Abstract
Peanut allergens can trigger a potent and sometimes dangerous immune response in an increasing number of people. The molecular structures of these allergens form the basis for understanding this response. This review describes the currently known peanut allergen structures and discusses how modifications both enzymatic and non-enzymatic affect digestion, innate immune recognition, and IgE interactions. The allergen structures help explain cross-reactivity among allergens from different sources, which is useful in improving patient diagnostics. Surprisingly, it was recently noted that related short peptide sequences among peanut allergens could also be a source of cross-reactivity. The molecular features of peanut allergens continue to inform predictions and provide new research directions in the study of allergic disease.
Keywords: Peanut allergy, Molecular basis, Peanut allergens, Major allergens, Minor allergens, Protein structures, Cross-reactivity, Molecular modifications
Introduction
Allergies to peanuts are a major public health concern. Recent data suggests that the incidence is increasing and currently 1–2 % of Americans, or nearly 30 million people, are allergic to peanuts [1]. Not surprisingly, the basis for peanut allergy has been the subject of extensive research. In this review, we hope to highlight new data focused on the molecular recognition of peanut allergens by the adaptive and innate immune system. This review will delve into two recent major topics: cross-reactivity among non-homologous peanut and nut allergens and molecular modifications to peanuts and their immunological consequences. We briefly discuss IgE epitopes in general, as this topic has recently been well reviewed[2–4]. We begin by discussing the molecular structures of the peanut allergens in order to set the stage for these topics.
Protein Structures of Peanut Allergens
Greater than 50 % of all plant food allergens can be categorized into just four structural protein families; prolamin superfamily, cupin superfamily, profilins, and Bet v-1-related proteins [5]. Almost all of these are either storage or plant defense-related proteins [6]. Peanuts harbor 12 allergens and multiple isoforms recognized by the Allergen Nomenclature Sub-Committee of the International Union of Immunological Societies, 70 % of which fall into these families. These 12 allergens, can be categorized into the four most common food allergen families: the Cupin superfamily (Ara h 1, 3), the Prolamin superfamily (Ara h 2, 6, 7, 9), the Profilin family (Ara h 5), and Bet v-1-related proteins (Ara h 8), as well as two additional families, Oleosin (Ara h 10,11) and Defensin (Ara h 12, 13). Currently, structural data exist for Ara h 1, 2, 3, 5, 6, and 8 [7–13]. We subdivided these structural descriptions into the major allergens that have the highest prevalence of IgE binding, and the minor allergens, which have less IgE-binding prevalence but significant cross reactivity with allergens from other sources.
Major Allergens
Allergens in a food are considered major if they are recognized by the serum IgE of greater than 50 % of the allergic population. The major allergens in peanuts are generally considered Ara h 1 and Ara h 3 that are members of the cupin superfamily of proteins, and Ara h 2 and Ara h 6 that are members of the prolamin superfamily. As can be inferred from the descriptions below, a remarkable amount of structural, biophysical, and bioinformatic information on these allergens has been obtained.
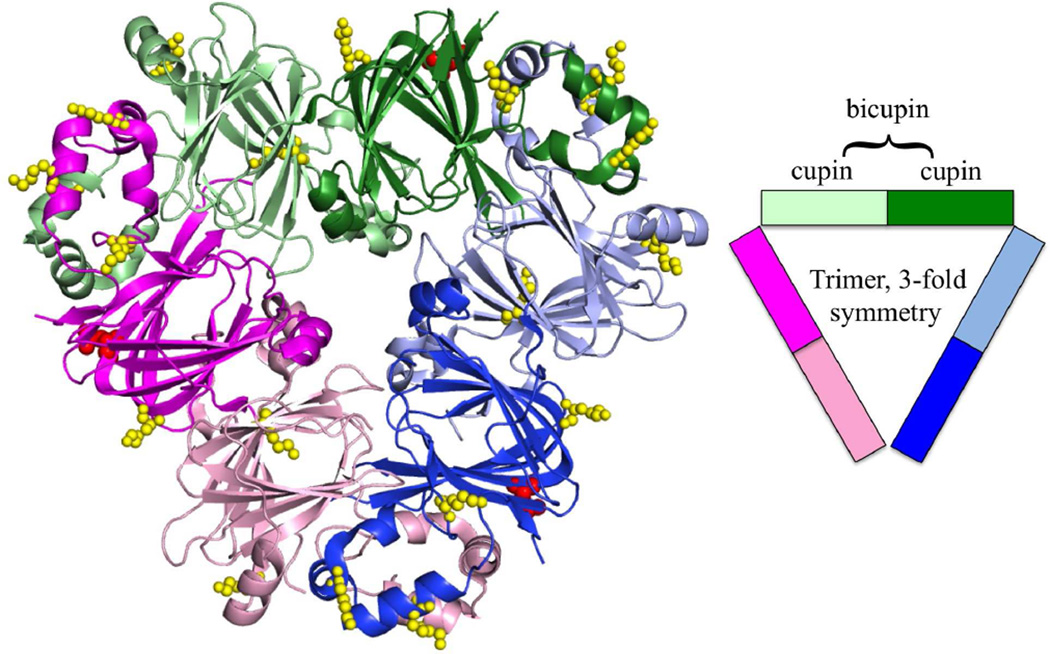
Cupins are a functionally diverse superfamily that can have low levels of sequence conservation yet maintain a high degree of structural similarity in a conserved β-barrel motif [14]. Ara h 1, a member of this superfamily, is a 65-kDa, 7S globulin or vicilin seed storage protein. Attempts by multiple groups to solve the structure of native protein were unsuccessful; however, attempts with recombinant expressed core domains resulted in two crystal structures [7, 8]. Ara h 1 forms a symmetrical trimer with a 3-fold axis running between the monomers (Fig. 1). Each monomer is comprised of two cupin domains (known as a bicupin) with small cavities flanked by α-helices. These two cupin domains share limited sequence identity (15 %) but are structurally conserved (r.m.s.d = 1.9 Å for 153 Cαs) and are thought to have evolved from a gene duplication event of an ancestral prokaryotic gene [15]. It has been suggested that the central cavities formed by the β-barrel may bind ligands [7]. Sequence variations between the two cavities leaves open the possibility that the ligands could be different. The cupin domains of the individual proteins are related by a pseudo-two-fold axis that is approximately perpendicular to the 3-fold axis. The α-helices that flank the cupin domain are found at the subunit interfaces and are involved in trimer formation. It is thought that Ara h 1 forms higher ordered oligomers consistent with trimer of trimers or tetramer of trimers based on small angle X-ray scattering (SAXS) data of the core domain and natural allergen as well as elution profiles from size-exclusion chromatography of native protein [7, 16]. As discussed below in the section on molecular modifications, oligomeric forms of Ara h 1 and other peanut allergens are covalently stabilized when peanuts are cooked, which may be related to the allergenicity of peanuts [17].
Figure 1.
