Abstract
The possible association between triclosan and bacterial susceptibility to antibiotic was examined among staphylococci and several species of gram-negative bacteria (GNB) isolated from the hands of individuals in a community setting. Hand cultures from individuals randomized to using either antibacterial cleaning and hygiene products (including a hand soap containing 0.2% triclosan) or nonantibacterial cleaning and hygiene products for a 1-year period were taken at baseline and at the end of the year. Although there was no statistically significant association between triclosan MICs and susceptibility to antibiotic, there was an increasing trend in the association the odds ratios (ORs) for all species were compared at baseline (OR = 0.65, 95% confidence interval [95%CI] = 0.33 to 1.27) versus at the end of the year (OR = 1.08, 95%CI = 0.62 to 1.97) and for GNB alone at baseline(OR = 0.66, 95%CI = 0.29 to 1.51) versus the end of year (OR = 2.69, 95%CI = 0.78 to 9.23) regardless of the hand-washing product used. Moreover, triclosan MICs were higher in some of the species compared to earlier reports on household, clinical, and industrial isolates, and some of these isolates had triclosan MICs in the range of concentrations used in consumer products. The absence of a statistically significant association between elevated triclosan MICs and reduced antibiotic susceptibility may indicate that such a correlation does not exist or that it is relatively small among the isolates that were studied. Still, a relationship may emerge after longer-term or higher-dose exposure of bacteria to triclosan in the community setting.
The emergence and spread of microorganisms with reduced susceptibility to antimicrobial agents is a major public health problem (24). Triclosan (2,4,4′-trichloro-2′-hydroxydiphenylether) is a bisphenol antimicrobial agent commonly incorporated into consumer products for skin care and oral hygiene and into plastic kitchen products and toys (7, 25). An increase in the use of triclosan-containing products within the community setting has prompted urging for further research into a possible association between reduced susceptibility to triclosan and to antibiotics in the community setting (13). Under laboratory conditions, bacteria selected for reduced susceptibility to triclosan can confer cross-resistance to antibiotics (8-10, 32, 35, 38). However, a clear association between elevated triclosan MICs and antibiotic resistance outside the laboratory environment has not been demonstrated (5, 8, 9, 12, 39, 40). The purposes of the present study were to provide community-based data on triclosan MICs among a large and diverse sample of bacterial flora isolated from hands and to determine whether there was an association between triclosan MICs and antibiotic susceptibility.
MATERIALS AND METHODS
Study design.
The data were derived from a one-year double-blinded and randomized home hygiene intervention trial conducted in upper Manhattan (19, 20). There were 238 households recruited at baseline, and 224 households remained at the end of the year. Half of the households were randomly assigned to use nonantibacterial products, including liquid soap for hand washing, and the other half used a similar-appearing liquid soap containing 0.2% triclosan. All products were readily available over the counter but were packaged in a blinded fashion for the present study. A detailed description of the randomization process and type of cleaning and hygiene products allocated has been discussed previously (20). The products were provided free of cost and were delivered to each household monthly. During the study period, the households were required to use only the assigned home hygiene products and were asked not to change any of their other normal hygiene practices. If a household was found to have or reported use of unassigned cleaning and hygiene products corresponding to the types of products allocated for the study period, they were dropped from the study. To monitor compliance, the products were weighed during each monthly household visit. In order to control for product use before study initiation, the data collector conducted a survey of the antibacterial cleaning and hygiene products used within the home at baseline.
Sample collection.
A hand culture from the primary caregiver within the home was obtained at baseline and 1 year later. The primary caregiver was defined as the family member that spent the most time in the household and provided the majority of the caregiving for the family within the home. Samples were taken after participants washed, rinsed, and dried both hands in their usual manner with the assigned liquid hand-washing product. A trained data collector randomly chose the hand to be cultured by using a coin flip.
After insertion of the hand into a sterile polyethylene bag containing 50 ml of culture medium (0.075 M phosphate buffer [pH 7.9] containing 0.1% polysorbate 80), the collector massaged the hand through the wall of the bag for 1 min. Post-hand-wash cultures were utilized since these cultures reflect any immediate influence attributed to use of either antibacterial or nonantibacterial hand-washing products on changes in bacterial species and better represent resident flora found on the hands (21, 29). The interviewers timed the subjects in seconds for the duration of their hand wash with the assigned product. In addition, the reported number of hand washes per day for each primary caregiver was recorded.
Bacterial strains and culture conditions.
The microbiological analysis, isolate identification, and antibiotic susceptibility testing were conducted at the Clinical Microbiology Laboratory at New York Presbyterian Hospital, Columbia University Medical Center, New York, N.Y. From the hand sampling solution, 10-fold dilutions (to 10−3) were prepared and spread onto the following plated media (Becton Dickinson Microbiology Systems, Sparks, Md.): 5% sheep blood agar to enumerate total bacteria by CFU, Columbia colistin-nalidixic acid agar for selective isolation of gram-positive cocci, MacConkey agar for selective isolation of gram-negative bacilli (GNB), and mannitol salt agar to select S. aureus. All plates were incubated at 35°C and observed for bacterial growth over a 48-h period. One representative colony of each phenotype was picked from each plate. GNB were speciated by using API 20NE and API 20E assays (bioMérieux, Hazelwood, Mo.) and staphylococci were identified by tube coagulase and Staphaurex (Murex Biotech Limited, Norcross, Ga.). Enterococci and micrococci were identified by using the MicroScan WalkAway 96 SI (Dade Behring, Deerfield, Ill.).
