultures.
B. subtilis 168 was grown in basal limitation medium (31). A 15-ml culture was inoculated at an A600 of 0.05 and incubated at 37°C and 200 rpm (Innova 4400 incubator shaker; New Brunswick Scientific, Edison, N.J.) and growth proceeded to an A600 of 0.25. At this optical density compounds were added from a 1,000× stock solution (Table 1). In parallel, control cultures were treated with the solvent for the respective compound. Cultures were further incubated under the conditions described above, and growth was monitoring by measuring the optical density at 600 nm at regular intervals. For RNA preparation, 10 ml of the culture was harvested by centrifugation at 3,000 × g at 10, 40, and 80 min posttreatment. Each datum point represents the results for three replicate cultures grown on individual days.
TABLE 1.
Compounds, their properties, and predicted MoA classes
| MoA class and compound | copt (μg/ml) | Solventa | Predictionb |
|---|---|---|---|
| Cell wall biosynthesis | |||
| Amoxicillin | 16 | DMSO | Folate |
| Cefalexin | 0.25 | 1 M NH4OH | Cell wall |
| Cefotaxime | 1 | Water | Cell wall |
| Cefoxitin | 0.5 | Water | Cell wall |
| Cycloserine | 16 | Water | Cell wall |
| Oxacillin | 0.25 | Water | Cell wall |
| Penicillin G | 4 | Water | Cell wall |
| Phosphomycin | 256 | Water | Cell wall |
| Ristocetin | 0.5 | DMSO | Cell wall |
| Vancomycin | 0.25 | Water | Cell wall |
| DNA topology | |||
| Ciprofloxacin | 0.5 | DMSO | Topo |
| Coumermycin A1 | 1 | DMSO | Membrane |
| Moxifloxacin | 0.25 | DMSO | Topo |
| Nalidixic acid | 8 | Water | Topo |
| Norfloxacin | 0.5 | DMSO | Topo |
| Novobiocin | 0.25 | Water | Topo |
| Fatty acid biosynthesis | |||
| Cerulenin | 4 | DMSO | Membrane |
| Triclosan | 1 | DMSO | Rejected |
| Folate biosynthesis | |||
| Dapsone | 64 | DMSO | Cell wall |
| Sulfacetamide | 64 | DMSO | Cell wall |
| Sulfamethizole | 1,024 | DMSO | Rejected |
| Trimethoprim | 1 | DMSO | Rejected |
| Membrane-active compounds and ionophoresc | |||
| Gramicidin A | 0.03 | DMSO | Membrane |
| Monensin | 0.125 | DMSO | Membrane |
| Nigericin | 0.008 | DMSO | Rejected |
| Nitrofurantoin | 2 | DMSO | Membrane |
| Polymyxin B | 64 | Water | Membrane |
| Triton X-114 | 64 | Water | Membrane |
| Protein biosynthesis | |||
| Chloramphenicol | 4 | DMSO | Protein |
| Clarithromycin | 0.25 | DMSO | Membrane |
| Clindamycin | 2 | Water | Protein |
| Erythromycin | 4 | DMSO | Protein |
| Fusidic acid | 0.06 | Water | Protein |
| Neomycin | 1 | Water | Protein |
| Puromycin | 64 | Water | Protein |
| Spectinomycin | 128 | Water | Protein |
| Tetracyclin | 0.5 | DMSO | Protein |
| Test compounds | |||
| Actinonin | 32 | DMSO | Rejected |
| Azaserine | 2 | Water | Topo |
| Doxorubicin | 16 | Water | Topo |
| Ethidium bromide | 1 | Water | Rejected |
| HCP | 0.03 | DMSO | Rejected |
| Hydrogen peroxide | 4 | Water | Topo |
| Rifampicin | 0.008 | DMSO | Rejected |
All compounds with the exception of sulfamethizole were added to the cultures from a 1,000× stock solution. Sulfamethizole was added from a 250× stock solution. DMSO, dimethyl sulfoxide.
For the reference compounds, prediction is the assignment in the cross validation (see Materials and Methods). For the test compounds, prediction is the prediction obtained by using the classifier trained with the complete reference data set. Topo, topoisomerase.
Ionophores and detergents were combined in the class membrane-active compounds in order to discriminate compounds with unspecific activity from compounds that act on specific protein targets.
For each compound we determined the optimal concentration (copt) for expression profiling experiments. To do so, the optical density of a culture, grown as described above, was monitored for 5 h. The criteria applied to deduce copt are outlined in the Results section.