Ara h 1 trimer. Ara h 1 of the cupin superfamily is a trimer of bicupins, colored by the cupin domains (PDB:3SMH). The individual bicupins are colored pink, green, and blue, with Nterminal domains lightly shaded and the C-terminal domains a darker shade. Highlighted on the structure are the identified sites of glycosylation colored red (47) and glycation colored yellow. As technology improves, more glycation sites and AGE modifications may be identified [59]
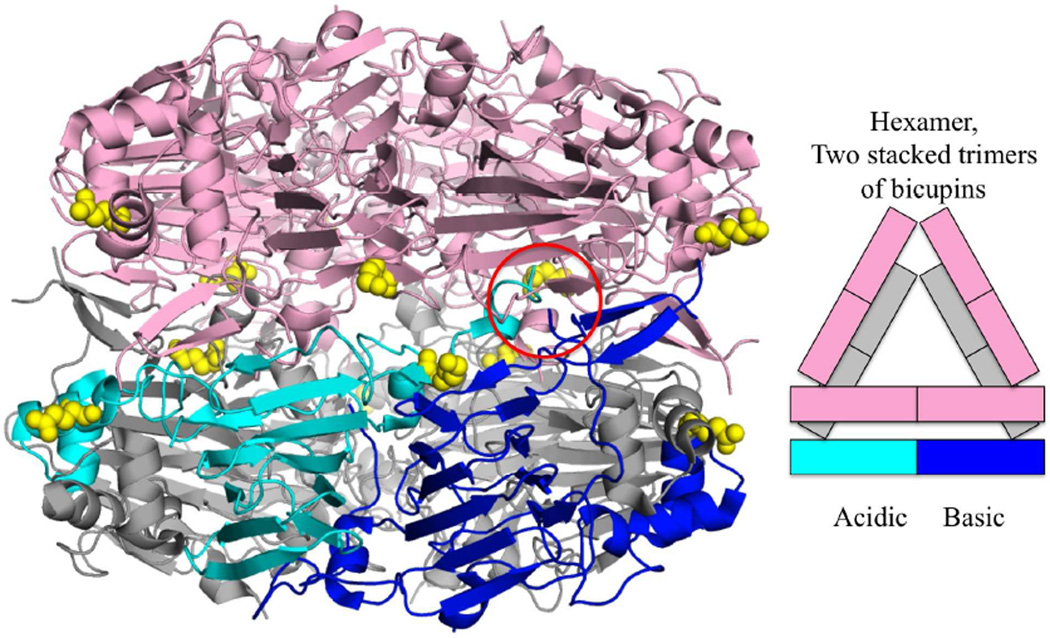
Ara h 3 is an 11S globulin or legumin in the cupin superfamily and shares 21 % sequence identity to Ara h 1. In addition to being a seed storage protein, Ara h 3 is also a trypsin inhibitor [18]. Unlike Ara h 1, Ara h 3 can be crystallized in its native form purified from dry peanut kernels [13]. Despite the low sequence identity, the crystal structure of Ara h 3 is very similar to that of Ara h 1 with an r.m.s.d. of 2.4 Å over 316 of the core residues. Ara h 3 forms a hexamer consisting of two Ara h 1-like trimers stacked head to head (Fig. 2). Ara h 3 is post-translationally modified by a proteolytic cleavage that occurs between the two cupin domains on a flexible loop. This cleavage appears to be required for hexamer formation as this loop needs to be removed for the two trimers to form the hexameric interface. The two cupin domains are known as the acidic and basic subunit and can be readily separated by isolectric focusing [19]. Certain cultivars of peanuts lacking the basic subunit of Ara h 3 have been studied as potentially less allergenic [20].
Figure 2.
Ara h 3 hexamer. Ara h 3 is a hexamer of two trimers of bicupins (PDB:3C3V). One trimer is colored pink and for the other: two bicupins are colored gray and the third dark blue for the basic N-terminal cupin domain and cyan for the acidic C-terminal cupin domain. The hexamer forms after cleavage of a peptide between the cupin domains (cleavage site circled in red). Sites of identified glycation are colored yellow [59]
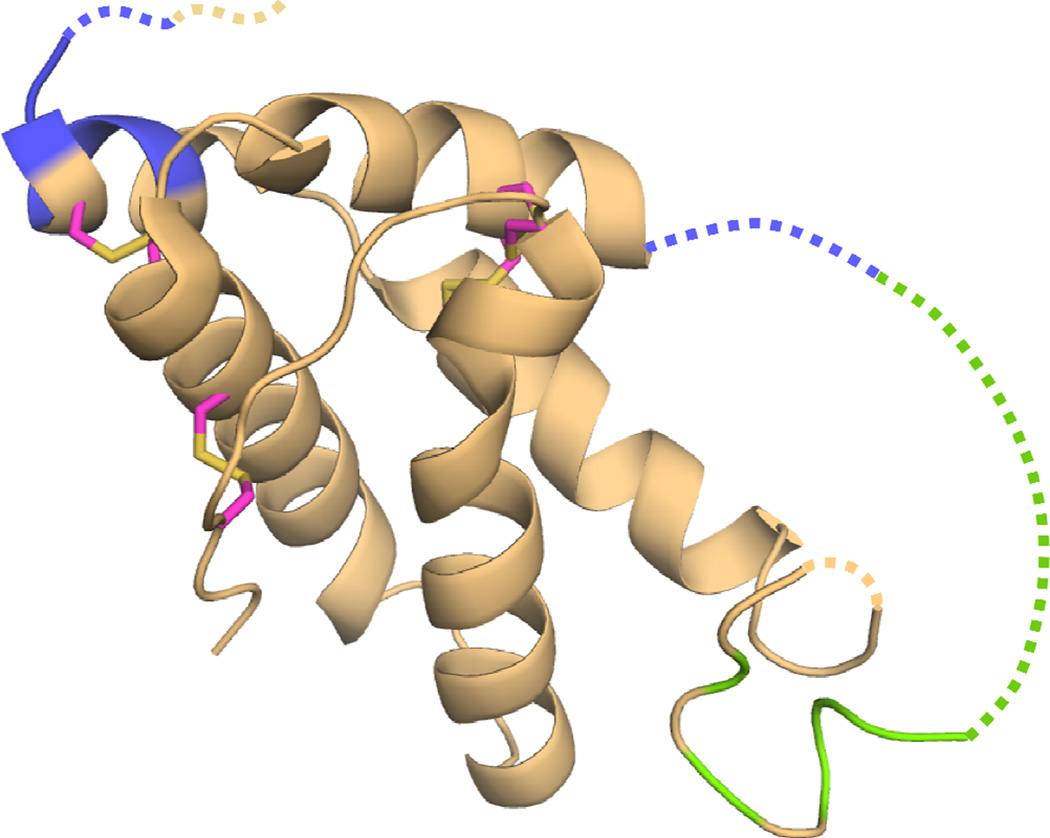
The prolamin superfamily consist of 2S albumins, cereal α-amylase, and trypsin inhibitors, as well as non-specific lipid transfer proteins (nsLTPs) [21]. These are cysteine-rich α-helical proteins of similar fold with multiple disulfide bonds that likely contribute to their resistance to proteolysis as well as to their high heat and pH stability [17, 21]. A recombinant maltose binding protein, (MBP)-Ara h 2 fusion protein, was used to solve the structure of Ara h 2, shown in Fig. 3 [9]. The crystal structure revealed Ara h 2 to be comprised of a five helical bundle with four disulfide bonds interconnecting the helices. Missing from the structure is a large, disordered loop of 31 residues connecting helices 2 and 3. Despite often being considered a 2S albumin, a search of the structural database revealed it to be structurally most similar to the α-amylase and trypsin inhibitors [9, 22]. This is consistent with previous reports of trypsin inhibition by Ara h 2 [23]. An NMR structure of recombinant Ara h 6 has also been determined [10]. Ara h 6 shares 59 % sequence identity to Ara h 2 and shares the same secondary and tertiary structural features (r.m.s.d. = 2.4 for 79 residues). While Ara h 2 is frequently cited as the most potent peanut allergen [24–26], it was only recently appreciated that Ara h 2 and Ara h 6 have highly similar allergenic activities [27, 28]. Given the highly similar physical and immunological characteristics, the two proteins probably should be considered collectively as related allergens.
Figure 3.