Although there was a variety of bacterial species found on the hands of the subjects (3, 22), only the most commonly isolated bacterial species (i.e., ≥38 isolates of a single species from combined baseline and end-of-year samples) were selected for analyses in the present study. The most prevalent GNB included Acinetobacter baumannii, Acinetobacter lwoffii, Enterobacter agglomerans, Enterobacter cloacae, Klebsiella pneumoniae, and Pseudomonas fluorescens-P. putida. The predominant staphylococcal species included Staphylococcus aureus, Staphylococcus warneri, Staphylococcus epidermidis, and Staphylococcus capitis.
Antibiotic and triclosan susceptibility testing.
All bacterial isolates were tested against a panel of antibiotics by using MicroScan WalkAway 96 SI. The standards recommended by the National Committee for Clinical Laboratory Standards (NCCLS) were used to classify the bacteria as resistant, intermediate, and susceptible to a particular antimicrobial agent (34). The specific antibiotics chosen for susceptibility testing against each GNB were based on the following criteria: (i) consistency with earlier research studies regarding a link between triclosan and antibiotic resistance and/or (ii) the clinical applicability of the antibiotics for a given species. Antibiotics were excluded for organisms harboring a corresponding intrinsic resistance. Accordingly, all GNB were tested against gentamicin, imipenem, and ciprofloxacin. Additional antibiotics tested included (i) amikacin and ticarcillin-clavulanate for A. baumannii and A. lwoffii, (ii) trimethoprim-sulfamethoxazole for E. agglomerans and E. cloacae, (iii) trimethoprim-sulfamethoxazole, piperacillin-tazobactam, and ceftriaxone for K. pneumoniae, and (iv) piperacillin-tazobactam and ceftazidime for P. fluorescens-P. putida. The staphylococcal species were tested against oxacillin to ascertain methicillin resistance.
Triclosan MICs were assessed at the Center for Adaptation Genetics and Drug Resistance at Tufts University School of Medicine, Boston, Mass., by using a modified NCCLS agar dilution method (34). Plates containing Mueller Hinton agar (Difco, Sparks, Md.) were prepared by using twofold increasing concentrations of triclosan (range, 0.0312 to 32 μg/ml) with a 5-mg/ml stock of triclosan in ethanol. By using a multipoint inoculator (Boekel, Inc., Feasterville, Pa.), ∼104 CFU of each logarithmically grown isolate was applied, and the inoculated plates were incubated aerobically for 24 h at 35°C. The lowest triclosan dilution that showed no visible growth indicated the MIC. The following strains were included as controls (with minimum and maximum triclosan MICs): wild-type Escherichia coli AG100 (0.0312 and 0.5 μg/ml [mode = 0.5]); AG100A, an acrAB deletion mutant (0.0312 and 0.125 μg/ml [mode = 0.0312 μg/ml]); and AGT11, containing a fabI mutation (16.0 and ≥ 32.0 μg/ml [mode ≥ 32.0 μg/ml]) (31). A randomly chosen subset of organisms for which the triclosan MICs were ≥32.0 μg/ml was retested as described above with agar containing a triclosan concentration range of 64.0 to 1,024.0 μg/ml by the methodology of Chuanchuen et al. (8) to examine whether the triclosan MICs approached the concentration used in the hand-washing soap. At these higher concentrations, drug precipitation (opacity) was observed.
Analytical considerations and methods.
For antibiotic susceptibility assays, strains were categorized as either susceptible or resistant (i.e., a combination of those designated either resistant or intermediate). S. aureus and coagulase-negative staphylococci (CNS) were categorized as susceptible or resistant to oxacillin. For the GNB, we categorized the susceptibility profile as resistant if the organism was either resistant or intermediate to ≥1 antibiotic. The proportion of isolates that were antibiotic resistant at baseline versus the end of the year were compared for each bacterial species by using chi-square and Fisher exact tests with SPSS v.10 (SPSS, Inc., Chicago, Ill.).
Given the sparse data regarding triclosan susceptibility testing and the lack of standardized breakpoints, we first examined the distribution of the triclosan MICs for each species and then calculated the 25th, 50th (median), and 75th percentiles. The proportions of isolates that exhibited a triclosan MIC above the median at baseline versus at the end of the year were compared for each bacterial species by using chi-square and Fisher exact tests with SPSS v.10.
Next, using the 75th- and 25th-percentile MICs as a cutoff, the organisms were classified as being associated with a high or low triclosan MIC (i.e., a high MIC ≥ 75% or a low MIC ≤ 25%). All species were combined to assess whether there was a relationship between high triclosan MICs and antibiotic resistance. Since the median and 25th- and 75th-percentile MICs for the S. aureus isolates at baseline were the same (i.e., the antibiotic MICs for almost all 30 isolates were 2.0 μg/ml), the low susceptibility values were designated as ≤1 μg/ml, reflecting a similar cutoff adopted in earlier literature (5, 40). In addition, there were only five isolates of S. capitis at baseline, and the MICs for all five were the same and so these strains were therefore not included in any further baseline analyses. The analyses were also conducted on the GNB and staphylococci as two separate groups in order to examine whether there were differences by genus and organism type.
