Abstract
Multiple myeloma is a plasma cell neoplasm with an extremely variable clinical course. Animal models are needed to better understand its pathophysiology and for preclinical testing of potential therapeutic agents. Hematopoietic cells expressing the hypermorphic Rad50s allele show hematopoietic failure, which can be mitigated by the lack of a transcription factor, Mef/Elf4. However, we find that 70% of Mef−/−Rad50s/s mice die from multiple myeloma or other plasma cell neoplasms. These mice initially show an abnormal plasma cell proliferation and monoclonal protein production, and then develop anemia and a decreased bone mineral density. Tumor cells can be serially transplanted and according to array CGH and whole exome sequencing, the pathogenesis of plasma cell neoplasms in these mice is not linked to activation of a specific oncogene, or inactivation of a specific tumor suppressor. This model recapitulates the systemic manifestations of human plasma cell neoplasms, and implicates cooperativity between the Rad50s and Mef/Elf4 pathways in initiating myelomagenic mutations that promote plasma cell transformation.
Multiple myeloma is characterized by a slowly progressive monoclonal expansion of plasma cells within the bone marrow, which in most cases is accompanied by a serum monoclonal gammopathy and clinical complications including anemia, multiple bone lesions, nephropathy and frequent infections1,2. Although outcomes for myeloma patients have greatly improved, multiple myeloma remains an incurable disease, despite the availability of newer treatment modalities1.
Several mouse models of multiple myeloma and plasma cell neoplasms have been reported3, including xenograft models of human myeloma cells and transplantation models from spontaneously arising or chemically induced murine plasma cell neoplasms4,5,6. In addition, several transgenic mice have been reported to develop multiple myeloma and plasma cell neoplasms7,8,9; these mice were genetically modified to trigger the increased expression of genes, such as c-Myc, XBP-1, and MafB, which have been implicated in the initiation or progression of myeloma in humans. However, these models imperfectly mimic the human disease and while they recapitulate aspects of its clinical and pathogenic features, they are driven by already known genes10.
Non-homologous end-joining (NHEJ) pathways, which mainly participate in the repair of DNA double strand breaks (DSBs), contribute to class switch recombination (CSR) in B cells11,12. The MRN complex, which contains Mre11, Rad50, and Nbs1 in mammalian cells, functions as a sensor of DNA DSBs, regulating DSB repair through homologous recombination and NHEJ pathways, activating ATM as well as ATR signaling13. The MRN complex plays multiple roles during CSR in B cells; and is essential for its integrity14. The MRN complex also accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions15, thereby playing a critical role in humoral immunity mediated by B cells. Mutations in the Mre11, Rad50, and Nbs1 genes have been reported in B-cell non-Hodgkin lymphoma and may relate to their pathogenesis16,17. NHEJ has been implicated in the development of multiple myeloma, with whole genome sequencing of multiple myeloma samples identifying a mutation in the coding region of the Mre11 gene18,19, and gene expression profiling of multiple myeloma cells showing increased expression of NHEJ-related genes, such as Rad50 and Xrcc420.
Previously, we reported that hematopoietic cells expressing the hypermorphic Rad50s allele show constitutive ATM activation, leading to cancer predisposition and progressive hematopoietic failure in Rad50s/s mice21,22. In our attempt to mitigate this hematopoietic failure, we crossed Rad50s/s mice with a variety of mice that lacked cell cycle regulatory genes that may control hematopoietic stem or progenitor cell (HSPC) quiescence and found that the absence of the transcription factor Mef/Elf4 led to the greatest rescue of the hematopoietic failure23,24,25. Based on our previous study, the Mef−/− Rad50s/s mice showed lower number of B cells, myeloid cells, NK cells, and HSPCs than wild type controls25; however, serendipitously, we observed that many Mef−/− Rad50s/s mice died with plasma cell hyperproliferation, which prompted us to generate and more intensively analyze doubly modified mice.
In this study, we have analyzed the phenotypic and genomic abnormalities present in the Mef−/− Rad50s/s mice, establishing a novel and transplantable mouse model of multiple myeloma and plasma cell neoplasms which mimics the human disease and is not attributed to the activation of a specific oncogene or inactivation of a specific tumor suppressor gene (other than Mef). We have begun to clarify the mechanisms by which the Mef−/− Rad50s/s mice develop plasma cell neoplasms. We believe this mouse model will be useful for further analyzing disease initiation and progression, and for further pre-clinical screening of anti-myeloma compounds.