Ara h 2 of the promalin family. Highlighted on Ara h 2 are peptides that were found to inhibit the IgE binding to Ara h 1 (colored green) and Ara h 3 (colored blue). Missing residues in the crystal structure are indicated with a dashed line. Disulfide connectivity is shown with magenta and yellow sticks (PDB:3OB4)
Minor Allergens
Minor allergens are recognized by the serum IgE of less than 50 % of the allergic population. The minor allergens in peanuts, for which there is structural information, include Ara h 5 from the profilin protein family, and Ara h 8 from the Bet v 1-like superfamily. These two structures were determined recently.
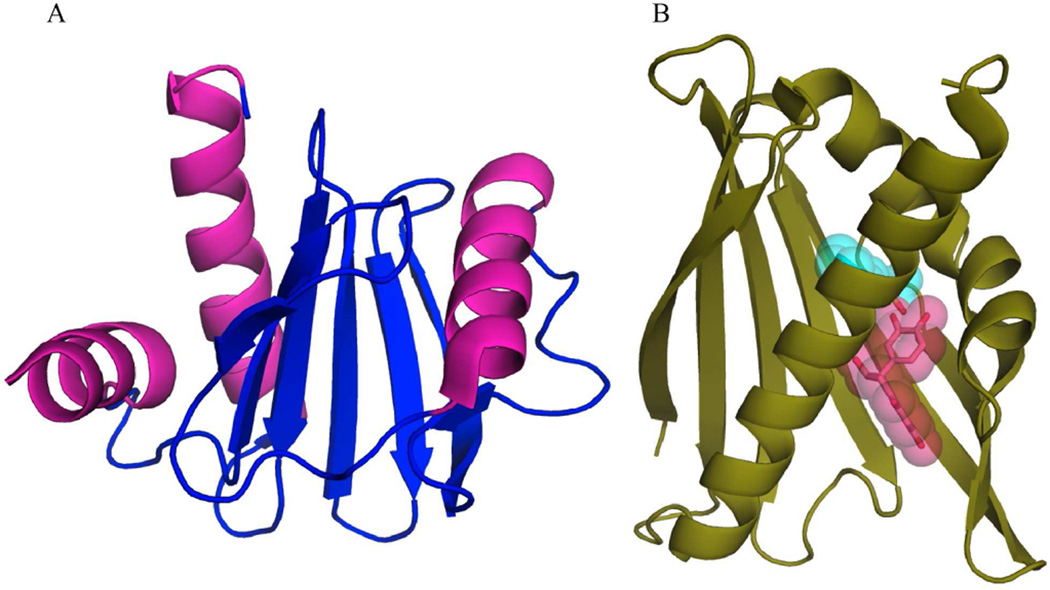
Unlike the aforementioned allergens, Ara h 5 in not a seed-storage protein, but rather belongs to the profilin family of proteins. Profilins are small, ~15-kDa, proteins found in all eukaryotic cells that interact with actin and are involved with a number of cellular processes such as cytoskeletal dynamics. In plants, they are involved in cell elongation, cell shape maintenance, polarized growth of root hair and flowering time [29, 30]. Figure 4 shows that the crystal structure of recombinant Ara h 5 is comprised of the canonical profilin α/β motif with a central anti-parallel β-sheet flanked by α-helices [11]. High sequence and structural conservation to profilins from other species such as the pollen allergen Bet v 2 and latex allergen Hev b 8 may explain why profilin panallergens show cross-reactivity from multiple sources [31].
Figure 4.
Ara h 5 and Ara h 8. Structures of Ara h 5 (a) from the profilin family and Ara h 8 (b) from the Bet v 1 related protein family. Ligands found in the active site of Ara h 8 from different structures (MES color cyan, and epicathecin color magenta) are rendered with a semitransparent surface (PDB:4ESP, 4M9W, and 4MA6, respectively)
Ara h 8 is a 17-kDa member of the pathogenesis-related protein 10 (PR-10) class of proteins, which belong to the Bet v 1-like superfamily. The functional roles of PR-10 proteins are not well understood, but some may play a role in a stress response/general defense mechanism as they can be induced by pathogen attack [32]. The structures of PR-10 proteins generally consist of a curved seven-stranded anti-parallel β-sheet flanked by three α-helices on one side creating a hydrophobic cavity with the ability to bind hydrophobic ligands [32]. Ara h 8 shares these structural features and has been shown to bind a number of biological compounds including flavonoids, suggesting a potential role as a flavonoid carrier protein [12]. Different crystal structures of recombinantly expressed Ara h 8 demonstrate binding of epicatechin as well as the laboratory buffer MES to the ligand-binding cavity (Fig. 4). These compounds are located in non-overlapping positions suggesting multiple ligands can bind simultaneously, similar to studies of ligand binding to the prototypical Bet v 1 [33]. Structural conservation of surface patches between Ara h 8 and the pollen allergen Bet v 1 may explain the IgE cross-reactivity between these panallergens [12].
IgE Epitopes
The utility of molecular structures of allergens in the study of allergy, and in particular the study of IgE epitopes, has been extensively reviewed [2–4]. The importance of the structures is primarily seen as a template for visualizing surface residues and consequently understanding the IgE interacting surface. As mentioned above, this can improve our understanding and make predictions about the potential for cross-reactivity. Presumably with knowledge of the epitopes, it may be possible to rationally design better diagnostic tools or hypoallergenic alternatives for safer and/or more effective immunotherapy. Aalberse and Crameri regard the latter suggestion as unlikely given the polyclonal nature of the IgE response, and the fact that usually rather drastic modifications to allergens are needed to significantly reduce reactivity [2]. However, they do suggest that another way in which epitope information may be useful is in discerning more fundamental information about the peculiarities of the IgE repertoire [2]. Several hypotheses have been presented concerning differences between IgE and IgG: IgE epitopes may be more cross-reactive and biased toward different surfaces [4]; IgE epitopes may cluster to certain regions [34]; and IgE paratopes may be more flexible [2]. While not proven and speculative in general, these ideas provide potentially interesting avenues for future research.
Cross-reactivity among peanut allergens
One of the biggest difficulties in food allergy diagnosis and detection involves allergen cross-reactivity. The phenomenon of cross-reactivity in allergy can be clinically relevant or irrelevant. It is common to observe positive specific IgE (sIgE) test results (by ELISA or by skin test) to foods that are tolerated. For example, peanut allergic individuals can demonstrate sIgE cross-reactivity to multiple nuts and or legumes, but have clinical symptoms to peanuts and tolerate none, one or more of the cross-reactive foods. In fact, approximately 50 % of peanut-allergic patients have positive skin prick tests to other legumes, but less than 5 % are clinically symptomatic upon ingestion of legumes [35]. Without a good medical history and a food challenge, it is increasingly difficult to assign accurate food avoidance diets and often leads to unnecessary blanket elimination diets [36]. These types of widespread dietary avoidance are very difficult for the patient and their families [36].
Historically, the general belief has been that cross-reactivity is only seen between proteins of the same family, mostly because of structural and sequential identity [6, 35, 37]. And, although it is well known and documented that cross-reactivity exists between proteins that share high homology in structure and sequence, recent studies that demonstrate IgE cross-reactivity between non-homologous protein families challenge this dogma in the field of food allergy [35, 38, 39]. In one study, a computational prediction method was used to search an allergen database for clinically cross-reactive epitopes based purely on physical and chemical properties of a previously known IgE binding site. This method does not rely on sequence alone and is therefore able to identify similar peptides with significantly divergent sequences that would not normally be found with the standard sequence-based search engines [39]. Originally, in 2005, potential cross-reactivity was suggested between known epitopes of Ara h 2 and Ara h 1, and among similar sequences within different regions of Ara h 1 using the prediction tool from the Structural Database for Allergic Proteins (SDAP) [40]. More recently, the epitopes that were predicted by SDAP to cross-react with known epitopes of Ara h 2 were empirically tested for IgE binding. A previously unidentified and highly cross-reactive IgE epitope was identified in Jug r 2, the walnut vicilin. The newly identified epitope was shown to inhibit IgE binding to Ara h 2 as well as a known Ara h 2 epitiope. A majority of the reactive peptides identified were shown to be exposed on the surface of the molecules. A second study tested the IgE cross-reactivity among the 4 major peanut allergens, Ara h 1, Ara h 2, Ara h 3, and Ara h 6, and peptides thereof with western blotting, competitive inhibition ELISA, and basophil-histamine release assays. All the allergens were able to significantly inhibit IgE binding to each other to various extents. Peptides from Ara h 2 that inhibited IgE binding to Ara h 1 and Ara h 3 are highlighted on the structure of Ara h 2 in Fig. 2. These combined results definitively demonstrate that the phenomenon of cross-reactivity in allergy is much more complex than originally imagined. Based on these studies and unpublished observations, we believe that IgE cross-reactivity among non-homologous proteins within one food and among different foods will prove to be relatively common.