Earlier analyses showed that no differences occurred in the total counts or types of organisms when subjects randomized to antibacterial versus nonantibacterial products were compared (19), but significant differences were observed in the types of organisms (GNB and staphylococci) isolated at baseline compared to those isolated at the end of the year, regardless of the randomization status (3). Therefore, all regression analyses were conducted separately at baseline and at the end of the year to accommodate the potential influence on triclosan MICs and antibiotic resistance resulting from species or strain changes over time. Logistic regression models with generalized estimating equations (GEE) were assessed by using STATA SE V.8 (STATA, College Station, Tex.) to adjust for intracluster correlation of organisms isolated within individual hand samples. The odds ratios (OR) or adjusted OR (aOR) and 95% confidence intervals (95%CI) were calculated from the GEE model regression estimates.
Antibiotic resistance was the outcome variable for all models. The main effect of interest was high triclosan MICs. Several other covariates were tested in the model to control for possible confounding: (i) assigned antibacterial product use (i.e., 0.2% triclosan versus plain soap), (ii) antibacterial product use prior to recruitment into the study, (iii) type of organism (GNB versus staphylococci), (iv) observed length of hand wash in seconds, and (v) reported number of hand washes per day. These covariates were selected for inclusion in the final model based on a change in the effect estimate (beta coefficient) of ≥10% when the model with high triclosan MICs alone was compared to the model including the covariate.
Sample size calculations.
Sample size calculations were conducted by using NCSS and PASS (NCSS, Kaysville, Utah). Since the total number of isolates was predetermined, we calculated the lowest detectable effect estimate for examining the association between elevated triclosan MICs and antibiotic resistance. For this calculation, we used the total number of test isolates, the proportion of antibiotic-resistant isolates with a low triclosan MIC (p1), a power of 80%, and a two-sided alpha level of 0.05 at baseline and at end of the year. Thus, at baseline with n = 220 and p1 = 0.26, we could detect an OR of ≥2.12. Similarly, at the end of the year (n = 290, p1 = 0.25), we could detect an OR of ≥2.12. The lowest detectable effect estimates for the GNB was an OR of ≥2.48 at baseline (n = 150, p1 = 0.26) and an OR of ≥4.87 at the end of the year (n = 93, p1 = 0.11). The lowest detectable effect estimate for the CNS was an OR of ≥3.91 at baseline (n = 70, p1 = 0.28) and an OR of ≥2.27 at the end of the year (n = 197, p1 = 0.30).
RESULTS
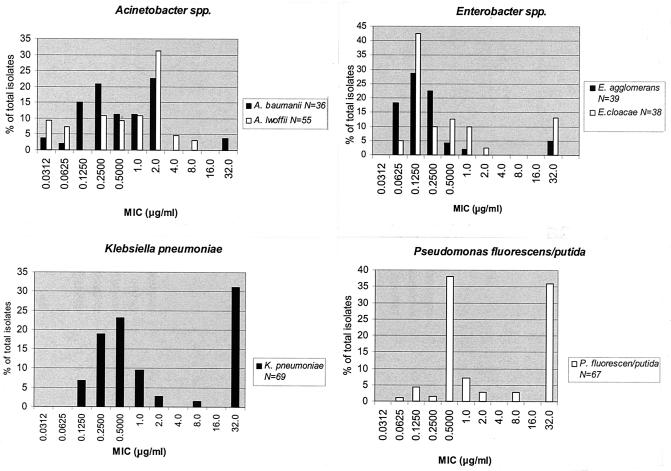
A total of 628 isolates were examined for their triclosan MICs and susceptibilities to selected antibiotics (Table 1). The organisms with the highest median triclosan MICs were K. pneumoniae at baseline and P. fluorescens-P. putida at the end of the year. Whereas the triclosan median MICs for the gram-negative species varied widely, the staphylococcus median values were very similar, except for that of S. aureus, which was 2 μg/ml at baseline and 0.03 μg/ml at the end of the year (Table 1). The proportions of isolates above the median triclosan MIC were compared for each species (Table 1). There was a significantly higher proportion of such isolates among the A. lwoffii isolates at the end of the year (39.4% [13 of 33]) compared to at baseline (0% [0 of 22]) (P < 0.001). For K. pneumoniae and S. aureus, the trend was reversed: K. pneumoniae, 50% (20 of 40) at baseline and 20.7% (6 of 29) at the end of year (P = 0.013) and, for S. aureus, 80% (24 of 30) at baseline and 0% (0 of 13) at the end of year (P < 0.001). There were no significant differences found for comparisons of triclosan MICs above and below the median value for any of the other species (all P > 0.05). For analyses of antibiotic resistance, there were no significant differences in the observed proportions when we compared baseline values to those at the end of the year (all P > 0.05), except for E. cloacae, which was significantly higher at baseline (36% [9 of 25]) than at the end of the year (0% [0 of 13]) (P = 0.016).
TABLE 1.