Results
Plasma cell neoplasms observed in the Mef −/− Rad50 s/s mice
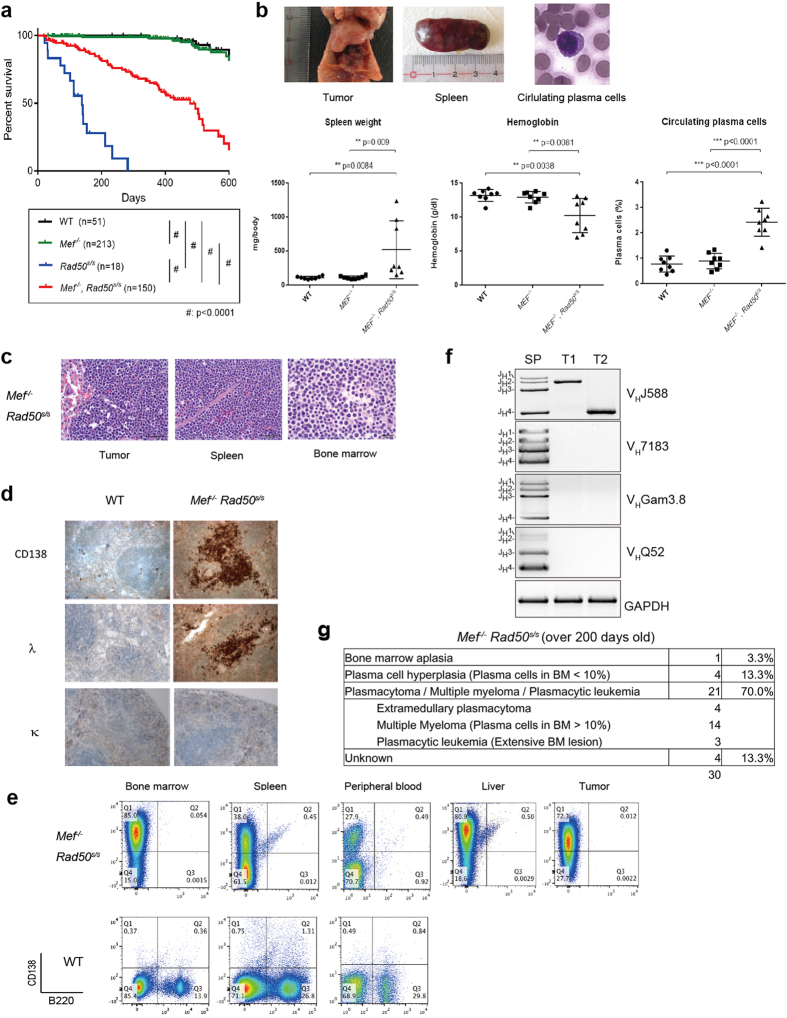
We analyzed the phenotypes of the Mef−/− Rad50s/s mice with wild type, Mef−/−, and Rad50s/s mice, and found that Mef−/− Rad50s/s mice have a longer survival than Rad50s/s mice, which is, nonetheless, much shorter than the survival of wild type mice (Fig. 1a). The median survival of the Mef−/− Rad50s/s mice was 478 days vs 138 days for the Rad50s/s mice, and when we examined Mef−/− Rad50s/s mice that were greater than 300 days old, many showed severe anemia, increased numbers of plasma cells in the peripheral blood, and/or tumor formation with splenomegaly (Fig. 1b, upper panels). We analyzed 10–12 month-old wild type, Mef−/−, and Mef−/− Rad50s/s mice, but not Rad50s/s mice, which die less than 10 months after birth (Fig. 1a), and found that Mef−/− Rad50s/s mice show statistically more anemia, circulating plasma cells, and splenomegaly, compared to the age-matched wild type and Mef−/− mice (Fig. 1b, lower panels). In addition, extramedullary tumors were observed in 3 of 15 Mef−/− Rad50s/s mice 300–500 day-old, but not in 20 age-matched wild type or 20 age-matched Mef−/− mice, with a significant difference (p = 0.023: wild type vs Mef−/− Rad50s/s, p = 0.023: Mef−/− vs Mef−/− Rad50s/s). Microscopically, we found an extensive infiltration of monotonous plasmacytoid cells throughout the spleen, bone marrow and peripheral blood (Fig. 1c). Immunohistochemical staining of the spleen showed that the plasmocyte-like cells were, in fact, B220 negative, CD138 positive plasma cells (Fig. 1d). We analyzed spleens, bone marrows, and tumors (if available) from 20 wild type, 20 Mef−/−, and 15 Mef−/− Rad50s/s mice between the age of 300 and 500 days old by immunohistochemistry. While aggregates of CD138+ B220− plasma cells were found in the spleen, bone marrow, and/or tumor in 12 Mef−/− Rad50s/s mice (80%), no plasma cell aggregates were observed in wild type or Mef−/− mice; this too represents a significant abnormality (p < 0.0001: Mef−/− Rad50s/s vs wild type, p < 0.0001: Mef−/− Rad50s/s vs Mef−/−). Although 95% of mouse B and plasma cells are normally κ light chain positive26, 4 of the 18 (22.2%) Mef−/− Rad50s/s mice that we analyzed by flow cytometry showed exclusive λ light chain positivity. On the other hand, 14 (77.8%) mice showed κ light chain positivity with λ exclusively negative. These imply an abnormal and likely monoclonal plasma cell proliferation (Supplementary Table S1 and Fig. 1d). When we analyzed cells from various Mef−/− Rad50s/s mouse tissues by flow cytometry, we found extensive involvement of multiple organs with CD138+ B220− plasma cells (Fig. 1e). These cells did not express cell surface CD3, CD4, or CD8, demonstrating that they are not T cells (data not shown). We also analyzed the immunoglobulin class in each mouse by flow cytometry and found that of the 19 mice analyzed, 16 showed IgG tumors, one mouse showed IgA, and 2 mice had no detectable immunoglobulin heavy chain (Supplementary Table S1).
Figure 1. Plasma cell neoplasms observed in the Mef−/− Rad50s/s mice.
(a) Kaplan-Meier curves showing the survival of wild type control, Mef−/−, Rad50s/s, and Mef−/− Rad50s/s mice. Mouse number of each group is demonstrated. (b) Macroscopic pathological findings of a representative 1-year-old Mef−/− Rad50s/s mouse and circulating plasma cells observed in Mef−/− Rad50s/s mice in the upper panels. Spleen weight (mg), hemoglobin concentration (g/dL), and frequency of circulating plasma cells in peripheral blood (%) were analyzed in 300–350-day-old wild type, Mef−/−, and Mef−/− Rad50s/s mice in the lower panel. P values between wild type and Mef−/− Rad50s/s mice, and Mef−/− and Mef−/− Rad50s/s mice are shown in each graph. (c) Histological images of tumor, spleen and bone marrow in a representative 1-year-old Mef−/− Rad50s/s mouse stained with hematoxylin and eosin at x400 magnification. (d) Immunohistochemical staining of the wild type and Mef−/− Rad50s/s spleens, using anti-CD138, anti-λ, and anti-κ antibodies. (e) Flow cytometric analysis of various tissues from 1-year-old Mef−/− Rad50s/s mice and age-matched wild type control mice. The profile of B220 and CD138 expression is shown. The number shows the frequency of each quadrant. (f) PCR detection of immunoglobulin gene rearrangements in tumor and normal spleen samples using VH family-specific forward primers and a reverse primer at JH4 to amplify variable-joining (VH-JH) regions of the IgH locus. An independent PCR assay amplifying a region of the GAPDH gene was performed for input control. A clear shift from multi-bands to mono-band can be observed in tumors only on the VHJ568 amplification, suggesting the monoclonality of the tumor. SP: spleen control from wild type mice; T1, T2: tumor from Mef−/− Rad50s/s mice. (g) Cause of death in Mef−/− Rad50s/s mice, as determined by pathology, immunohistochemistry, and flow cytometry.