Understanding this “non-homologous” cross-reactivity may be related to the severity of the patient response. Sensitization to a single peanut allergen correlated with less severe reactions to peanuts compared to patients that were sensitized to multiple peanut allergens [41–43]. We hypothesize that those patients sensitized to multiple peanut allergens are recognizing the IgE epitopes that are cross-reactive among the non-homologous proteins, leading to a more severe response.
Molecular Modifications
To understand the allergen-immune system recognition and response further, the added complexities of modifications to allergens and their potential role in cross-reactivity needs to be addressed. Two categories of molecular modifications to peanut allergens have previously been described, enzymatic and non-enzymatic. Enzymatic modifications include glycosylation while non-enzymatic modifications arise from food processing primarily in the form of advanced glycation end products, or AGEs.
Enzymatic
Plant allergens are frequently glycosylated. The two main O-linked sugar motifs are xylose and core-3-linked fucose, which are both found in nearly all plants [44]. Therefore, any IgE antibodies against these glycans could potentially interact with a huge variety of glycosylated plant proteins. It was recognized in 1981 that carbohydrate epitopes were a source of cross-reactivity between plant and insect allergens [45]. These glyco-epitopes became known as cross-reactive carbohydrate determinants (CCDs) and can make it difficult to correctly diagnose the important sensitizing allergen source in a clinical setting [46]. Among peanut allergens only Ara h 1 is known to be glycosylated and at a single site (Fig. 1) [47], which implies that anti-CCD IgE will only bind one site per peptide chain. This further implies that, for two IgE molecules to cross-link on the surface of mast cells to initiate symptoms via the CCD, one of two scenarios must occur [44]. Cross-linking could occur through different antibodies recognizing the CCD and another protein epitope or, if the protein forms multimers like Ara h 1, two anti-CCD IgE could potentially initiate symptoms. This is an interesting case where the anti-CCD IgE could stimulate mast cells. Fortunately for most patients, no clinical symptoms accompany anti-glycan IgE, probably due to the presence of soluble anti-glycan IgG molecules that serve as decoys to prevent IgE crosslinking [48, 49]. Anti-carbohydrate antibodies are generally not considered important in allergic disease; however, similar carbohydrates on helminthes can have potent effects [50, 51].
Indeed, the carbohydrate determinants on Ara h 1 were demonstrated to have immunomodulatory properties [52]. Ara h 1 glycosylation is high in mannose and occasionally contains xylose moieties [47]. These carbohydrates interact with various receptors on dendritic cells (DC), which play an important sentinel role in the innate immune response. The Ara h 1 stimulation of DC via the lectin receptors MR and DC-SIGN has been demonstrated to induce cytokines known to bias the immune response towards an allergic or Th2 type response [52, 53]. Hence, the gycosylation state of the peanut allergens stimulates the innate immune cells to signal that an allergic-type response is warranted by downstream T-cells. The properties of immune stimulation via C-type lectin receptors have been extensively studied in the case of dust mite allergens, as recently reviewed [54].
Non-enzymatic
Proteins can also be modified by carbohydrates in a non-enzymatic mechanism known as the Maillard reaction. The formation of AGEs occurs when sugars react primarily with free amines and undergo an Amadori rearrangement [55]. The modifications are most common on lysines and are less frequently observed on arginines, the N-terminus, and cysteines [56]. In addition, stable covalent cross-links can be formed between the aforementioned residues. This process is accelerated by higher temperatures (i.e. cooking): dry roasting versus boiling can increase the number of AGE modifications by greater than 10-fold [57]. It is important to note that these modifications are spontaneous and occur at room temperature, although at a slower rate compared to cooking temperatures. Indeed, AGE modifications can be detected in raw peanuts to varying degrees [58, 59]. Therefore, it may be more prudent to utilize recombinant allergens in studies designed to isolate the effect of AGEs instead of comparing raw versus roasted peanuts.
In terms of peanuts, several studies have characterized the AGE modifications on peanut allergens. Early studies utilized antibodies specific for certain types of AGEs to demonstrate that Ara h 1 and Ara h 3 are more commonly modified than Ara h 2 [58]. More recent studies have utilized mass spectrometry (MS) to specifically identify modified residues and to characterize the modifications (Figs. 1, 2) [59–61]. While providing more detailed atomic information, several technical challenges make this a difficult process. First, the modifications on arginine residues prevent digestion of the allergens with commonly used trypsin-related proteases. As an aside, a similar phenomenon occurs using in vitro models of gastric digestion, further suggesting that the refractory nature of peanut allergens to digestion may play a role in sensitization [17, 23, 62]. Second, modified allergens, and proteins in general, were difficult to extract and purify from roasted peanuts [63], and therefore difficult to detect by MS and required extraction with urea [61] or multiple chromatography steps [59]. Nevertheless, commonly modified peptides have been identified. Knowing the exact molecular weight and common fragmentation patterns are useful for MS detection of trace amounts of peanut allergen in prepared food, which could improve the accuracy and safety of food labeling for allergic individuals [61].
Molecular Modifications, Allergy and Immunology
AGE modifications on peanuts are suggested to skew the immune response towards allergy. The mechanism for this was demonstrated to be stimulation of receptors like RAGE (Receptor for Advanced Glycation End products) and scavenger receptor class A type I and II (SR-AI/II) [64–66]. Two independent studies have demonstrated that the stimulation of dendritic cells with AGE-modified OVA compared to control OVA leads to activation of more IL-4- [65] or IL-5 [66]-producing T-cells than IFN-γ-producing T-cells. Both results suggest a Th2 bias. Further studies in Caco-2 cells, which are a model for intestinal epithelia, demonstrated that RAGE activation by AGEs stimulated MAP-kinases [67]. More recently, AGE-modified Ara h 1 was demonstrated to influence the proliferation of Caco-2 cells, in a manner dependent on the incubation time and temperature, indicating the possibility that specific AGE modifications may be important for influencing the pro-inflammatory network [68].
Besides peanut allergy, there appears to be a role for RAGE in other allergic diseases. RAGE knockout mice surprisingly develop a similar adaptive immune response to dust mite extract as normal mice, but do not develop asthma symptoms such as airway hypersensitivity, eosinophilic inflammation, and airway remodeling [69]. The same study further demonstrated that the use of soluble RAGE (sRAGE) as a decoy for the ligands of membrane-bound RAGE had similar results to the RAGE knockout mouse, indicating a possible tolerizing role for sRAGE [69]. Bronchioalveolar lavage of patients with neutrophilic asthma and chronic obstructive pulmonary disease (COPD) similarly showed a lack of sRAGE showing how dysregulation of the soluble versus membrane-bound RAGE is altered in disease [70]. Different organs express different forms of RAGE: the epidermis primarily makes soluble forms of RAGE while the lung primarily expresses membrane-bound RAGE [71–73]. This heterogeneity leads to some intriguing suggestions about the strategic manipulation of RAGE in immunotherapy. It may be that epicutaneous immunotherapy (EPIT) with roasted peanuts would generate more sRAGE than oral immunotherapy, thereby reducing the risk of inflammation or further sensitization. Recently, using a mouse model of peanut allergy, the safety of EPIT was demonstrated along with encouraging data regarding a tolerogenic immune profile [74]; however, the role of RAGE was not evaluated. It should be noted that the EPIT requires intact skin, suggesting that epithelial-derived factors are likely important. More research is clearly needed to understand whether or not RAGE was an important factor, and whether the deliberate manipulation of RAGE can be utilized to induce tolerance.