Triclosan MICs and antibiotic susceptibility
| Organism and sampling point | No. of isolatesa | Triclosan MIC (μg/ml) at percentile: |
Antibiotic resistance (%) | ||
|---|---|---|---|---|---|
| 25th | 50th | 75th | |||
| A. baumannii | |||||
| Baseline | 10 | 1.00 | 1.50 | 2.00 | 1.0 |
| 1 yr | 26 | 0.13 | 0.25 | 1.25 | 18.4 |
| A. lwoffiib | |||||
| Baseline | 22 | 0.50 | 2.00 | 2.00 | 9.1 |
| 1 yr | 33 | 0.06 | 1.00 | 2.00 | 15.2 |
| E. agglomerans | |||||
| Baseline | 21 | 0.13 | 0.13 | 0.25 | 0 |
| 1 yr | 18 | 0.06 | 0.13 | 0.25 | 11.1 |
| E. cloacaec | |||||
| Baseline | 25 | 0.13 | 0.50 | 1.00 | 36 |
| 1 yr | 13 | 0.13 | 0.13 | 0.25 | 0 |
| K. pneumoniaeb | |||||
| Baseline | 40 | 0.50 | 16.50 | >32.00 | 15 |
| 1 yr | 29 | 0.25 | 0.50 | 1.00 | 6.9 |
| P. fluorescens-P. putida | |||||
| Baseline | 15 | 0.50 | 0.50 | >32.00 | 45.3 |
| 1 yr | 14 | 0.22 | 8.00 | >32.00 | 42.9 |
| S. aureusb | |||||
| Baseline | 30 | 2.00 | 2.00 | 2.00 | 6.7 |
| 1 yr | 13 | 0.03 | 0.03 | 0.27 | 7.7 |
| S. capitis | |||||
| Baseline | 5 | 0.06 | 0.06 | 0.06 | 40 |
| 1 yr | 36 | 0.06 | 0.06 | 2.00 | 11.1 |
| S. epidermidis | |||||
| Baseline | 20 | 0.03 | 0.09 | 2.00 | 30 |
| 1 yr | 53 | 0.03 | 0.06 | 0.25 | 34 |
| S. warneri | |||||
| Baseline | 28 | 0.06 | 0.06 | 0.25 | 39.3 |
| 1 yr | 127 | 0.06 | 0.06 | 0.25 | 33.1 |
There were totals of 171 GNB strains at baseline and 133 GNB strains at the end of the year and totals of 53 CNS strains at baseline and 216 strains at the end of the year.
There was a significantly higher proportion of A. lwoffii for which the triclosan MICs were greater than the median at the end of the year compared to the baseline (P = 0.001) as determined by Fisher exact test. There were significantly higher proportions of K. pneumoniae and S. aureus for which the triclosan MICs were greater than the median at baseline compared to the end of the year (P = 0.013 and P < 0.001, respectively) as determined by the chi-square or Fisher exact tests. There were no significant differences when we compared the proportion of triclosan MIC values greater than the median at baseline versus that at the end of the year for any of the other organisms (all P > 0.05).
There was a significantly higher proportion of antibiotic-resistant E. cloacae at baseline compared to at the end of the year (P = 0.016) as determined by using the Fisher exact test. There were no significant differences in antibiotic resistance for any of the other species when we compared baseline levels to those at the end of the year (all P > 0.05).
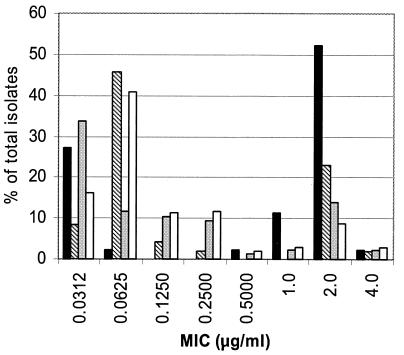
For each species, the proportion of isolates at each triclosan MIC is shown in Fig. 1 and 2. All GNB, except for A. lwoffii, had at least one isolate for which the triclosan MIC was ≥32.0 μg/ml (Fig. 1). The triclosan MICs were ≥2 and ≥32.0 μg/ml for 38% (26 of 69) and 31% (21 of 69), respectively, of the K. pneumoniae isolates. Approximately 36% (24 of 67) of the P. fluorescens-P. putida isolates were associated with triclosan MICs of ≥32.0 μg/ml. The triclosan MIC for 67% (29 of 43) of the S. aureus isolates was ≥1 μg/ml (Fig. 2). All other staphylococcus species had at least one isolate for which the triclosan MIC was ≥2 μg/ml.
FIG. 1.
Triclosan susceptibilities of representative species found on the hands of both antibacterial and nonantibacterial user groups combined at both baseline and the end of the year. Bars represent the percent of all isolates at the MIC indicated. N, number of isolates tested. The lowest and highest dilutions represent triclosan concentrations of ≤0.0312 and ≥32.0 μg/ml, respectively.
FIG. 2.
Triclosan susceptibilities (percentage of isolates at designated MIC) for S. aureus, S. capitis, S. epidermidis, and A. warneri found on the hands of both antibacterial and nonantibacterial user groups combined at both baseline and the end of the year. The lowest dilution represents an MIC of ≤0.0312 μg/ml. The triclosan MIC was >4 μg/ml for none of the species. Bars: ▪, S. aureus (n = 43); ▧, S. capitis (n = 42); ░⃞, S. epidermidis (n = 73); □, S. warneri (n = 155). n, number of isolates of each species.
A subset of GNB for which the MIC was ≥32.0 μg/ml (three K. pneumoniae, one E. cloacae, one A. baumannii, and two P. fluorescens) were examined by using an agar incorporation method that reportedly allows assessment at higher concentrations of triclosan (9). All isolates in the subset grew on agar containing triclosan at 1,024 μg/ml.