For the clonality analysis, we performed V(D)J sequencing to examine the clonality of the plasma cell infiltration found in the Mef−/− Rad50s/s mice, using PCR to amplify the multiple variable-joining (VH-JH) regions of the IgH locus. We used the tumors from the Mef−/− Rad50s/s mice and the spleen control samples for this analysis, though we were not able to collect enough numbers of purified tumor cells from block samples because of technical limitations. While the PCR products from spleen tissue demonstrated multiple bands, which represent polyclonality, the PCR products from the tumor samples clearly showed a single band, only derived from the VHJ588 family, representing monoclonality (Fig. 1f). Monoclonal bands from three tumor samples were cloned and sequenced, and we confirmed that all (10 out of 10 sequenced) had the same monoclonal VH, DH, and JH usage with modest numbers of the same somatic mutations in the VH regions (Table 1). In addition, we performed PCR-based D-JH rearrangement PCR assays on tumor samples from several different Mef−/− Rad50s/s mice and found clonal but distinct D-JH rearrangements in the different mice (Supplementary Figure S1). Furthermore, to confirm the clonality of the plasma cells in Mef−/− Rad50s/s tumors, we looked for V(D)J rearrangement at the IgH locus by Southern blotting using a JH4-Eμ probe and found monoclonal VDJ rearrangement bands only in Mef−/− Rad50s/s tumors (Supplementary Figure S2); the control, wild type splenic B cells showed multiple VDJ rearrangement bands. Taken together, these data suggest that the plasma cell infiltrates and tumors in the Mef−/− Rad50s/s mice are monoclonal in nature and of post-germinal center origin.
Table 1. Mutation analysis of VDJ region in Mef−/− Rad50s/s plasma cell tumor samples.
| Mice UID | Tumor | VH | DH | JH | No.Mutationsat VH | Mutations atVH andAmion Acid Changes | Percent Mutationsat VH |
|---|---|---|---|---|---|---|---|
| 33 | MM | IGHV1-50*01, IGHV1-59*01 or IGHV1S40*01 | IGHD2-3*01 | IGHJ4*01 | 1 | c227 > g, A76 > G | 1.0 |
| 112 | MM | IGHV1-67*01 | IGHD4-1*01 | IGHJ3*01 | 2 | c227 > g, A76 > G a232 > t, M78 > L | 2.1 |
| 217 | PCT | IGHV1-67*01 | IGHD3-2*02 | IGHJ3*01 | 7 | c227 > g, A76 > G | 7.3 |
| a232 > t, M78 > L | |||||||
| g25 > c, S85 > T | |||||||
| a256 > t, T86 > S | |||||||
| a263 > t, Y88 > F | |||||||
| a290 > g, E97 > G | |||||||
| g291 > a, E97 > G |
The first two columns show mouse UID and the phenotype of the mice (MM, multiple myeloma; PCT, solitary plasmacytoma). The VH, DH, and JH usage and mutations were scored by comparing each sequence with the germline sequences at the IMGT server.
We determined the cause of death in many of the Mef−/− Rad50s/s mice (~85%), and found that ~15% of the Mef−/− Rad50s/s mice had severe bone marrow failure, while the remaining ~70% developed plasma cell dyscrasias, initially a syndrome resembling monoclonal gammopathy of unknown significance (MGUS), which unlike the human disorder, invariably progressed to multiple myeloma and even plasmacytic leukemia in some mice (Fig. 1g and Supplementary Table S1).
Progressive plasma cell neoplasms found in the Mef −/− Rad50 s/s mice
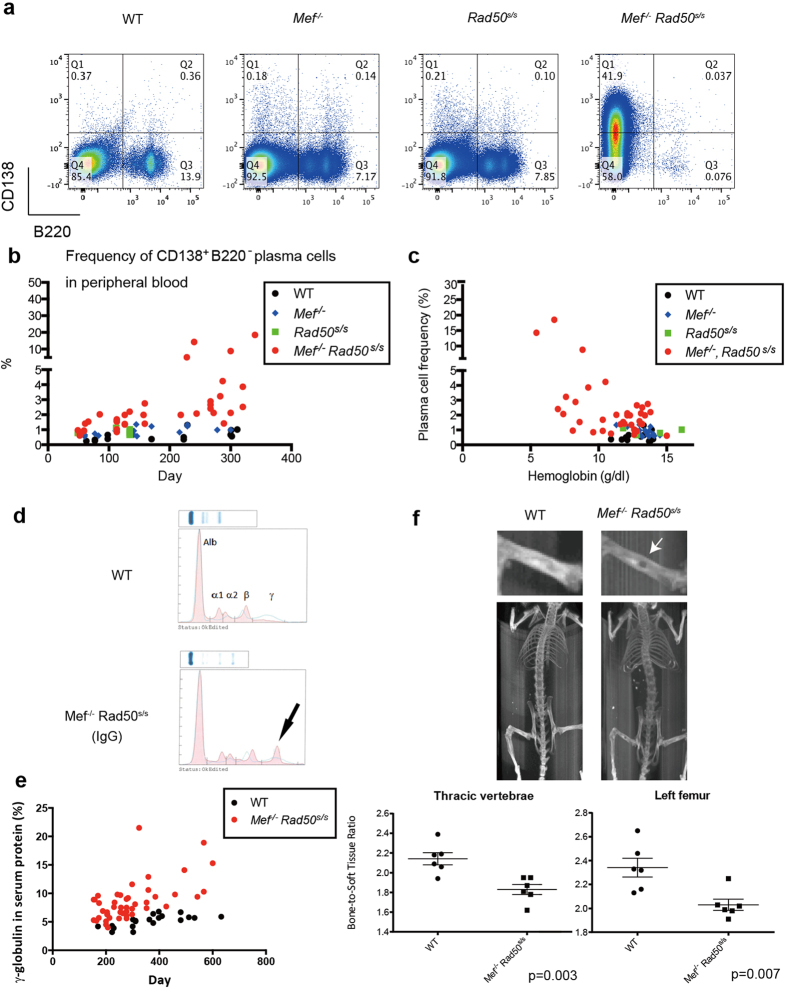