RNA preparation and labeling.
After cells were harvested, cell pellets were immediately resuspended in 450 μl of lysis buffer (1% [vol/vol] β-mercaptoethanol and 1 mM EDTA in RLT buffer [Qiagen, Hilden, Germany]). This suspension was transferred to a FastPrep tube (Qbiogene, Carlsbad, Calif.) prefilled with 400 μl of glass beads (diameter, 106 μm; acid washed; Sigma), 500 μl of citric acid-saturated phenol, and 200 μl of chloroform. The tube was processed in a FP120 FastPrep cell disrupter (Qbiogene) for 45 s at 6.5 m/s. The slurry was then transferred to a Phase-Lock tube (2 ml, heavy; Eppendorf, Hamburg, Germany) and spun for 10 min in a microcentrifuge. Ethanol (250 μl) was added to the supernatant, and the RNA was purified from this mixture on an RNeasy column (Qiagen), according to the instructions of the manufacturer. At this stage three replicates of each sample in equal amounts were pooled to give a total RNA amount of 100 μg. The prepurified and pooled RNAs were digested with RNase-free DNase I (Roche, Basel, Switzerland), and cleanup was performed with RNeasy columns (Qiagen). Quality control of each RNA sample included spectrophotometric analysis and formaldehyde gel electrophoresis. Furthermore, each RNA was subjected to reverse transcription-PCR. Amplification of the transcript in the presence of reverse transcriptase served as an indicator of the integrity of the RNA, while reverse transcription-PCR without the addition of reverse transcriptase was performed to verify the absence of any genomic DNA.
Labeling of RNA was performed as described previously (21), but 2 μg of total RNA and random hexamers were used. As an additional step, a chasing reaction was performed by adding 1 μl of labeling buffer containing 10 mM deoxynucleoside triphosphates and 50 U of reverse transcriptase for 15 min after the labeling reaction.
Expression profiling.
DNA fragments for open reading frames (ORFs) were computationally selected within the first third (proximal to the 5′ end) of the coding region. Each fragment was checked for its uniqueness; up to 80% similarity with any other sequence on the genome was the maximum allowed. No DNA stretch greater than 24 nucleotides was identical to any other such stretch (with the exception of ORFs shorter than 140 bp, which were used in their entirety). Two or three fragments were generated for ORFs longer than 3,000 bp.
Each fragment was prepared by annealing two 80-base oligonucleotides designed to overlap at their 3′ ends to form a 20-bp duplex. Rational and specific features of these arrays are described below (see Results). An extension reaction was then performed to yield a full-length product of 140 bp. Briefly, each pair of oligonucleotides was annealed in 20 mM Tris (pH 8.8)-10 mM KCl-10 mM (NH4)2SO4 and extended overnight at 65°C in the presence of 1.5 mM (each) deoxynucleoside triphosphate, 10 mM MgSO4, 5× Q solution (Qiagen), and 8 U of Bst DNA polymerase (New England Biolabs, Beverly, Mass.). Spotting, hybridization, scanning, data processing, and data storage were performed as described previously (21).
Data analysis.
Experimental data are publicly available at the website www.gpc-biotech.com/supplementary_material.htm. The logarithmic (base 10) ratios of the expression signals were calculated for each compound treatment on the basis of the signal for its corresponding control, i.e., that of the mock-treated sample. The log ratios for the three time points were arranged in one vector, called the feature vector, for each compound. Thus, the number of features (n = 12,615) corresponds to the number of DNA fragments spotted on the nylon membrane (n = 4,205) times the number of time points (n = 3).
The feature vectors of all compounds were hierarchically clustered by an agglomerative method (22). The similarities between feature vectors were measured by determination of the Euclidian distance. The similarities between clusters were measured by the complete linkage method, which is implemented in the Spotfire DecisionSite for Functional Genomics (Spotfire AB, Gothenburg, Sweden).
The MoA class was predicted with a support vector machine (SVM) (27, 33). A linear kernel was used. SVMs allow discrimination between two classes. Therefore, for each MoA class a single classifier that discriminates that class from all other classes had to be built. Each classifier returns the quasiprobability (vote) that the input belongs to the corresponding class. The outputs of all classifiers were combined to a final predictor, such that the predicted class is the class with the maximum vote. If the maximum vote was less than 0.8, the input was rejected; thus, no classification was made.