Categorizing the triclosan MIC distributions into the highest and lowest percentile groups (75th and 25th) yielded 220 isolates available at baseline and 290 isolates at the end of the year for analysis in the GEE logistic regression models (Table 2). In addition to the examination of the relationship between high triclosan MICs and antibiotic resistance, several cofactors were assessed in the models, including: (i) assigned antibacterial product use (i.e., 0.2% triclosan versus plain soap), (ii) antibacterial product use prior to recruitment into the study, (ii) type of organism (GNB versus staphylococci), (iv) observed length of hand wash in seconds, and (v) reported number of hand washes per day.
TABLE 2.
Association between high triclosan MICsa and antibiotic resistance: OR and aOR estimates
| Organism and sampling point (n) | Odds of antibiotic resistance among species for which triclosan MICs were high |
|||
|---|---|---|---|---|
| OR (95%CI) | P | aOR (95%CI) | P | |
| All species combined | ||||
| Baseline (220) | 0.65 (0.33-1.27) | 0.20 | - | - |
| End of yr (290)b | 0.92 (0.53-1.59) | 0.76 | 1.08 (0.62-1.97) | 0.73 |
| GNB only | ||||
| Baseline (150)c | 0.71 (0.32-1.55) | 0.39 | 0.66 (0.29-1.51) | 0.33 |
| End of yr (93)d | 2.31 (0.75-7.12) | 0.14 | 2.69 (0.78-9.23) | 0.12 |
| Staphylococci only | ||||
| Baseline (70)e | 0.64 (0.21-1.95) | 0.43 | 0.69 (0.20-2.42) | 0.56 |
| End of yr (197)c | 0.74 (0.38-1.45) | 0.38 | 0.70 (0.35-1.38) | 0.30 |
“High triclosan MICs” refers to organisms that were in the upper 75th percentile MIC distribution. For each model, cofactors that were assessed included assigned soap user group (antibacterial versus plain), use of antibacterial cleaning and hygiene products in the home before study initiation, observed length of hand wash, and reported number of hand washes per day. For the models examining the species combined, we also assessed the type of species (GNB or staphyloccoci) as a cofactor. These variables were only included in the final model if there was a >10% change in the regression coefficient compared to the model examining the triclosan level alone. n, number of isolates.
The aOR is controlling for the type of species (i.e., either GNB or staphylococci), the reported number of hand washes per day, and prior antibacterial cleaning and hygiene product use.
The aOR is controlling for the observed number of seconds hands were washed.
The aOR is controlling for the reported number of handwashes per day.
The aOR is controlling for the observed number of seconds hands were washed and assigned antibacterial soap use.
At baseline, there was no significant association between high triclosan MICs and antibiotic resistance among all species combined (resistant GNB and methicillin [oxacillin]-resistant staphylococci) (OR = 0.65, 95% CI = 0.33 to 1.27). This association did not change even after we controlled for antibacterial product use prior to the study, the use of assigned hand-washing soap containing triclosan, the type of organism tested (GNB versus staphylococci), or the hand-washing practices (see Table 2).
After 1 year of product use there was also no significant association between triclosan MICs and antibiotic resistance among all species combined (aOR = 1.08, 95% CI = 0.62 to 1.97). This estimate was adjusted for the type of species, the reported number of hand washes per day, and the reported number of hand washes prior to antibacterial product use, since there was a >10% change when each of these factors was added to the regression model. Next, we examined the relationship between high triclosan MICs and antibiotic resistance among GNB and staphylococci separately. Although the effect estimate was not significant, there was a 2.7-fold higher odds of observing antibiotic resistance among GNB species for which triclosan MICs were high than among those for which MICs were low (see Table 2). The odds of observing methicillin (oxacillin) resistance among all staphylococci combined (CNS and S. aureus), given high triclosan MICs at the end of the year, was similar to the baseline value (aOR = 0.70, 95% CI = 0.35 to 1.38) (Table 2). There were similar odds of observing methicillin resistance among only CNS species associated with high triclosan MICs versus those associated with low triclosan MICs at the end of the year (OR = 0.69, 95% CI = 0.34 to 1.38). Since there were few isolates of methicillin-resistant S. aureus (MRSA) and methicillin-susceptible S. aureus, we could not examine the association among these species separately.
DISCUSSION
We examined a community-based sample of bacterial isolates obtained from the hands of individuals (randomized to using or not using antibacterial cleaning and hygiene products) in order to assess the relationship between triclosan MICs and antibiotic susceptibility. This sample of isolates was among the largest and most diverse tested for triclosan MICs and corresponding antibiotic susceptibility within the community setting.
Prior studies that have reported data on the same species examined here utilized samples from either a home setting, culture type collections, clinical settings, or environmental sources (4, 5, 11, 23, 28, 39, 41). In a household setting, Cole et al. found that the triclosan MICs for three S. aureus isolates obtained from hands ranged from ≤0.004 to 0.064 μg/ml and for environmental isolates ranged from ≤0.004 to 0.128 μg/ml (11). In the present study, the MICs were considerably higher, ranging from 0.03 to 4.00 μg/ml, with 67% being >1 μg/ml. The median triclosan MIC for the CNS isolates studied here was 0.06 μg/ml, with a maximum MIC of 4 μg/ml, which is 10-fold higher than the maximum values previously reported. Clinical isolates of S. aureus (n = 32) have been reported for which the triclosan MICs were between 0.025 and 1 μg/ml (with 9.4% at 1.0 μg/ml) (40), between ≤0.12 and 32.0 μg/ml for 186 isolates (with 7.5% at ≥1.0 μg/ml) (5), and 1 to 4.0 μg/ml in 10 of 232 (4.3%) MRSA isolates (4). In contrast, the triclosan MICs for six strains of S. aureus isolates from national culture collections (American Type Culture Collection and National Collection of Type Cultures) ranged from 0.01 to 0.1 μg/ml, with a median value of 0.01 μg/ml (41). Thus, even at baseline in our study, the median triclosan MIC for S. aureus, known to be susceptible to triclosan at low concentrations, was 2 μg/ml, a level which exceeds those for earlier isolates, which likely had had extensive triclosan exposure (23, 40).