Multiple studies suggest that AGE-modified peanut allergens are more readily recognized by patients [17, 58, 75, 76]. However, when assessing relative IgE binding, two factors are important to consider: patient exposure and cooking/extraction protocols. First, raw peanuts are rarely consumed, so it is expected that few people are exposed to completely unmodified peanut allergens. Thus, their IgE will be biased to detect AGE-modified allergens. Second, as mentioned earlier, extraction methods can strongly influence the allergen content. The soluble extraction of Ara h 1 from peanuts is maximal when dry roasted for 15 min: shorter roasting times resulted in less extracted Ara h 1 and much longer times (25–30 min) produced more denatured protein [77]. Early studies that compared IgE binding to peanut allergens derived from different preparative techniques, such as boiling, frying, and roasting, produced mixed conclusions [75, 78]. A more recent and thorough examination of protein content in the soluble and insoluble fractions after different cooking techniques confirms that peanuts are not that different from other foods, in that boiling is the only method that reduces the number of AGEs [57, 63]. The differences among the different studies may be explained by different extraction techniques and the use of IgE as the detection measurement [63].
It is, however, unlikely that the AGEs are primarily what is recognized by IgE. Supporting this supposition is a recent paper that compared the IgE recognition of recombinant Ara h 1 over a time course of heating in the presence of sugars to create AGEs [59]. The IgE binding of five patients was similar to the total protein content over the whole time course, and increased slightly with more AGE modifications. Therefore, some common modifications are likely recognized by IgE, but the strong recognition of the unmodified rAra h 1 indicates that the allergen is primarily what is being recognized. When the binding of IgE obtained from three of the patients was tested against a control allergen, Bos d 6, which had been AGE-modified in the same protocol, no IgE binding was detected, indicating again that the allergen is more important than the AGE modifications and that the AGEs are recognized within the context of the protein [59]. Since AGEs are present in nearly all cooked foods, it would seem to be extremely dangerous to have IgE antibodies specifically against AGEs, analogous to the discussion above about having antibodies against common plant carbohydrates.
If the AGE modifications are so common in cooking, are there important health effects in other foods? The importance of dietary AGEs in general are extensively debated in the literature. In animal models, there is a clear connection between low AGE diets and the inhibition of atherosclerosis and the prevention of diabetic nephropathy [79]. However, there are conflicting studies as to the consequences of dietary AGEs in humans. Some studies describe AGEs as ‘glycotoxins’ and encourage reducing AGEs in the human diet [57]. In contrast, a recent meta-analysis of human trials involving AGE-restricted diets concluded that there is insufficient evidence to encourage this dietary restriction in healthy, diabetic, or renal impaired individuals [80]. The review further notes that all of the studies indicating a beneficial effect of AGE-restricted diets emerged from one research group, and all of the studies could benefit from better study design and standardized measurements to facilitate better comparisons [80].
The reason that peanuts generate such potent reactions is unlikely to have a single causative factor but is probably a combination of unfortunate events that work in concert. The AGE modifications and the trypsin inhibitory properties of Ara h 2 and Ara h 3 reduce proteolysis. This leads to more peanut protein surviving digestion, and, therefore, more whole protein entering the gut. The surviving proteins or fragments thereof are likely to maintain structural elements [81]. This probably stimulates the immune system by both the adaptive immune system via IgE binding and the innate immure response through lectin receptors and receptors such as RAGE recognizing the glycosylation and glycation modifcations, respectively.
Conclusions
The structural features of allergens and the protein families provide important information about detection, diagnosis, and the design of therapeutic tools in allergy. Recent data demonstrate that IgE-reactivity across protein families, i.e. among non-homologous proteins, is also important in peanut allergy, and may correlate with the most severe symptoms.. The glycosylation of peanut allergens is unlikely to be important for IgE antibody binding, but rather may be important for the stimulation of innate immunity via dectin or lectin receptors. Similarly, the glycation state (addition of advanced glycation end products) is suggested to affect innate immune stimulation, digestion of the peanut allergens, and antibody recognition of the allergens.
Acknowledgments
The authors wish to thank Drs. Robert London, Michael Fessler, and Jason Williams for critical readings of the manuscript. This research was supported by Research Project Number Z01-ES102885-01 and ZIA- ES102645 in the Intramural Research Program of the National Institute of Environmental Health Sciences, National Institutes of Health.
Footnotes
Compliance with Ethics Guidelines
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Conflict of Interest
Geoffrey A. Mueller, Soheila J. Maleki, and Lars C. Pedersen declare that they have no conflict of interest.
References
- 1.Liu AH, Jaramillo R, Sicherer SH, Wood RA, Bock SA, Burks AW, et al. National prevalence and risk factors for food allergy and relationship to asthma: results from the National Health and Nutrition Examination Survey 2005–2006. J Allergy Clin Immunol. 2010 Oct;126(4):798–806. e13. doi: 10.1016/j.jaci.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aalberse RC, Crameri R. IgE-binding epitopes: a reappraisal. Allergy. 2011 Oct;66(10):1261–1274. doi: 10.1111/j.1398-9995.2011.02656.x. Thought-provoking summary of IgE antibodies.
- 3. Dall'antonia F, Pavkov-Keller T, Zangger K, Keller W. Structure of allergens and structure based epitope predictions. Methods. 2013 Jul 23; doi: 10.1016/j.ymeth.2013.07.024. Excellent review of the structural biology of allergens and in particular the mapping the IgE epitopes.
- 4.Pomes A. Relevant B Cell Epitopes in Allergic Disease. Int Arch Allergy Imm. 2010;152(1):1–11. doi: 10.1159/000260078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radauer C, Breiteneder H. Evolutionary biology of plant food allergens. J Allergy Clin Immun. 2007 Sep;120(3):518–525. doi: 10.1016/j.jaci.2007.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Breiteneder H, Radauer C. A classification of plant food allergens. J Allergy Clin Immun. 2004 May;113(5):821–830. doi: 10.1016/j.jaci.2004.01.779. [DOI] [PubMed] [Google Scholar]
- 7. Chruszcz M, Maleki SJ, Majorek KA, Demas M, Bublin M, Solberg R, et al. Structural and immunologic characterization of Ara h 1, a major peanut allergen. J Biol Chem. 2011 Nov 11;286(45):39318–39327. doi: 10.1074/jbc.M111.270132. First structures of Ara h 1 and Ara h 3.
- 8. Cabanos C, Urabe H, Tandang-Silvas MR, Utsumi S, Mikami B, Maruyama N. Crystal structure of the major peanut allergen Ara h 1. Mol Immunol. 2011 Oct-Nov;49(1–2):115–123. doi: 10.1016/j.molimm.2011.08.004. First structures of Ara h 1 and Ara h 3.