The classifiers were trained with the feature vectors for reference compounds. The quality of the predictor was tested by means of a “leave-one-out” strategy: (i) one of the compounds was left out of the training set, (ii) the predictor was trained with the remaining compounds, and (iii) the predictor was applied to the compound left out and the predicted class was compared to the known class for that compound. These steps were repeated for all compounds.
This procedure gives an unbiased estimation of the error rate of the final predictor. The SVM, the multiclass classifier, and the cross-validation procedure are implemented in the Matlab toolbox PRTools (The Mathworks GmbH, Aachen, Germany; Delft University of Technology, Delft, The Netherlands [http://www.ph.tn.tudelft.nl/prtools/]).
Spectrophotometric assay of FabI.
FabI activity was assayed spectrophotometrically by monitoring the decrease in absorption at 340 nm by using an adaptation of the spectrophotometric assay described previously (12). Standard reaction mixtures contained 4 mM crotonyl coenzyme A, 21 μg of homogeneous B. subtilis FabI (13), 100 μM NADH, and 0.1 M sodium phosphate (pH 7.5) in a final volume of 300 μl. The reactions were performed at 24°C in semimicro quartz cuvettes. The change in optical density was continuously monitored for 1 min, and the reaction rate was calculated from the slope of the trace. Triclosan and HCP were added to the final concentrations indicated in Fig. 3 from serially diluted stock solutions in dimethyl sulfoxide. The dimethyl sulfoxide concentration in all assays was maintained at 1.66%, which did not significantly affect the FabI activity. Under these experimental conditions, the specific activity of FabI in the absence of drugs was 0.385 nmol/min/μg. Each datum point represents the mean of duplicate assays, and individual values were within 7% of the average.
FIG. 3.
(A) Spectrophotometric assay of FabI showing the effects of triclosan (○) and HCP (•) on the initial rate of the FabI reaction; (B) formation of a high-affinity complex between FabI, [3H]NAD+, and either triclosan (○) or HCP (•).
Analysis of [3H]NAD+ binding to FabI.
Triclosan-dependent complex formation with FabI and NAD+ was measured as described previously (12) by using a polyvinylidene difluoride membrane to separate free [3H]NAD+ from FabI-bound [3H]NAD+.
RESULTS
MoA database of expression profiles.
In living cells, environmental changes eventually lead to specific changes in patterns of gene expression. A collection of genome-wide gene expression profiles generated following treatment with antibacterial compounds of known MoAs can therefore potentially serve as a MoA database for the generation of information on the MoAs of novel antibacterial compounds. In this prototype study we used 37 well-characterized antibacterial compounds covering several known MoA classes (Table 1). The three largest classes comprise compounds that inhibit protein biosynthesis, cell wall biosynthesis, and class II topoisomerases. Other classes include compounds that inhibit fatty acid biosynthesis and folic acid biosynthesis, as well as ionophores and detergents.
Production of gene expression data.
The approach outlined herein requires a robust experimental platform. We generated a genome array carrying all predicted ORFs of B. subtilis 168. Double-stranded 140-bp DNA fragments were generated by annealing and extending oligonucleotides with overlapping 3′ ends. All extension products were spotted on nylon membranes (8 by 12 cm) in duplicate. The quality of the arrays was tested by hybridization of B. subtilis genomic DNA (1, 26). A total of 99.6% of all ORFs showed a signal significantly (more than 3 standard deviations) above the background signal. Cell culturing, RNA preparation, labeling, and hybridization were standardized and largely automated. All steps were performed in triplicate, resulting in six datum points (duplicate spots on three membranes) for each gene that were used to calculate the expression levels and the respective standard errors (see Material and Methods).
Preanalyses revealed that the compound concentration is of crucial importance for data quality. The best results were obtained with subinhibitory concentrations, i.e., at concentrations that are just low enough not to affect the growth of the organism (data not shown). The copt of each compound could be deduced from the growth curves of the drug-treated cultures. copt is the highest concentration that fulfills three criteria: (i) the optical density of a culture after 1 h of compound treatment is no more than 15% less than the optical density of the control culture; (ii) the optical density at 600 nm reaches a minimum of 1.0 after 5 h; and (iii) during these 5 h the optical density increases steadily, i.e., does not decrease at any time point. Cultures were harvested for RNA preparation following 10, 40, and 80 min of drug treatment, since these three time points delivered the most meaningful results in preliminary experiments (data not shown).
MoA classification.
The simplest approach used to study the MoA of an antibacterial compound by means of gene expression profiling is to investigate the biology of the transcriptional responses elicited by the compound (7). However, this strategy is limited to cases in which conclusions about the MoA can be drawn directly from the annotation of deregulated genes.