We analyzed the frequency of plasma cells in the bone marrow of younger mice (aged 150 to 200 days old) by flow cytometry, and while the bone marrow of the wild type, Mef−/−, and Rad50s/s mice contained <1% CD138+ B220− plasma cells (0.38 ± 0.10%, 0.47 ± 0.26%, and 0.67 ± 0.38%, respectively, n = 4 each), the Mef−/− Rad50s/s bone marrows contained significantly larger numbers of plasma cells (14.43 ± 3.41%, n = 4), at a time when the mice showed no signs of disease (Fig. 2a and Supplementary Figure S3). Similarly, the peripheral blood of the Mef−/− Rad50s/s mice had higher plasma cell frequencies and more absolute numbers of plasma cells than the other genotypes, with the plasma cell frequency increasing significantly with age (p value is 0.028, comparing the slopes of linear regression by AVCOVA: wild type vs Mef−/− Rad50s/s mice) (Fig. 2b). The Mef−/− Rad50s/s mice also had lower hemoglobin concentrations than the other mice (aged 200–300 days old, p = 0.0323: Mef−/− Rad50s/s vs wild type, p = 0,0156: Mef−/− Rad50s/s vs Mef−/−, n = 6 each), and the severity of the anemia significantly correlated with the number of plasma cells in the peripheral blood (p value is 0.015, comparing the slopes of linear regression by AVCOVA: wild type vs Mef−/− Rad50s/s mice) (Fig. 2c). We looked for monoclonal protein secretion, and found monoclonal peaks (“M spikes”) on the serum protein electrophoresis of all Mef−/− Rad50s/s mice with multiple myeloma or plasmacytic leukemia that were over 300 days old (Fig. 2d and Supplementary Table S1). We also performed serum protein electrophoresis, on younger Mef−/− Rad50s/s mice: 12 mice were less than 100 days, 8 were 100–200 days, and 9 were 200–300 days old. These mice were not anemic and they displayed no symptoms; none of the mice less than 100 days old had a monoclonal peak, while two (25%) of the 100–200-day-old mice and four (44%) of the 200–300-day-old mice showed monoclonal peaks. This suggests that clonal plasma cells gradually expand over time and produce more M protein. Similarly, serum γ-globulin levels were also significantly higher in the Mef−/− Rad50s/s mice, than the control mice, and γ-globulin levels increased as the mice aged (p value is 0.037, comparing slopes of linear regression by AVCOVA: wild type vs Mef−/− Rad50s/s mice) (Fig. 2e). The micro-vessel density in the Mef−/− Rad50s/sbone marrow was significantly higher than the bone marrow of the wild type controls (p = 0.010) (Supplementary Figure S4).
Figure 2. Progressive plasma cell neoplasms found in the Mef−/− Rad50s/s mice.
(a) The B220 and CD138 expression profile of bone marrow cells obtained from 6-month-old wild type control, Mef−/−, Rad50s/s, and Mef−/− Rad50s/s mice. The number shows the frequency of cells in each quadrant. (b) The frequency of CD138+ B220− plasma cells in the peripheral blood is plotted vs the age of the mice. P value is 0.028, comparing the slopes of linear regression by ANCOVA: wild type vs Mef−/− Rad50s/s mice. (c) The hemoglobin concentration (g/dL) is plotted against the frequency of CD138+ B220− plasma cells in the peripheral blood. P value is 0.015, comparing the slopes of linear regression by AVCOVA: wild type vs Mef−/− Rad50s/s mice. (d) Serum protein electrophoresis of 1-year-old wild type and Mef−/− Rad50s/s mice. The M-spike is indicated by the arrow. (e) The γ-globulin percentage of total serum protein is plotted vs the age of the corresponding mouse. P value is 0.037, comparing the slopes of linear regression by AVCOVA: wild type vs Mef−/− Rad50s/s mice. (f) Detection of osteolytic lesions by X-ray analysis of Mef−/− Rad50s/s mice (upper panels). The solitary osteolytic lesion is marked by the white arrow. The ratio of the bone density of the thoracic vertebrae and left femur, to the soft tissue density is calculated for the wild type and Mef−/− Rad50s/s mice (lower panels) (n = 6 each group). P values are 0.003 (thoracic vertebrae) and 0.007 (left femur), respectively.