- 9.Mueller GA, Gosavi RA, Pomes A, Wunschmann S, Moon AF, London RE, et al. Ara h 2: crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy. 2011 Jul;66(7):878–885. doi: 10.1111/j.1398-9995.2010.02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann K, Schweimer K, Reese G, Randow S, Suhr M, Becker WM, et al. Structure and stability of 2S albumin-type peanut allergens: implications for the severity of peanut allergic reactions. Biochem J. 2006 May 1;395(3):463–472. doi: 10.1042/BJ20051728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Fu TJ, Howard A, Kothary MH, McHugh TH, Zhang YZ. Crystal Structure of Peanut (Arachis hypogaea) Allergen Ara h 5. J Agr Food Chem. 2013 Feb 20;61(7):1573–1578. doi: 10.1021/jf303861p. [DOI] [PubMed] [Google Scholar]
- 12.Hurlburt BK, Offermann LR, McBride JK, Majorek KA, Maleki SJ, Chruszcz M. Structure and function of the peanut panallergen Ara h 8. J Biol Chem. 2013 Nov 19; doi: 10.1074/jbc.M113.517797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin T, Guo F, Chen YW, Howard A, Zhang YZ. Crystal structure of Ara h 3, a major allergen in peanut. Mol Immunol. 2009 May;46(8–9):1796–1804. doi: 10.1016/j.molimm.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 14.Dunwell JM. Cupins: A new superfamily of functionally diverse proteins that include germins and plant storage proteins. Biotechnol Genet Eng. 1998;15:1–32. doi: 10.1080/02648725.1998.10647950. [DOI] [PubMed] [Google Scholar]
- 15.Dunwell JM, Purvis A, Khuri S. Cupins: the most functionally diverse protein superfamily? Phytochemistry. 2004 Jan;65(1):7–17. doi: 10.1016/j.phytochem.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 16.Van Boxtel EL, Van Beers MMC, Koppelman SJ, Van den Broek LAM, Gruppen H. Allergen Ara h 1 occurs in peanuts as a large oligomer rather than as a trimer. J Agr Food Chem. 2006 Sep 20;54(19):7180–7186. doi: 10.1021/jf061433+. [DOI] [PubMed] [Google Scholar]
- 17.Maleki SJ, Chung SY, Champagne ET, Raufman JP. The effects of roasting on the allergenic properties of peanut proteins. J Allergy Clin Immun. 2000 Oct;106(4):763–768. doi: 10.1067/mai.2000.109620. [DOI] [PubMed] [Google Scholar]
- 18.Dodo HW, Viquez OM, Maleki SJ, Konan KN. cDNA clone of a putative peanut (Arachis hypogaea L.) trypsin inhibitor has homology with peanut allergens Ara h 3 and Ara h 4. J Agr Food Chem. 2004 Mar 10;52(5):1404–1409. doi: 10.1021/jf034765c. [DOI] [PubMed] [Google Scholar]
- 19.Guo B, Liang X, Chung SY, Maleki SJ. Proteomic screening points to the potential importance of Ara h 3 basic subunit in allergenicity of peanut. Inflamm Allergy Drug Targets. 2008 Sep;7(3):163–166. doi: 10.2174/187152808785748182. [DOI] [PubMed] [Google Scholar]
- 20.Guo BZ, Liang XQ, Chung SY, Holbrook CC, Maleki SJ. Proteomic analysis of peanut seed storage proteins and genetic variation in a potential peanut allergen. Protein Peptide Lett. 2008 Jul;15(6):567–577. doi: 10.2174/092986608784966877. [DOI] [PubMed] [Google Scholar]
- 21.Breiteneder H, Mills ENC. Plant food allergens - structural and functional aspects of allergenicity. Biotechnol Adv. 2005 Sep;23(6):395–399. doi: 10.1016/j.biotechadv.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008 Dec 1;24(23):2780–1. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maleki SJ, Viquez O, Jacks T, Dodo H, Champagne ET, Chung SY, et al. The major peanut allergen, Ara h 2, functions as a trypsin inhibitor, and roasting enhances this function. J Allergy Clin Immun. 2003 Jul;112(1):190–195. doi: 10.1067/mai.2003.1551. [DOI] [PubMed] [Google Scholar]
- 24.Koppelman SJ, Wensing M, Ertmann M, Knulst AC, Knol EF. Relevance of Ara h1, Ara h2 and Ara h3 in peanut-allergic patients, as determined by immunoglobulin E Western blotting, basophil-histamine release and intracutaneous testing: Ara h2 is the most important peanut allergen. Clin Exp Allergy. 2004 Apr;34(4):583–590. doi: 10.1111/j.1365-2222.2004.1923.x. [DOI] [PubMed] [Google Scholar]
- 25.Palmer GW, Dibbern DA, Burks AW, Bannon GA, Bock SA, Porterfield HS, et al. Comparative potency of Ara h 1 and Ara h 2 in immunochemical and functional assays of allergenicity. Clin Immunol. 2005 Jun;115(3):302–312. doi: 10.1016/j.clim.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 26.Stanley JS, King N, Burks AW, Huang SK, Sampson H, Cockrell G, et al. Identification and mutational analysis of the immunodominant IgE binding epitopes of the major peanut allergen Ara h 2. Arch Biochem Biophys. 1997 Jun 15;342(2):244–253. doi: 10.1006/abbi.1997.9998. [DOI] [PubMed] [Google Scholar]
- 27.Chen XN, Wang Q, El-Mezayen R, Zhuang YH, Dreskin SC. Ara h 2 and Ara h 6 Have Similar Allergenic Activity and Are Substantially Redundant. Int Arch Allergy Imm. 2013;160(3):251–258. doi: 10.1159/000341642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koid AE, Chapman MD, Hamilton RG, Van Ree R, Versteeg SA, Dreskin SC, et al. Ara h 6 Complements Ara h 2 as an Important Marker for IgE Reactivity to Peanut. J Agr Food Chem. 2013 Dec 11; doi: 10.1021/jf4022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Birbach A. Profilin, a multi-modal regulator of neuronal plasticity. Bioessays. 2008 Oct;30(10):994–1002. doi: 10.1002/bies.20822. [DOI] [PubMed] [Google Scholar]
- 30.Ramachandran S, Christensen HEM, Ishimaru Y, Dong CH, Chao-Ming W, Cleary AL, et al. Profilin plays a role in cell elongation, cell shape maintenance, and flowering in arabidopsis. Plant Physiol. 2000 Dec;124(4):1637–1647. doi: 10.1104/pp.124.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hauser M, Roulias A, Ferreira F, Egger M. Panallergens and their impact on the allergic patient. Allergy Asthma Clin Immunol. 2010 Jan;6(1):1. doi: 10.1186/1710-1492-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandes H, Michalska K, Sikorski M, Jaskolski M. Structural and functional aspects of PR-10 proteins. Febs J. 2013 Mar;280(5):1169–1199. doi: 10.1111/febs.12114. [DOI] [PubMed] [Google Scholar]
- 33.Kofler S, Asam C, Eckhard U, Wallner M, Ferreira F, Brandstetter H. Crystallographically Mapped Ligand Binding Differs in High and Low IgE Binding Isoforms of Birch Pollen Allergen Bet v 1. Journal of molecular biology. 2012 Sep 7;422(1):109–123. doi: 10.1016/j.jmb.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flicker S, Steinberger P, Norderhaug L, Sperr WR, Majlesi Y, Valent P, et al. Conversion of grass pollen allergen-specific human IgE into a protective IgG(1) antibody. Eur J Immunol. 2002 Aug;32(8):2156–2162. doi: 10.1002/1521-4141(200208)32:8<2156::AID-IMMU2156>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 35.Sicherer SH, Wood RA, Immunology SA. Allergy Testing in Childhood: Using Allergen-Specific IgE Tests. Pediatrics. 2012 Jan;129(1):193–197. doi: 10.1542/peds.2011-2382. [DOI] [PubMed] [Google Scholar]
- 36.Gupta RS, Dyer AA, Jain N, Greenhawt MJ. Childhood Food Allergies: Current Diagnosis, Treatment, and Management Strategies. Mayo Clin Proc. 2013 May;88(5):512–526. doi: 10.1016/j.mayocp.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Radauer C, Breiteneder H. Pollen allergens are restricted to few protein families and show distinct patterns of species distribution. J Allergy Clin Immun. 2006 Jan;117(1):141–147. doi: 10.1016/j.jaci.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 38. Bublin M, Kostadinova M, Radauer C, Hafner C, Szepfalusi Z, Varga EM, et al. IgE cross-reactivity between the major peanut allergen Ara h 2 and the nonhomologous allergens Ara h 1 and Ara h 3. J Allergy Clin Immun. 2013 Jul;132(1):U118–U212. doi: 10.1016/j.jaci.2013.01.022. This represents a new paradigm in the cross-reactivity of peanut allergens. It will be interesting to see if this generalizes to other allergen groups besides nuts and legumes.