The triclosan MICs determined here for other species, such as K. pneumoniae, also differed from those of previous reports (23, 41). For example, the MICs for K. pneumoniae isolates gathered from industrial sources (where triclosan exposure was likely) ranged from 0.1 to 1 μg/ml (23). In a household study, one strain with an MIC of 1.025 μg/ml was isolated from a household tub (11). In contrast, the triclosan MICs for 38 and 33% of the K. pneumoniae isolates in our study were ≥2 and ≥64 μg/ml, respectively. For a few other gram-negative species, such as E. cloacae and A. baumannii, the range of triclosan MICs in the present study is consistent with isolates gathered from another sample of homes (11).
Since the proportions of isolates with triclosan MICs above the median were the same or higher at baseline compared to the end of the year among several of the species examined in the present study, it is possible that the prior widespread use of triclosan-containing products may have influenced the overall susceptibility of species within this community setting. At present, the clinical relevance of high triclosan MICs is unknown (15).
Assessing triclosan susceptibility is problematic because of its limited solubility in inorganic solvents, which precludes attaining the concentrations found in most consumer products (i.e., 0.1 to 0.2% triclosan = 1,000 to 2,000 μg/ml). Nonetheless, we reexamined a subset of organisms with high MICs (≥32.0 μg/ml) and found that none of the isolates (A. baumannii, E. agglomerans, E. cloacae, K. pneumoniae, and P. fluorescens-P. putida) were inhibited on agar plates containing concentrations of 1,024 μg/ml, suggesting that they can survive the triclosan concentrations used in some consumer products. In our laboratory experiments, such concentrations produced various levels of opacity in the test media, revealing triclosan insolubility, and these strains produced zones of clearing similar to the zones of triclosan degradation reported by Meade et al. (33). In another study, a triclosan-selected E. coli mutant could survive two- to fourfold longer than the wild type in broth supplemented with a triclosan-based soap. Some researchers have proposed that it is the residual concentrations of triclosan in the environment that may play a role in selecting for organisms with reduced susceptibility (14, 25, 26, 36). Since bacteria on the hand normally reside in clumps, squames, fluid, and sometimes biofilms, a gradient of exposure to triclosan among hand flora is likely (6). Triclosan has been considered to be relatively stable in the environment and has been isolated from environmental sources such as rivers and also in breast milk (1, 18, 27, 37). Therefore, the ambient exposure to low concentrations of triclosan, which most likely occurs within the environment and over longer time periods, could lead to reduced susceptibility to triclosan within the community setting.
Prior reports examining small numbers of S. aureus and MRSA isolates from the clinical and industrial setting found no significant association between triclosan MICs and antibiotic resistance (5, 11, 23, 39). Although we also noted no statistically significant association, there was a trend in that direction when we compared the odds at baseline to the those at the end of the year for all species combined and also for GNB.
Such associations have been noted in laboratory studies (10, 12, 30). One study reported isoniazid resistance in Mycobacterium smegmatis, selected by triclosan via mutations in the InhA, which is the same target for triclosan (30). Triclosan-resistant clones of P. aeruginosa were associated with increased MICs of clinically relevant antibiotics such as ciprofloxacin (10). Multidrug efflux pumps in P. aeruginosa confer high levels of intrinsic triclosan resistance to antibiotics (9). In fact, the triclosan MICs for P. aeruginosa mutants overexpressing these pumps were in the range of concentrations used in consumer cleaning and hygiene products (9). Moreover, research has shown that several species, such as E. coli and S. aureus, share the same triclosan growth-inhibitory target, namely, FabI (the homolog of InhA) (16, 17, 31).
The lack of an association between triclosan susceptibility and antibiotic resistance in community isolates compared to laboratory findings may be a reflection of different experimental procedures that are not reproduced by the exposure conditions of bacteria in a community. Since our study design relied upon data from only a single isolate from each phenotypically similar population, we could have missed members of subpopulations which might have demonstrated resistance. It is also possible that such a relationship may be below the statistical detection level provided by our sample size or may require a longer time period to emerge. Also, unlike laboratory studies, we could not compare the same organism at baseline and at the end of the study. Since earlier analyses showed significantly different types of species found at baseline compared to the end of the year (3), we conducted cross-sectional analyses at these two time points. It is likely that the species genotypes changed intermittently over the 1-year study period; therefore, our analyses within species types may reflect various genotypes carried on the hands of the primary caregiver over time.
Hygiene has a measurable impact on reducing the burden of infections in the developing world, as well as in specialized populations and settings in the United States (2). However, there has been little evidence that the use of a 0.2% triclosan soap affords any benefit in the reduction of infectious symptoms, bacterial counts, or types of bacteria on the hands of individuals within the household setting in the developed world (3, 19, 20). On the other hand, based on the present study, the general levels of decreased susceptibility to triclosan seem to be increasing in the community, regardless of whether triclosan-containing products are used in the home or not. The eventual clinical implications of this decreased susceptibility warrant continued surveillance. Therefore, it is important to investigate further the impacts from the prolonged use of such products within the home environment, where organisms with reduced susceptibilities to triclosan already dwell.