We looked for the kinds of end organ damage that is observed in human multiple myeloma patients2, and analyzed the bone mineral density of the vertebra and femurs of the Mef−/− Rad50s/s mice using micro-CT. The micro-CT showed that 4 of the 6 Mef−/− Rad50s/smice (over 300 days old) had focal, lytic bone lesions, while none of the 6 wild type littermate controls had lytic lesions. We also calculated the ratio of the bone density to the soft tissue density of the thoracic vertebrae and left femurs in a variety of mice and observed significantly lower bone density in the Mef−/− Rad50s/s mice than in the control mice (P = 0.003 and 0.007 for the thoracic vertebrae and the left femur, respectively); thus Mef−/− Rad50s/s mice suffer from diffuse osteoporosis (Fig. 2f) and lytic bone lesions, similar to human multiple myeloma patients27. We also performed tartrate-resistant acid phosphatase and hematoxylin-eosin staining of the bones to detect osteoclasts in the Mef−/− Rad50s/s and wild type mice. The Mef−/− Rad50s/s mice showed significantly more tartrate-positive osteoclasts in the femur than did wild type mice (p = 0.013) (Supplementary Figure S5), findings that are also consistent with that seen in human multiple myeloma. We examined the kidneys of Mef−/− Rad50s/s mice, by Congo red fluorescence staining and found two of 12 Mef−/− Rad50s/s mice with amyloid deposition in their glomerulus, as demonstrated by apple-green birefringence (Supplementary Figure S6). These findings illustrate how Mef−/− Rad50s/s mice recapitulate the biological and clinical features of human multiple myeloma and plasma cell neoplasms. We also examined the chemosensitivity of the malignant plasma cells ex vivo and found that melphalan inhibited Mef−/− Rad50s/s plasma cell proliferation to a greater degree than control plasma cells (Supplementary Figure S7a).
Transplantability of Mef −/− Rad50 s/s plasma cell neoplasms
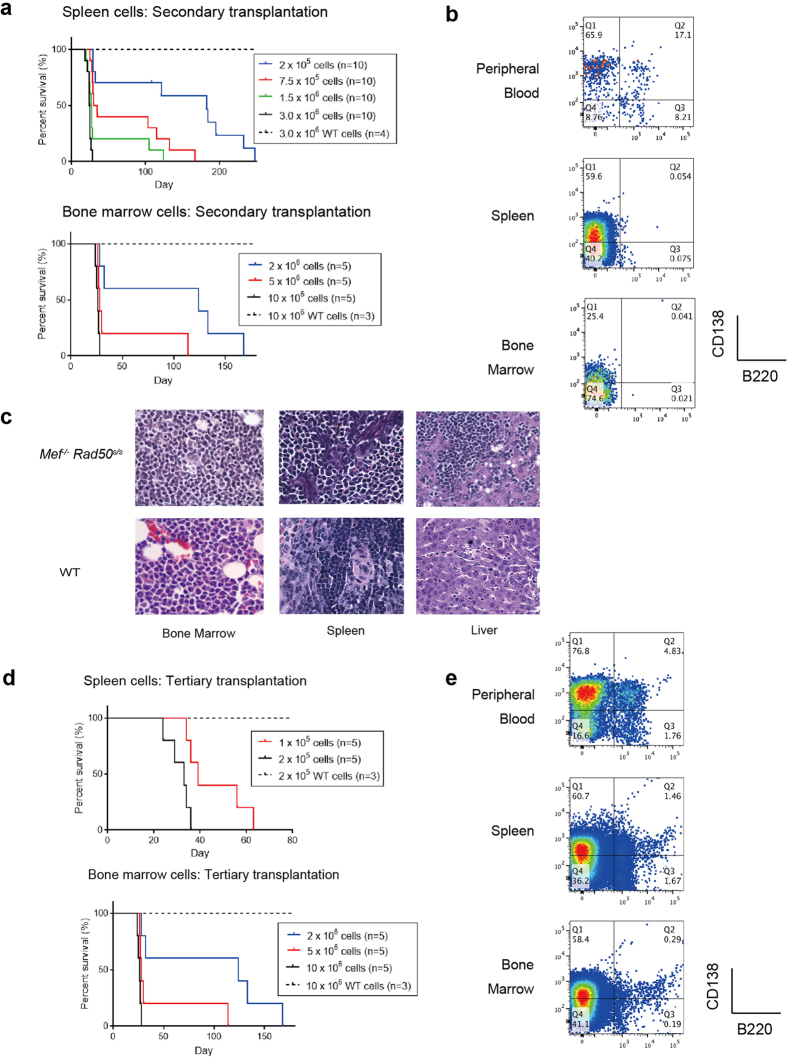
We next studied if the plasma cell neoplasms that we observe in Mef−/− Rad50s/s mice can be transplanted. Spleen and bone marrow cells from tumor-carrying Mef−/− Rad50s/s mice (400–450 days old) were injected into sub-lethally irradiated (4.75Gy) recipient mice in a dose-escalating manner. The recipient mice died within 25–35 days (Fig. 3a). Flow cytometric analysis, and histologic sectioning, demonstrated that the recipient mice suffered from the same plasma cell neoplasms as the original mice (Fig. 3b,c). We measured the bone density in four recipients of 2 × 105 Mef−/− Rad50s/s spleen cells and found a modest but significant decrease from 1.908 ± 0.062 to 1.695 ± 0.052 (p = 0.0043), suggesting that the decreased bone density is indeed driven by the plasma cell neoplasm. We also performed tertiary transplantation, using spleen or bone marrow cells collected from 3-week-old recipient mice that had received neoplastic Mef−/− Rad50s/s plasma cells. All sub-lethally irradiated recipient mice that received 2 × 105 spleen cells or 1 × 105 bone marrow cells again died within 25–35 days (Fig. 3d), of the same plasma cell disease (Fig. 3e). Fewer cells were needed for the tertiary transplant than the secondary transplant (Fig. 3d), indicating an enrichment for disease-initiating cells with each sequential transplantation.
Figure 3. Transplantability of Mef−/− Rad50s/s plasma cell neoplasms.
(a) The Kaplan-Meier curves showing survival after the secondary transplantation of a dose escalating number of spleen and bone marrow cells from tumor-carrying Mef−/− Rad50s/s mice or from wild type control mice. (b) The profile of B220 and CD138 expression in peripheral blood, spleen and bone marrow cells obtained from recipients 3 weeks after the secondary transplantation of Mef−/− Rad50s/s spleen cells. The number shows the frequency of cells in each quadrant. (c) The histology of the bone marrow, spleen and liver are shown in recipients 3 weeks after the secondary transplantation of Mef−/− Rad50s/s or wild type spleen cells, at x600 magnification. (d) The Kaplan-Meier curves showing survival after the tertiary transplantation of a dose escalating number of neoplastic spleen and bone marrow cells or wild type cells from control mice. (e) The profile of B220 and CD138 expression in peripheral blood, spleen and bone marrow cells obtained from recipients, 3 weeks after the tertiary transplantation of Mef−/− Rad50s/s spleen cells. The number shows the frequency of cells in each quadrant.