- 39.Maleki SJ, Teuber SS, Cheng H, Chen D, Comstock SS, Ruan S, et al. Computationally predicted IgE epitopes of walnut allergens contribute to cross-reactivity with peanuts. Allergy. 2011 Dec;66(12):1522–1529. doi: 10.1111/j.1398-9995.2011.02692.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schein CH, Ivanciuc O, Braun W. Common physical-chemical properties correlate with similar structure of the IgE epitopes of peanut allergens. J Agr Food Chem. 2005 Nov 2;53(22):8752–8759. doi: 10.1021/jf051148a. [DOI] [PubMed] [Google Scholar]
- 41.Astier C, Morisset M, Roitel O, Codreanu F, Jacquenet S, Franck P, et al. Predictive value of skin prick tests using recombinant allergens for diagnosis of peanut allergy. J Allergy Clin Immun. 2006 Jul;118(1):250–256. doi: 10.1016/j.jaci.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 42.Flinterman AE, Knol EF, Lencer DA, Bardina L, Jager CFDH, Lin J, et al. Peanut epitopes for IgE and IgG4 in peanut-sensitized children in relation to severity of peanut allergy. J Allergy Clin Immun. 2008 Mar;121(3):737–743. doi: 10.1016/j.jaci.2007.11.039. [DOI] [PubMed] [Google Scholar]
- 43.Peeters KABM, Koppelman SJ, van Hoffen E, van der Tas CWH, Jager CFD, Penninks AH, et al. Does skin prick test reactivity to purified allergens correlate with clinical severity of peanut allergy? Clinical and Experimental Allergy. 2007 Jan;37(1):108–115. doi: 10.1111/j.1365-2222.2006.02628.x. [DOI] [PubMed] [Google Scholar]
- 44.Altmann F. The role of protein glycosylation in allergy. Int Arch Allergy Imm. 2007;142(2):99–115. doi: 10.1159/000096114. [DOI] [PubMed] [Google Scholar]
- 45.Aalberse RC, Koshte V, Clemens JGJ. Immunoglobulin-E Antibodies That Crossreact with Vegetable Foods, Pollen, and Hymenoptera Venom. J Allergy Clin Immun. 1981;68(5):356–364. doi: 10.1016/0091-6749(81)90133-0. [DOI] [PubMed] [Google Scholar]
- 46.Malandain H. IgE-reactive carbohydrate epitopes--classification, cross-reactivity, and clinical impact. Eur Ann Allergy Clin Immunol. 2005 Apr;37(4):122–128. [PubMed] [Google Scholar]
- 47.van Ree R, Cabanes-Macheteau M, Akkerdaas J, Milazzo JP, Loutelier-Bourhis C, Rayon C, et al. beta(1,2)-xylose and alpha(1,3)-fucose residues have a strong contribution in IgE binding to plant glycoallergens. Journal of Biological Chemistry. 2000 Apr 14;275(15):11451–11458. doi: 10.1074/jbc.275.15.11451. [DOI] [PubMed] [Google Scholar]
- 48.vanderVeen MJ, vanRee R, Aalberse RC, Akkerdaas J, Koopelman SJ, Jansen HM, et al. Poor biologic activity of cross-reactive IgE directed to carbohydrate determinants of glycoproteins. J Allergy Clin Immun. 1997 Sep;100(3):327–334. doi: 10.1016/s0091-6749(97)70245-8. [DOI] [PubMed] [Google Scholar]
- 49.Kochuyt AM, Van Hoeyveld EM, Stevens EAM. Prevalence and clinical relevance of specific immunoglobulin E to pollen caused by sting-induced specific immunoglobulin E to cross-reacting carbohydrate determinants in Hymenoptera venoms. Clinical and Experimental Allergy. 2005 Apr;35(4):441–447. doi: 10.1111/j.1365-2222.2005.02217.x. [DOI] [PubMed] [Google Scholar]
- 50.Dell A, Haslam SM, Morris HR, Khoo KH. Immunogenic glycoconjugates implicated in parasitic nematode diseases. Bba-Mol Basis Dis. 1999 Oct 8;1455(2–3):353–362. doi: 10.1016/s0925-4439(99)00064-2. [DOI] [PubMed] [Google Scholar]
- 51.van Die I, Gomord V, Kooyman FNJ, van den Berg TK, Cummings RD, Vervelde L. Core alpha 1 ->3-fucose is a common modification of N-glycans in parasitic helminths and constitutes an important epitope for IgE from Haemonchus contortus infected sheep. Febs Letters. 1999 Dec 10;463(1–2):189–193. doi: 10.1016/s0014-5793(99)01508-2. [DOI] [PubMed] [Google Scholar]
- 52.Shreffler WG, Castro RR, Kucuk ZY, Charlop-Powers Z, Grishina G, Yoo S, et al. The major glycoprotein allergen from Arachis hypogaea, Ara h 1, is a ligand of dendritic cell-specific ICAM-grabbing nonintegrin and acts as a Th2 adjuvant in vitro. J Immunol. 2006 Sep 15;177(6):3677–3685. doi: 10.4049/jimmunol.177.6.3677. [DOI] [PubMed] [Google Scholar]
- 53.Royer PJ, Emara M, Yang CX, Al-Ghouleh A, Tighe P, Jones N, et al. The Mannose Receptor Mediates the Uptake of Diverse Native Allergens by Dendritic Cells and Determines Allergen-Induced T Cell Polarization through Modulation of IDO Activity. J Immunol. 2010 Aug 1;185(3):1522–1531. doi: 10.4049/jimmunol.1000774. [DOI] [PubMed] [Google Scholar]
- 54.Wills-Karp M. Allergen-specific pattern recognition receptor pathways. Curr Opin Immunol. 2010 Dec;22(6):777–782. doi: 10.1016/j.coi.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hodge JE. The Amadori Rearrangement. Adv Carbohyd Chem. 1955;10:169–205. doi: 10.1016/s0096-5332(08)60392-6. [DOI] [PubMed] [Google Scholar]
- 56.Rabbani N, Thornalley PJ. Glycation research in amino acids: a place to call home. Amino Acids. 2012 Apr;42(4):1087–1096. doi: 10.1007/s00726-010-0782-1. [DOI] [PubMed] [Google Scholar]
- 57.Uribarri J, Woodruff S, Goodman S, Cai WJ, Chen X, Pyzik R, et al. Advanced Glycation End Products in Foods and a Practical Guide to Their Reduction in the Diet. J Am Diet Assoc. 2010 Jun;110(6):911–916. doi: 10.1016/j.jada.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chung SY, Champagne ET. Association of end-product adducts with increased IgE binding of roasted peanuts. J Agr Food Chem. 2001 Aug;49(8):3911–3916. doi: 10.1021/jf001186o. [DOI] [PubMed] [Google Scholar]
- 59.Mueller GA, Maleki SJ, Johnson K, Hurlburt BK, Cheng H, Ruan S, et al. Indentification of Maillard reaction products on panut allergens that influence binding to the receptor for advanced glycation end products. Allergy. 2013 doi: 10.1111/all.12261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chassaigne H, Norgaard JV, van Hengel AJ. Proteomics-based approach to detect and identify major allergens in processed peanuts by capillary LC-Q-TOF (MS/MS) J Agr Food Chem. 2007 May 30;55(11):4461–4473. doi: 10.1021/jf063630e. [DOI] [PubMed] [Google Scholar]