We investigated two data analysis strategies for obtaining MoA information independent of the functional annotation of affected genes, namely, clustering and classification.
Clustering is an unsupervised method, in the sense that it does not require the assignment of the reference compounds to MoA classes before analysis. We used an agglomerative clustering method to build a hierarchical tree of the compounds under investigation (see the supplemental materialat www.gpc-biotech.com/supplementary_material.htm). One would expect that compounds with similar MoAs would show similar gene expression responses and would be located in the same part of the tree. This is indeed the case for most of the compounds, but not for all of them.
During clustering the similarity between two compounds is calculated globally; that is, the expression of all genes is taken into account. However, it is likely that only a small set of these genes is regulated as a consequence of the primary compound-target interaction (the primary MoA). The majority of the regulated genes may represent unselective, secondary effects. For this reason we did not consider unsupervised clustering as a satisfactory stand-alone method for assignment of a MoA to novel compounds.
We next tested the classification strategy, an analysis strategy that applies a priori knowledge of the MoA classes of the reference compounds. This approach consists of the following steps: (i) MoA classes are defined on the basis of the known MoAs of the reference compounds (Table 1), (ii) a predictor based on gene expression data is built for each MoA class, (iii) the predictor is validated, and (iv) the expression data generated with compounds of unknown MoAs are used to assign a MoA from the predefined classes. If the MoA of a novel compound is not represented within the database, no MoA is assigned (i.e., the compound is rejected).
Among the many classification methods described in the literature (3, 8, 10), we used SVMs. SVMs were first introduced by Vapnik et al. (32) and are also described elsewhere (27). They are particularly suited for the type of classification problem presented here. SVMs have been used in the past for gene expression-based classification of tissues (3, 6, 32) and genes (4).
By using SVM, a predictor for MoA classification was calculated according to the MoA classes defined in Table 1 (see Material and Methods). The quality of the predictor was tested by means of a leave-one-out strategy, in which each compound was removed from the data set one by one and treated as a compound of unknown MoA (see Materials and Methods and Table 1 for details). The best success rates for correct MoA classification were achieved for the MoA classes cell wall biosynthesis, topoisomerase, membrane activity and ionophores, and protein biosynthesis. Nine of the 10 inhibitors of cell wall biosynthesis were classified correctly. Amoxicillin was misclassified as an inhibitor of folic acid biosynthesis. Similarly, there was only one misclassification, that for clarithromycin, for the nine compounds that inhibit protein biosynthesis. Of the compounds that inhibit type II topoisomerase, only coumermycin A1 was misclassified. Interestingly, MoA classification was not successful for the MoA classes fatty acid biosynthesis and folate biosynthesis. It is very likely that these MoA classes were underrepresented in our compound list and that a minimum of five to six compounds per MoA class is required to generate a robust MoA predictor.
In order to obtain more information about the quality of the predictor, we investigated a few test compounds that are directly or indirectly related to the MoAs of the reference compounds (Table 1).
Actinonin inhibits the deformylation of N-formylmethionine of newly synthesized peptides (9). Interestingly, actinonin was not classified as a protein biosynthesis inhibitor but was rejected (i.e., it was not assigned to any of the MoA reference classes). This indicates that a compound that inhibits a process closely associated with protein biosynthesis can be differentiated from agents that act on the central process of protein biosynthesis (i.e., on the ribosome).
Most interestingly, three compounds known to cause DNA damage, namely, azaserine, doxorubicin, and hydrogen peroxide, were classified as topoisomerase inhibitors.
Doxorubicin and hydrogen peroxide both produce reactive oxygen species (17, 25). For azaserine, two MoAs are described in the literature: (i) it triggers the onset of DNA repair through its action as a carboxymethylating agent (20), and (ii) it inhibits purine biosynthesis through its action as a glutamine analogue (18). Four quinolones and two coumarin antibiotics were used as part of this study. These two chemical classes act on the same target (type II topoisomerase); however, they act with different mechanisms. Quinolones interrupt the cleavage and resealing cycle during the type II topoisomerase-catalyzed introduction of negative supercoils into DNA, thereby causing double-stranded breaks (7, 14). Coumarins bind to the ATP binding site of the enzyme and decrease the affinity of type II topoisomerase for this nucleotide, leaving the DNA largely intact (7, 34).