We also performed in vivo treatment of tertiary transplanted mice that had received 2 × 105 spleen cells from the secondary recipients of tumor-carrying Mef−/− Rad50s/s mice or age-matched wild type control mice, administered melphalan (2.5 mg/kg, day1–5) or bortezomib (0.5 mg/kg, day 1–4), and following their survival (Supplementary Figure S7b). Both melphalan and bortezomib, which are standard anti-myeloma drugs, significantly prolonged the survival of the mice that had received neoplastic Mef−/− Rad50s/s spleen cells, compared to the control vehicle (n = 6, p = 0.0016 and p = 0.0011, respectively). Chesi et al. reported that Bortezomib prolonged the survival of secondary or tertiary transplanted Vκ*MYC mice and observed a similar prolongation of survival28. Thus, our results of in vivo treatment of plasma cell neoplasms is comparable to that obtained using a distinct, transgenic mouse model of multiple myeloma.
Pathophysiology of Mef −/− Rad50 s/s plasma cell neoplasms
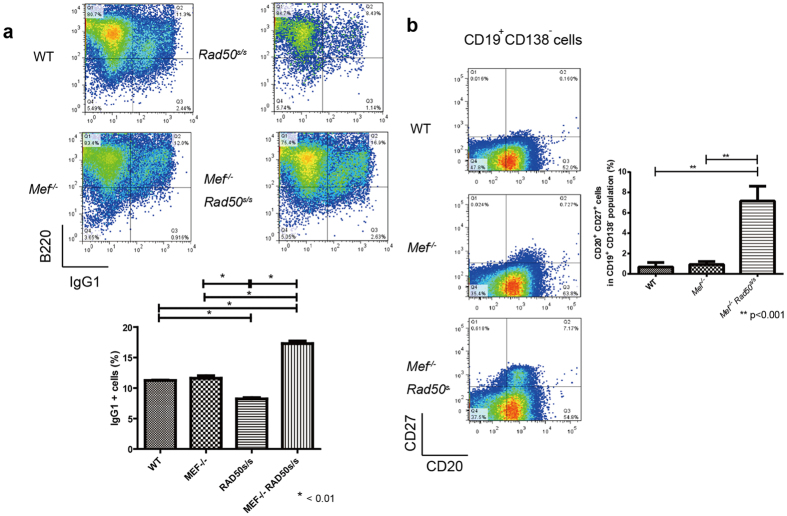
To address the underlying mechanisms by which the Mef−/− Rad50s/s mice develop plasma cell neoplasms, we analyzed the CSR capability of Mef−/− Rad50s/s B cells, using flow cytometric analysis of IgG1+ B cells after a 4 day ex vivo stimulation with anti-CD40 Ab (1 μg/mL) and IL-4 (100 ng/mL). Mef−/− Rad50s/s B cells showed more IgG1+ cells poststimulation than did the control, Mef−/−, or Rad50s/s B cells, demonstrating their enhanced CSR capacity (Fig. 4a). We performed V(D)J sequencing of Mef−/− Rad50s/s IgG1+ “normal” B cells following a 4 day ex vivo stimulation with anti-CD40 Ab and IL-4; the PCR products from these cells were polyclonal without specific V(D)J patterns, indicating normal ex vivo class switching.
Figure 4. Pathophysiology of Mef−/− Rad50s/s plasma cell neoplasms.
(a) Analysis of ex vivo B cell class switch recombination (CSR) of 6-month-old wild type control, Mef−/−, Rad50s/s, and Mef−/− Rad50s/s mice. IgG1+B cells present after a 96-hour stimulation with anti-CD40 Ab (1 μg/mL) and IL-4 (100 ng/mL), were analyzed by flow cytometry (upper panels). The histogram shows the percentage of stimulated IgG1+ B cells from wild type control, Mef−/−, Rad50s/s, and Mef−/− Rad50s/s mice (n = 4) (lower panel). (b) The profile of CD20 and CD27 expression on the splenic CD19+ CD138− B cells of 6-month-old wild type control, Mef−/−, and Mef−/− Rad50s/s mice (left panels). The histogram shows the percentage of CD20+ CD27+ cells in the splenic CD19+ CD138− B cells from wild type control, Mef−/−, and Mef−/− Rad50s/s mice (n = 4) (right panel).
When we phenotypically analyzed the splenic B cells in 200-day-old, apparently healthy control wild type, Mef−/−, Mef−/− Rad50s/s mice, we found many more CD20+ CD27+ CD19+ CD138− cells in the Mef−/− Rad50s/s spleen (Fig. 4b), cells that are thought to represent post germinal center memory B cells29. Mef deficient mice had more splenic B cells than control mice30, yet Mef deficiency itself had little impact on CSR or the size of the memory B cell compartment. Thus, Mef deficiency and Rad50s mutations work synergistically to induce plasma cell transformation.