Acknowledgments
This study was supported by Home Hygiene Practices and Infection Transmission in Households (3 RO1, NR05251-02s1), funded by National Institute of Nursing Research, National Institutes of Health. We also acknowledge the Robert Wood Johnson Health and Society Scholar Program.
REFERENCES
- 1.Adolfsson-Erici, M., M. Pettersson, J. Parkkonen, and J. Sturve. 2002. Triclosan, a commonly used bactericide found in human milk and in the aquatic environment in Sweden. Chemosphere 46:1485-1489. [DOI] [PubMed] [Google Scholar]
- 2.Aiello, A. E., and E. L. Larson. 2002. What is the evidence for a causal link between hygiene and infections? Lancet Infect. Dis. 2:103-110. [DOI] [PubMed] [Google Scholar]
- 3.Aiello, A. E., L. V. Lee, P. Della-Latta, S. Lin, and E. Larson. 2002. Types of microbial flora on hands of subjects and an environmental surface in homes randomized to antimicrobial or non-antimicrobial cleaning and hygiene products. 40th Annual Meeting of the Infectious Disease Society of America, Chicago, Ill. IDSA, Alexandria, Va.
- 4.Al-Doori, Z., D. Morrison, G. Edwards, and C. Gemmell. 2003. Susceptibility of MRSA to triclosan. J. Antimicrob. Chemother. 51:185-186. [DOI] [PubMed] [Google Scholar]
- 5.Bamber, A. I., and T. J. Neal. 1999. An assessment of triclosan susceptibility in methicillin-resistant and methicillin-sensitive Staphylococcus aureus. J. Hosp. Infect. 41:107-109. [DOI] [PubMed] [Google Scholar]
- 6.Baquero, F., C. Patron, R. Canton, and M. Martinez Ferrer. 1991. Laboratory and in-vitro testing of skin antiseptics: a prediction for in-vivo activity? J. Hosp. Infect. 18(Suppl. B):5-11. [DOI] [PubMed] [Google Scholar]
- 7.Bhargava, H. N., and P. A. Leonard. 1996. Triclosan: applications and safety. Am. J. Infect. Control. 24:209-218. [DOI] [PubMed] [Google Scholar]
- 8.Chuanchuen, R., K. Beinlich, T. T. Hoang, A. Becher, R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2001. Cross-resistance between triclosan and antibiotics in Pseudomonas aeruginosa is mediated by multidrug efflux pumps: exposure of a susceptible mutant strain to triclosan selects nfxB mutants overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 45:428-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chuanchuen, R., R. R. Karkhoff-Schweizer, and H. P. Schweizer. 2003. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am. J. Infect. Control. 31:124-127. [DOI] [PubMed] [Google Scholar]
- 10.Chuanchuen, R., C. T. Narasaki, and H. P. Schweizer. 2002. The MexJK efflux pump of Pseudomonas aeruginosa requires OprM for antibiotic efflux but not for efflux of triclosan. J. Bacteriol. 184:5036-5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, E. C., R. M. Addison, J. R. Rubino, K. E. Leese, P. D. Dulaney, M. S. Newell, J. Wilkins, D. J. Gaber, T. Wineinger, and D. A. Criger. 2003. Investigation of antibiotic and antibacterial agent cross-resistance in target bacteria from homes of antibacterial product users and nonusers. J. Appl. Microbiol. 95:664-676. [DOI] [PubMed] [Google Scholar]
- 12.Cookson, B. D., H. Farrelly, P. Stapleton, R. P. Garvey, and M. R. Price. 1991. Transferable resistance to triclosan in MRSA. Lancet 337:1548-1549. [DOI] [PubMed] [Google Scholar]
- 13.Council of Scientific Affairs. 2000. Report 2 of the council on scientific affairs: use of antimicrobials in consumer products 2 (A-00). American Medical Association, Chicago, Ill.