We next analyzed the MoA class of the topoisomerase inhibitors by hierarchical clustering. Figure 1A shows that the two MoA subgroups described above (coumarins and quinolones) can be clearly distinguished by this approach. The results obtained by inclusion of the test compounds hydrogen peroxide, doxorubicin, and azaserine in this analysis indicate that these compounds are part of the quinolone cluster. Interestingly, the two radical-forming agents doxorubicin and hydrogen peroxide separate from azaserine in the clustering (Fig. 1B).
FIG. 1.
(A) Hierarchical clustering of all topoisomerase inhibitors subjected to expression profiling in this study; (B) hierarchical clustering, as in panel A, with the additional compounds hydrogen peroxide, doxorubicin, and azaserine.
In summary, these findings allowed us to conclude that (i) the two mechanisms of inhibition exerted by coumarins and quinolones on a single target (type II topoisomerase) can be clearly distinguished and (ii) the gene expression signatures generated following treatment with DNA-damaging agents of the types azaserine, doxorubicin, and hydrogen peroxide are closely related to the ones induced by quinolones. This finding must be taken into account when compounds of unknown MoA are classified with this predictor. Interestingly, azaserine also elicits in bacterial reporter strains a strong signal indicative of inhibition of class II topoisomerases (16). It should be noted, however, that ethidium bromide (a DNA-intercalating agent) was rejected (i.e., it was not classified in any of the MoA classes represented in the predictor). This finding indicates that certain types of DNA stress are not classified as topoisomerase inhibition.
HCP.
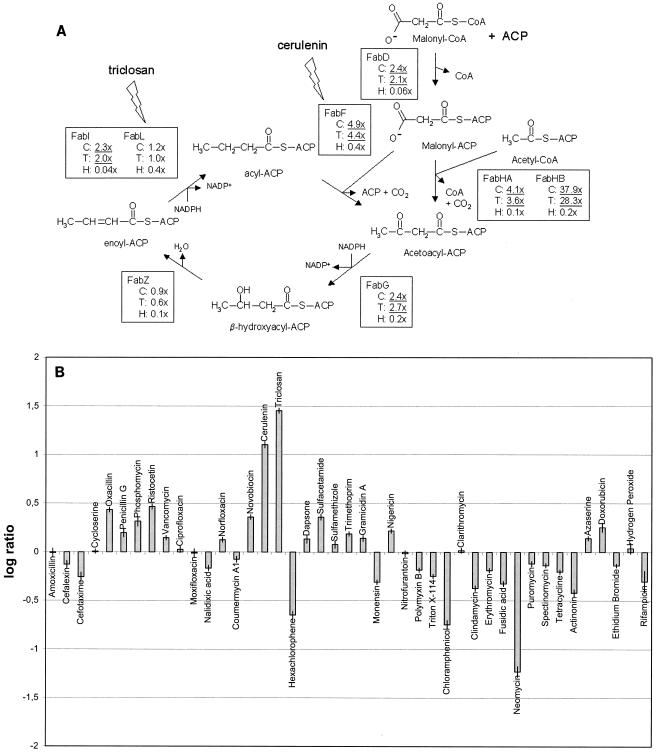
In the course of our analyses of the data, we discovered that one of the compounds, HCP, did not behave as expected. Initially, HCP was included as a member of the class of fatty acid biosynthesis inhibitors due to its known in vitro effect on FabI (11) (Fig. 2A). We did not obtain conclusive data from the classification using SVMs (Table 1), probably because the MoA group contained too few compounds to calculate a robust predictor. However, two additional observations furthered our interest in HCP: (i) in all clustering analyses that we performed, HCP was clearly separated from triclosan and cerulenin (two other well-characterized inhibitors of the fatty acid biosynthesis pathway [see the supplementalmaterial at www.gpc-biotech.com/supplementary_material.htm]), and (ii) triclosan and cerulenin both induced the expression of genes encoding enzymes of their target pathway (fatty acid biosynthesis; Fig. 2A). In particular, the fabHB (fabH2) gene, which encodes β-keto-acyl-ACP synthases, was induced on the order of 30-fold, a response that proved to be very selective for fatty acid biosynthesis inhibitors (Fig. 2B). None of these responses were observed following treatment with HCP (Fig. 2), indicating that in vivo HCP does not act via fatty acid biosynthesis inhibition.
FIG. 2.