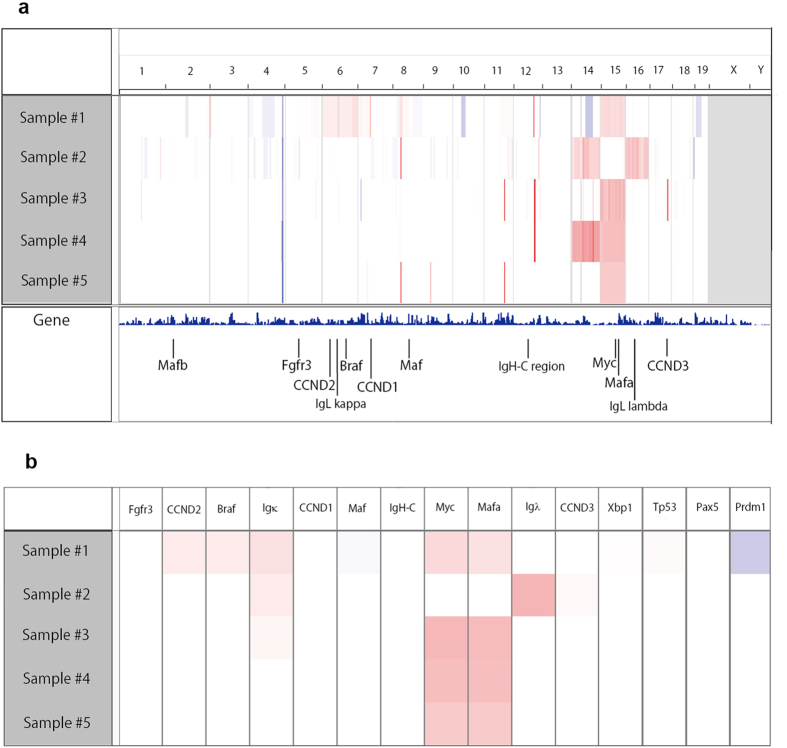
We analyzed the level of apoptosis in splenic plasma cells isolated from wild type and Mef−/− Rad50s/s 200-day-old mice, and found no difference in the frequency of apoptotic plasma cells (data not shown). We also compared Bcl2 and Bax mRNA expression in wild type plasma cells (n = 4), and Mef−/− Rad50s/s plasma cell tumors (n = 8) and found no significant differences, using a qPCR assay (Supplementary Figure S8). Thus, Mef−/− Rad50s/s plasma cells have no apparent change in their apoptotic threshold. Amplification, or dysregulated expression, of the c-Myc gene is thought to be important for the development of multiple myeloma and plasma cell neoplasms31,32, so we used array CGH to examine plasma cell samples from five independent Mef−/− Rad50s/s mice. Four samples showed high level amplification of chromosome 15, which includes the c-Myc gene (Fig. 5a), but we did not find translocation of c-Myc to the Ig locus by FISH analysis (data not shown).
Figure 5. Array CGH of plasma cell neoplasms in Mef−/− Rad50s/s mice.
(a) Array CGH data from 5 different tumor samples in Mef−/− Rad50s/s mice. Horizontal marks demonstrate the location of the chromosomes. Red and blue colors mean amplified and decreased locations, respectively. The locations of several myeloma-related genes are indicated below. (b) Gene amplification related to human myelomagenesis analyzed from 5 Mef−/− Rad50s/s tumor samples by array CGH. Red and blue colors mean amplified and decreased locations, respectively.
Of the other recurrently translocated genes observed in myeloma patients (CCND1, CCND3, MafB, Maf, Fgfr, and Mmset)33, we found CCND3 amplification in one sample (Fig. 5b). Nonetheless, CCND3 expression did not vary between wild type plasma cells (n = 4) and the Mef−/− Rad50s/s plasma cell tumors (n = 8) using qPCR assays. In contrast, CCND1 expression was significantly higher in the Mef−/− Rad50s/s plasma cell tumors than in wild type plasma cells (Supplementary Figure S9a,b). Increased expression of CCND genes is universally observed in MGUS and multiple myeloma, which can disrupt the E2F/RB pathway10,34. Taken together, these findings mirror the gene expression profiling studies that have compared human plasma cell neoplasm samples and normal human plasma cells35,36,37.
Myc expression in Mef −/− Rad50 s/s plasma cell neoplasms
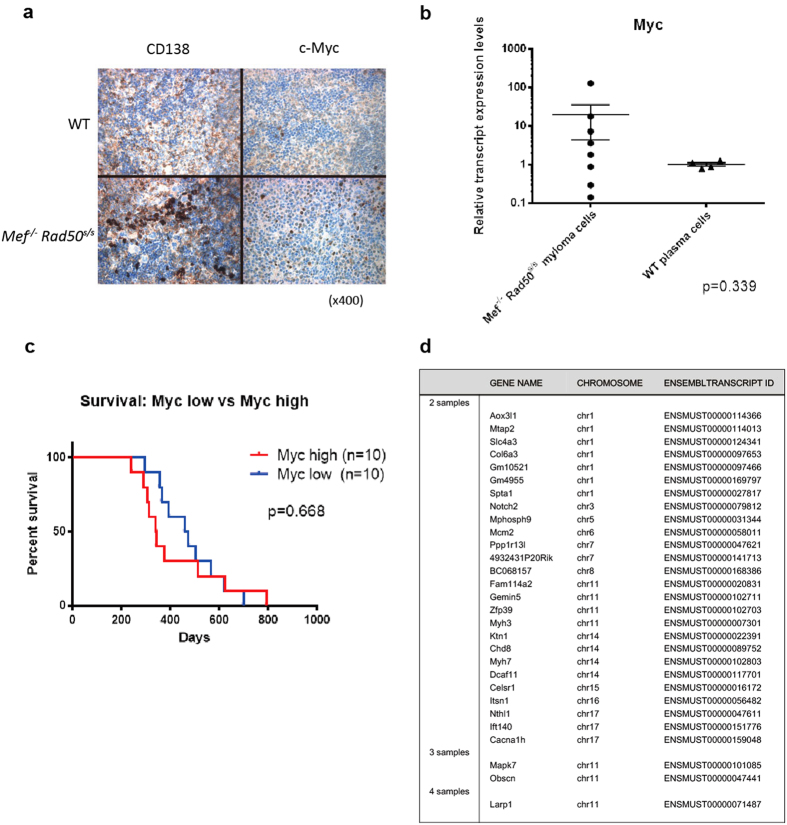
We also analyzed cell surface CD138 expression and intranuclear Myc expression in the spleen cells of eight affected Mef−/− Rad50s/smice (400–450 days old) by immunohistochemistry (Fig. 6a and Supplementary Figure S10). Five samples (62.5%) showed high Myc expression, while three showed no detectable Myc expression, even though CD138 was highly expressed. We also examined Myc mRNA expression levels in the Mef−/− Rad50s/s plasma cell tumors, using qPCR, and found variable Myc expression (both increased and decreased expression) compared to wild type plasma cells (Fig. 6b).