- 61.Hebling CM, McFarland MA, Callahan JH, Ross MM. Global Proteomic Screening of Protein Allergens and Advanced Glycation Endproducts in Thermally Processed Peanuts. J Agr Food Chem. 2012 Oct 16; doi: 10.1021/jf303554t. [DOI] [PubMed] [Google Scholar]
- 62.Petersen A, Rennert S, Kull S, Becker WM, Notbohm H, Goldmann T, et al. Roasting And Lipid Binding Provide Allergenic And Proteolytic Stability To The Peanut Allergen Ara H 8. Biol Chem. 2013 Sep 21; doi: 10.1515/hsz-2013-0206. [DOI] [PubMed] [Google Scholar]
- 63.Schmitt DA, Nesbit JB, Hurlburt BK, Cheng HP, Maleki SJ. Processing Can Alter the Properties of Peanut Extract Preparations. J Agr Food Chem. 2010 Jan 27;58(2):1138–1143. doi: 10.1021/jf902694j. [DOI] [PubMed] [Google Scholar]
- 64.Ilchmann A, Burgdorf S, Scheurer S, Waibler Z, Nagai R, Wellner A, et al. Glycation of a food allergen by the Maillard reaction enhances its T-cell immunogenicity: role of macrophage scavenger receptor class A type I and II. J Allergy Clin Immunol. 2010 Jan;125(1):175–183. e1–e11. doi: 10.1016/j.jaci.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 65.Buttari B, Profumo E, Capozzi A, Facchiano F, Saso L, Sorice M, et al. Advanced glycation end products of human beta(2) glycoprotein I modulate the maturation and function of DCs. Blood. 2011 Jun 9;117(23):6152–6161. doi: 10.1182/blood-2010-12-325514. [DOI] [PubMed] [Google Scholar]
- 66.Hilmenyuk T, Bellinghausen I, Heydenreich B, Ilchmann A, Toda M, Grabbe S, et al. Effects of glycation of the model food allergen ovalbumin on antigen uptake and presentation by human dendritic cells. Immunology. 2010 Mar;129(3):437–445. doi: 10.1111/j.1365-2567.2009.03199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zill H, Gunther R, Erbersdobler HF, Folsch UR, Faist V. RAGE expression and AGE-induced MAP kinase activation in Caco-2 cells. Biochem Bioph Res Co. 2001 Nov 16;288(5):1108–1111. doi: 10.1006/bbrc.2001.5901. [DOI] [PubMed] [Google Scholar]
- 68.Teodorowicz M, Fiedorowicz E, Kostyra H, Wichers H, Kostyra E. Effect of Maillard reaction on biochemical properties of peanut 7S globulin (Ara h 1) and its interaction with a human colon cancer cell line (Caco-2) Eur J Nutr. 2013 Jan 20; doi: 10.1007/s00394-013-0494-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Milutinovic PS, Alcorn JF, Englert JM, Crum LT, Oury TD. The Receptor for Advanced Glycation End Products Is a Central Mediator of Asthma Pathogenesis. Am J Pathol. 2012 Oct;181(4):1215–1225. doi: 10.1016/j.ajpath.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sukkar MB, Wood LG, Tooze M, Simpson JL, McDonald VM, Gibson PG, et al. Soluble RAGE is deficient in neutrophilic asthma and COPD. Eur Respir J. 2012 Mar;39(3):721–729. doi: 10.1183/09031936.00022011. [DOI] [PubMed] [Google Scholar]
- 71.Cheng CM, Tsuneyama K, Kominami R, Shinohara H, Sakurai S, Yonekura H, et al. Expression profiling of endogenous secretory receptor for advanced glycation end products in human organs. Modern Pathol. 2005 Oct;18(10):1385–1396. doi: 10.1038/modpathol.3800450. [DOI] [PubMed] [Google Scholar]
- 72.Chuah YK, Basir R, Talib H, Tie TH, Nordin N. Receptor for Advanced Glycation End Products and Its Involvement in Inflammatory Diseases. Int J Inflam. 2013;2013:403460. doi: 10.1155/2013/403460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hudson BI, Carter AM, Harja E, Kalea AZ, Arriero M, Yang HJ, et al. Identification, classification, and expression of RAGE gene splice variants. Faseb J. 2008 May;22(5):1572–1580. doi: 10.1096/fj.07-9909com. [DOI] [PubMed] [Google Scholar]
- 74.Mondoulet L, Dioszeghy V, Puteaux E, Ligouis M, Dhelft V, Letourneur F, et al. Intact skin and not stripped skin is crucial for the safety and efficacy of peanut epicutaneous immunotherapy (EPIT) in mice. Clin Transl Allergy. 2012;2(1):22. doi: 10.1186/2045-7022-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Beyer KB, Morrow E, Li XM, Bardina L, Bannon GA, Burks AW, et al. Effects of cooking methods on peanut allergenicity. J Allergy Clin Immun. 2001 Jun;107(6):1077–1081. doi: 10.1067/mai.2001.115480. [DOI] [PubMed] [Google Scholar]
- 76.Maleki SJ, Casillas AM, Kaza U, Wilson BA, Nesbit JB, Reimoneqnue C, et al. Differences among heat-treated, raw, commercial peanut extracts by skin testing and immunoblotting. Ann Allerg Asthma Im. 2010 Dec;105(6):451–457. doi: 10.1016/j.anai.2010.09.025. [DOI] [PubMed] [Google Scholar]
- 77.Pomes A, Butts CL, Chapman MD. Quantification of Ara h 1 in peanuts: why roasting makes a difference. Clin Exp Allergy. 2006 Jun;36(6):824–830. doi: 10.1111/j.1365-2222.2006.02490.x. [DOI] [PubMed] [Google Scholar]
- 78.Mondoulet L, Paty E, Drumare MF, Ah-Leung S, Scheinmann P, Willemot RM, et al. Influence of thermal processing on the allergenicity of peanut proteins. J Agr Food Chem. 2005 Jun 1;53(11):4547–4553. doi: 10.1021/jf050091p. [DOI] [PubMed] [Google Scholar]
- 79.Uribarri J, Cai W, Peppa M, Goodman S, Ferrucci L, Striker G, et al. Circulating glycotoxins and dietary advanced glycation endproducts: Two links to inflammatory response, oxidative stress, and aging. J Gerontol a-Biol. 2007 Apr;62(4):427–433. doi: 10.1093/gerona/62.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kellow NJ, Savige GS. Dietary advanced glycation end-product restriction for the attenuation of insulin resistance, oxidative stress and endothelial dysfunction: a systematic review. Eur J Clin Nutr. 2013 Mar;67(3):239–248. doi: 10.1038/ejcn.2012.220. [DOI] [PubMed] [Google Scholar]
- 81.Nesbit JB, Hurlburt BK, Schein CH, Cheng HP, Wei H, Maleki SJ. Ara h 1 structure is retained after roasting and is important for enhanced binding to IgE. Mol Nutr Food Res. 2012 Nov;56(11):1739–1747. doi: 10.1002/mnfr.201100815. [DOI] [PubMed] [Google Scholar]