(A) Deregulation of fatty acid biosynthesis genes after 80 min of treatment with three antibacterial compounds. The level of deregulation is indicated as the fold change in comparison to that for the respective controls. C, cerulenin; T, triclosan; H, HCP; CoA, coenzyme A. Underlined numbers indicate the levels of deregulation that were statistically significant. The target enzymes of triclosan and cerulenin are indicated. (B) Deregulation of the fabHB gene after 80 min of treatment. Changes in the expression levels are shown as log ratios in comparison to the levels for the respective controls.
It has been reported elsewhere that effective in vivo inhibitors of FabI enzymes require the formation of a stable ternary complex with the enzyme and its cofactor, NAD+ (12, 13). Furthermore, FabI inhibition by HCP has not been characterized for the B. subtilis enzyme. Thus, we investigated the inhibition of purified FabI from B. subtilis by triclosan and HCP biochemically. Figure 3A shows that triclosan and HCP both inhibit B. subtilis FabI in vitro. However, it is evident that, in contrast to triclosan, HCP does not form a stable ternary complex with FabI and NAD+ (Fig. 3B).
In summary, these data lead us to conclude that HCP does not exert its growth- inhibitory effect on B. subtilis cells via inhibition of fatty acid biosynthesis but, rather, does so by an unknown mechanism.
DISCUSSION
The aim of this study was to generate a prototype reference database for assignment of antibacterial compounds to MoAs. For this purpose we generated and analyzed the gene expression profiles for B. subtilis cells following treatment with 37 antibacterial compounds with known MoAs (the reference compounds). We tested this data set for its potential to predict the MoAs of antibacterial compounds and to generate other information relevant to antibacterial drug discovery.
Following data production we investigated several statistical data analysis approaches in order to assess the power of the data set to predict MoAs. The most promising results were obtained by using the MoA classifications obtained with an SVM. SVM is distinguished from clustering, in that SVM applies a priori knowledge of the MoA classes of the reference compounds. In a test experiment with the reference compounds (the leave-one-out validation), the success rate of MoA classification was best for the MoA classes cell wall biosynthesis, DNA topology, membrane activity and ionophores, and protein biosynthesis. These MoA groups represent the largest groups in the data set. In particular, the high rate of correct classification for cell wall biosynthesis inhibitors should be noted. As has been reported previously, it is very difficult to identify discriminative responses for this pathway by conventional methods (2, 30). The SVM-based MoA predictor presented herein has a high likelihood of correctly assigning an unknown inhibitor of this pathway. We have no direct explanation for the reason why one compound in each of the classes cell wall biosynthesis, DNA topology, and protein biosynthesis was misclassified. This finding may simply reflect the limitations of the present database, and precision will improve with increasing numbers of compounds per class. This is in agreement with our observation that the MoA classifications were not successful for the MoA classes in the data set with small numbers of compounds (fatty acid biosynthesis and folate biosynthesis). Thus, it is likely that the resolution of the SVM approach is limited by the number of compounds in each class with a defined MoA. For example, the MoA class cell wall biosynthesis addresses the entire pathway, whereas the MoA class DNA topology represents the inhibition of one enzyme (type II topoisomerase). The resolution of the SVM classification of any pathway could be increased by analysis of a larger number of compounds and by definition of subclasses.
We did not analyze compounds with unknown MoAs. However, we investigated several test compounds for which the MoAs, directly or indirectly, relate to the MoAs represented by the reference compounds. This analysis revealed clear strengths of the SVM-based classification approach, but it also revealed some limitations. Actinonin, an antibacterial with a MoA closely associated with protein biosynthesis (deformylation inhibitor), could clearly be distinguished from protein biosynthesis inhibitors acting directly on the ribosome. However, three test compounds known to cause DNA damage (stress), namely, azaserine, doxorubicin, and hydrogen peroxide, were classified as topoisomerase inhibitors (Table 1). Clustering analysis of these compounds together with the reference compounds for the MoA class DNA topology resulted in the grouping of the compounds with the quinolones. This finding indicates that the corresponding MoA predictor was not absolutely selective for quinolones but indicates DNA stress in a broader sense. This finding must be taken into account when compounds of unknown MoAs are analyzed. The notable fact that the intercalating agent ethidium bromide was not classified as DNA topology shows that the predictor does not assign all types of DNA stress to this class.