- 14.Gilbert, P., and A. J. McBain. 2003. Potential impact of increased use of biocides in consumer products on prevalence of antibiotic resistance. Clin. Microbiol. Rev. 16:189-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert, P., A. J. McBain, and S. F. Bloomfield. 2002. Biocide abuse and antimicrobial resistance: being clear about the issues. J. Antimicrob. Chemother. 50:137-140. [DOI] [PubMed] [Google Scholar]
- 16.Heath, R. J., J. Li, G. E. Roland, and C. O. Rock. 2000. Inhibition of the Staphylococcus aureus NADPH-dependent enoyl-acyl carrier protein reductase by triclosan and hexachlorophene. J. Biol. Chem. 275:4654-4659. [DOI] [PubMed] [Google Scholar]
- 17.Heath, R. J., N. Su, C. K. Murphy, and C. O. Rock. 2000. The enoyl-[acyl-carrier-protein] reductases FabI and FabL from Bacillus subtilis. J. Biol. Chem. 275:40128-40133. [DOI] [PubMed] [Google Scholar]
- 18.Kolpin, D. W., E. T. Furlong, M. T. Meyer, E. M. Thurman, S. D. Zaugg, L. B. Barber, and H. T. Buxton. 2002. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999-2000: a national reconnaissance. Environ. Sci. Technol. 36:1202-1211. [DOI] [PubMed] [Google Scholar]
- 19.Larson, E., A. Aiello, L. V. Lee, P. Della-Latta, C. Gomez-Duarte, and S. Lin. 2003. Short- and long-term effects of handwashing with antimicrobial or plain soap in the community. J. Community Health 28:139-150. [DOI] [PubMed] [Google Scholar]
- 20.Larson, E., Lin, SX, Gomez-Duarte, C, Della-Latta, P. 2004. Effect of antibacterial home cleaning and handwashing products on infectious disease symptoms. Ann. Intern. Med. 140:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson, E. L. 1981. Persistent carriage of gram-negative bacteria on hands. Am. J. Infect. Control. 9:112-119. [DOI] [PubMed] [Google Scholar]
- 22.Larson, E. L., C. Gomez-Duarte, L. V. Lee, P. Della-Latta, D. J. Kain, and B. H. Keswick. 2003. Microbial flora of hands of homemakers. Am. J. Infect. Control. 31:72-79. [DOI] [PubMed] [Google Scholar]
- 23.Lear, J. C., J. Y. Maillard, P. W. Dettmar, P. A. Goddard, and A. D. Russell. 2002. Chloroxylenol- and triclosan-tolerant bacteria from industrial sources. J. Ind. Microbiol. Biotechnol. 29:238-242. [DOI] [PubMed] [Google Scholar]
- 24.Levy, S. 2002. The antibiotic paradox: how the misuse of antibiotics destroys their curative powers. Perseus Publishing, New York, N.Y.
- 25.Levy, S. B. 2001. Antibacterial household products: cause for concern. Emerg. Infect. Dis. 7:512-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy, S. B. 2000. Antibiotic and antiseptic resistance: impact on public health. Pediatr. Infect. Dis. J. 19:S120-S122. [DOI] [PubMed] [Google Scholar]
- 27.Lindstrom, A., I. J. Buerge, T. Poiger, P. A. Bergqvist, M. D. Muller, and H. R. Buser. 2002. Occurrence and environmental behavior of the bactericide triclosan and its methyl derivative in surface waters and in wastewater. Environ. Sci. Technol. 36:2322-2329. [DOI] [PubMed] [Google Scholar]
- 28.McBain, A. J., R. G. Bartolo, C. E. Catrenich, D. Charbonneau, R. G. Ledder, B. B. Price, and P. Gilbert. 2003. Exposure of sink drain microcosms to triclosan: population dynamics and antimicrobial susceptibility. Appl. Environ. Microbiol. 69:5433-5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride, M. E., L. F. Montes, W. J. Fahlberg, and J. M. Knox. 1974. Microbial flora of nurses' hands. II. Qualitative differences in occupational groups. Int. J. Dermatol. 13:197-204. [DOI] [PubMed] [Google Scholar]
- 30.McMurry, L. M., P. F. McDermott, and S. B. Levy. 1999. Genetic evidence that InhA of Mycobacterium smegmatis is a target for triclosan. Antimicrob. Agents Chemother. 43:711-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol. Lett. 166:305-309. [DOI] [PubMed] [Google Scholar]
- 32.McMurry, L. M., M. Oethinger, and S. B. Levy. 1998. Triclosan targets lipid synthesis. Nature 394:531-532. [DOI] [PubMed] [Google Scholar]
- 33.Meade, M. J., R. L. Waddell, and T. M. Callahan. 2001. Soil bacteria Pseudomonas putida and Alcaligenes xylosoxidans subsp. denitrificans inactivate triclosan in liquid and solid substrates. FEMS Microbiol. Lett. 204:45-48. [DOI] [PubMed] [Google Scholar]
- 34.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 35.Parikh, S. L., G. Xiao, and P. J. Tonge. 2000. Inhibition of InhA, the enoyl reductase from Mycobacterium tuberculosis, by triclosan and isoniazid. Biochemistry 39:7645-7650. [DOI] [PubMed] [Google Scholar]
- 36.Schweizer, H. P. 2001. Triclosan: a widely used biocide and its link to antibiotics. FEMS Microbiol. Lett. 202:1-7. [DOI] [PubMed] [Google Scholar]
- 37.Singer, H., S. Muller, C. Tixier, and L. Pillonel. 2002. Triclosan: occurrence and fate of a widely used biocide in the aquatic environment: field measurements in wastewater treatment plants, surface waters, and lake sediments. Environ. Sci. Technol. 36:4998-5004. [DOI] [PubMed] [Google Scholar]
- 38.Slayden, R. A., R. E. Lee, and C. E. Barry III. 2000. Isoniazid affects multiple components of the type II fatty acid synthase system of Mycobacterium tuberculosis. Mol. Microbiol. 38:514-525. [DOI] [PubMed] [Google Scholar]
- 39.Suller, M. T., and A. D. Russell. 1999. Antibiotic and biocide resistance in methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococcus. J. Hosp. Infect. 43:281-291. [DOI] [PubMed] [Google Scholar]
- 40.Suller, M. T., and A. D. Russell. 2000. Triclosan and antibiotic resistance in Staphylococcus aureus. J. Antimicrob. Chemother. 46:11-18. [DOI] [PubMed] [Google Scholar]
- 41.Vischer, W. A., and J. Regos. 1974. Antimicrobial spectrum of Triclosan, a broad-spectrum antimicrobial agent for topical application. Zentbl. Bakteriol. Orig. A 226:376-389. [PubMed] [Google Scholar]