HCP was included as a reference compound in the study, based on reports (11) that this compound inhibits an enzyme of the fatty acid biosynthesis pathway in vitro (Fig. 2A). The MoA class fatty acid inhibition comprised only three compounds (triclosan, cerulenin, and HCP), which was too small for successful classification by use of SVM. However, the two known fatty acid biosynthesis inhibitors in the study (triclosan and cerulenin) yielded a response diagnostic for their MoAs by inducing expression of several genes encoding fatty acid biosynthesis enzymes (Fig. 2A). This effect has been demonstrated (28, 29), and a transcription factor, FapR, that controls the expression of these genes in response to these drugs has been identified (29). In this study it was evident that HCP did not elicit such a response. This finding, together with the biochemical data (Fig. 3), leads to the conclusion that HCP exerts its antibacterial activity by a mechanism other than fatty acid biosynthesis inhibition. The data described herein did not give us any hints about the unknown in vivo MoA of HCP. However, the knowledge that an anticipated MoA based on in vitro data is not confirmed in vivo is relevant to drug discovery and represents value in itself.
In summary, we have shown that the approach presented in this study produced information that has the potential to support priority decisions in an antibacterial drug discovery process: (i) a high rate of success in MoA assignment was achieved by the classification approach, (ii) two different MoAs for a single target could be distinguished (i.e., coumarins versus quinolones), and (iii) evidence that the in vivo MoA of HCP differs from the in vitro MoA was generated. As discussed above, we also observed several limitations. The major limitation is the number of compounds that is necessary for successful MoA classification. Our data indicate that a minimum of five to six compounds per class is required. It will therefore be difficult to identify a sufficient number of reference compounds for certain MoA classes. Nevertheless, in the light of the positive aspects outlined above, we believe that the approach described herein may well be used to prioritize compound candidates in drug discovery settings.
Acknowledgments
We thank Tanja Trosch, Christina Fischer, and the members of the gene expression profiling team for excellent technical assistance and Ruairi Friel and Dana Torrey for helpful comments on the manuscript.
The research in the laboratory of C. O. Rock was supported by National Institutes of Health grant GM 34496, Cancer Center (CORE) support grant CA 21765, and the American Lebanese Syrian Associated Charities.
REFERENCES
- 1.Amon, P., and I. Ivanov. 2003. Genomic DNA labeling for hybridization with DNA arrays. BioTechniques 34:700-702. [DOI] [PubMed] [Google Scholar]
- 2.Bandow, J. E., H. Brotz, L. I. Leichert, H. Labischinski, and M. Hecker. 2003. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 47:948-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Dor, A., L. Bruhn, N. Friedman, I. Nachman, M. Schummer, and Z. Yakhini. 2000. Tissue classification with gene expression profiles. J. Comput. Biol. 7:559-583. [DOI] [PubMed] [Google Scholar]
- 4.Brown, M. P., W. N. Grundy, D. Lin, N. Cristianini, C. W. Sugnet, T. S. Furey, M. Ares, Jr., and D. Haussler. 2000. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc. Natl. Acad. Sci. USA 97:262-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Finkelstein, D., R. Ewing, J. Gollub, F. Sterky, J. M. Cherry, and S. Somerville. 2002. Microarray data quality analysis: lessons from the AFGC project. Plant Mol. Biol. 48:119-131. [DOI] [PubMed] [Google Scholar]
- 6.Furey, T. S., N. Cristianini, N. Duffy, D. W. Bednarski, M. Schummer, and D. Haussler. 2000. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics 16:906-914. [DOI] [PubMed] [Google Scholar]
- 7.Gmuender, H., K. Kuratli, K. Di Padova, C. P. Gray, W. Keck, and S. Evers. 2001. Gene expression changes triggered by exposure of Haemophilus influenzae to novobiocin or ciprofloxacin: combined transcription and translation analysis. Genome Res. 11:28-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golub, T. R., D. K. Slonim, P. Tamayo, C. Huard, M. Gaasenbeek, J. P. Mesirov, H. Coller, M. L. Loh, J. R. Downing, M. A. Caligiuri, C. D. Bloomfield, and E. S. Lander. 1999. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531-537. [DOI] [PubMed] [Google Scholar]
- 9.Haas, M., D. Beyer, R. Gahlmann, and C. Freiberg. 2001. YkrB is the main peptide deformylase in Bacillus subtilis, a eubacterium containing two functional peptide deformylases. Microbiology 147:1783-1791. [DOI] [PubMed] [Google Scholar]
- 10.Hastie, T., R. Tibshirani, and J. H. Friedman. 2001. The elements of statistical learning. Springer-Verlag, Heidelberg, Germany.
- 11.Heath, R. J., J. Li, G. E. Roland, and C. O. Rock. 2000. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J. Biol. Chem. 275:4654-4659. [DOI] [PubMed] [Google Scholar]
- 12.Heath, R. J., J. R. Rubin, D. R. Holland, E. Zhang, M. E. Snow, and C. O. Rock. 1999. Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J. Biol. Chem. 274:11110-11114. [DOI] [PubMed] [Google Scholar]
- 13.Heath, R. J., N. Su, C. K. Murphy, and C. O. Rock. 2000. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J. Biol. Chem. 275:40128-40133. [DOI] [PubMed] [Google Scholar]
- 14.Hooper, D. C. 1999. Mode of action of fluoroquinolones. Drugs 58(Suppl. 2):6-10. [DOI] [PubMed] [Google Scholar]
- 15.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 16.Hutter, B., C. Fischer, A. Jacobi, C. Schaab, and H. Loferer. 2004. Panel of Bacillus subtilis reporter strains indicative for various modes of action. Antimicrob. Agents Chemother. 48:2588-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostoryz, E. L., and D. M. Yourtee. 2001. Oxidative mutagenesis of doxorubicin-Fe(III) complex. Mutat. Res. 490:131-139. [DOI] [PubMed] [Google Scholar]
- 18.Kubitschek, H. E., and R. J. Sepanski. 1982. Azaserine: survival and mutation in Escherichia coli. Mutat. Res. 94:31-38. [DOI] [PubMed] [Google Scholar]
- 19.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 20.Lawson, T. 1989. Nicotinamide and selenium stimulate the repair of DNA damage produced by N-nitrosobis (2-oxopropyl) amine. Anticancer Res. 9:483-486. [PubMed] [Google Scholar]
- 21.Machl, A. W., C. Schaab, and I. Ivanov. 2002. Improving DNA array data quality by minimising ′neighbourhood' effects. Nucleic Acids Res. 30:e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirkin, B. 1996. Mathematical classification and clustering. In P. Pardalos and R. Horst (ed.), Nonconvex optimization and its applications, vol. 11. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 23.Novak, J. P., R. Sladek, and T. J. Hudson. 2002. Characterization of variability in large-scale gene expression data: implications for study design. Genomics 79:104-113. [DOI] [PubMed] [Google Scholar]
- 24.Planet, P. J., R. DeSalle, M. Siddall, T. Bael, I. N. Sarkar, and S. E. Stanley. 2001. Systematic analysis of DNA microarray data: ordering and interpreting patterns of gene expression. Genome Res. 11:1149-1155. [DOI] [PubMed] [Google Scholar]
- 25.Potter, A. J., K. A. Gollahon, B. J. Palanca, M. J. Harbert, Y. M. Choi, A. H. Moskovitz, J. D. Potter, and P. S. Rabinovitch. 2002. Flow cytometric analysis of the cell cycle phase specificity of DNA damage induced by radiation, hydrogen peroxide and doxorubicin. Carcinogenesis 23:389-401. [DOI] [PubMed] [Google Scholar]
- 26.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schölkopf, B., and A. J. Smola. 2002. Learning with kernels: support vector machines, regularization, optimization, and beyond. The MIT Press, Cambridge, Mass.
- 28.Schujman, G. E., K. H. Choi, S. Altabe, C. O. Rock, and D. de Mendoza. 2001. Response of Bacillus subtilis to cerulenin and acquisition of resistance. J. Bacteriol. 183:3032-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schujman, G. E., L. Paoletti, A. D. Grossman, and D. de Mendoza. 2003. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev. Cell 4:663-672. [DOI] [PubMed] [Google Scholar]
- 30.Shaw, K. J., N. Miller, X. Liu, D. Lerner, J. Wan, A. Bittner, and B. J. Morrow. 2003. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J. Mol. Microbiol. Biotechnol. 5:105-122. [DOI] [PubMed] [Google Scholar]
- 31.Stülke, J., R. Hanschke, and M. Hecker. 1993. Temporal activation of β-glucanase synthesis in Bacillus subtilis is mediated by the GTP pool. J. Gen. Microbiol. 139:2041-2045. [DOI] [PubMed] [Google Scholar]
- 32.Valentini, G. 2002. Gene expression data analysis of human lymphoma using support vector machines and output coding ensembles. Artif. Intell. Med. 26:281-304. [DOI] [PubMed] [Google Scholar]
- 33.Vapnik, V. N. 1999. The nature of statistical learning theory. Springer-Verlag, New York, N.Y.
- 34.Wigley, D. B. 1995. Structure and mechanism of DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 24:185-208. [DOI] [PubMed] [Google Scholar]