Figure 6. Myc expression in Mef−/− Rad50s/s mice.
(a) Immunohistochemical analysis of WT and Mef−/− Rad50s/s spleen sections stained with CD138 and c-Myc, observed at x400 magnification. (b) Myc transcript expression levels between WT plasma cells (n = 4) vs Mef−/− Rad50s/s plasma cell tumors (n = 8) using qPCR. P value is 0.339. (c) The survival of two distinct Mef−/− Rad50s/s mouse groups, classified by the level of Myc transcript expression. The Myc high group has more than twice the Myc transcript level of WT plasma cells, while the Myc low group has less than twice the Myc transcript level of WT plasma cells (n = 10 each group). P value is 0.668 by Log-rank test. (d) The result of the exome sequencing for Mef−/− Rad50s/s plasma cell tumor samples. List of the somatically mutated genes, found in 2, 3, or 4 different tumor samples. 4 Mef−/− Rad50s/s tumor and 5 WT tail samples were used for the exome sequencing.
We classified Mef−/− Rad50s/s tumor-bearing mice into two groups based on Myc mRNA levels, and examined their survival: we found no differences in the survival between the high Myc group (Myc expression 2 x greater than wild type plasma cells) and the low Myc group (where Myc expression was less than 2 x the wild type plasma cells). (Figure 6c). Thus, the Mef−/− Rad50s/s intracellular milieu generated plasma cell neoplasms with a variety of Myc expression levels, which were comparably aggressive. The plasma cell neoplasms that occur in the Mef−/− Rad50s/smice, also mimic what is found in human plasma cell neoplasms, where Myc overexpression is seen in some but not all patients31. We also measured Irf4, Prdm1, and Xbp-1 expression levels in the Mef−/− Rad50s/s plasma cell tumors, and found that the expression of these transcripts was not increased compared to wild type plasma cells (Supplementary Figure S9c–e). This too is similar to what is seen when human plasma cell neoplasms and normal plasma cells are compared36,37.
Gene mutations detected by whole exome sequencing found in Mef −/− Rad50 s/s plasma cell neoplasms
We performed whole exome sequencing of four plasma cell tumor samples obtained from Mef−/− Rad50s/s mice, using tail samples from five wild-type littermate mice as the germline controls. We found on average 204 exon mutations per sample (range 169–269). One mutated gene (Larp1) was seen in all four samples, two mutated genes (Obscn and Mapk7) were found in 3 of the 4 samples, and 26 mutated genes were found in 2 of the 4 samples (Fig. 6d). These included mutations in the following genes: Sptn1, Mphosph9, and Obscn, all of which have also been observed in human myeloma samples using whole genome sequencing19. We validated the mutations in the Larp1 gene, which were found in 4 samples from Mef−/− Rad50s/s mice and the mutations in the Mapk7 gene, which were found in 3 samples from Mef−/− Rad50s/s mice, confirming mutations in Larp1 for the amino acid changes, A170P, G304W, N593T and S807I, and mutations in Mapk7 for the amino acid changes, K107R, L167I, V237E, and L368V. Of the 569 genes where we found mutations, 55 genes were identified as having somatic mutations that affect protein-coding regions in human myeloma samples19 (Supplementary Table S2). Compared with the currently available genome sequencing data of human multiple myeloma, the plasma cell neoplasms derived from Mef−/− Rad50s/s mice possess a wider range of exon mutations, which may imply the absence of one dominant oncogene that drives pathogenesis of the observed plasma cell neoplasms19. To identify biologically relevant mutations, we performed functional annotation clustering, using DAVID software, and found ABC transporter, NF-κB signaling, Notch signaling, and focal adhesion signaling clusters to be significantly disturbed (Supplementary Table S3). This suggests that NF-κB signaling pathway genes among others, drive the pathogenesis of Mef−/− Rad50s/s driven plasma cell neoplasm, as these genes are also significantly mutated in human multiple myeloma samples19.
Discussion
The Mef−/− Rad50s/s mouse is a novel model of human multiple myeloma and plasma cell neoplasms; an abnormal proliferation of plasma cells is seen initially, accompanied by a monoclonal serum protein, mimicking MGUS or smoldering myeloma. However, the mice then develop progressive anemia and osteoporosis, indicative of full-blown myeloma. While MGUS is generally recognized as a premalignant condition that progresses to multiple myeloma at a rate of about 1 percent per year38, disease progression in this mouse model occurs with a much greater frequency. In fact, nearly all mice that do not succumb to hematopoietic failure, develop advanced multiple myeloma, or a related plasma cell neoplasm, with time. Thus, this mouse will be useful for studying MGUS and also plasmacytic leukemia, as nearly all of the tertiary transplant recipient mice die of plasmacytic leukemia.
Based on our observations, we hypothesize that the enhanced CSR seen in Mef−/− Rad50s/s mice facilitates the accumulation of post-germinal center memory B cells, which together with the genomic instability induced by Rad50s triggers oncogenic mutations leading to clonogenic plasma cell proliferation39. Genomic instability is induced during CSR by activation-induced cytidine deaminase (AID), which has been identified as an enzyme required for somatic hypermutation and CSR40. AID has oncogenic activity in post germinal center neoplasms8,41, thus further studies are needed to clarify whether the development of plasma cell neoplasms in Mef−/− Rad50s/s mice is dependent on AID activity. Subsequent mutations, or perhaps epigenetic events, may then trigger the progression to multiple myeloma, or a related plasma cell neoplasm. Clearly, our data suggest that the development of a plasma cell disease in the Mef−/− Rad50s/s mice is not necessarily linked to Myc overexpression, which is similar to the situation with human plasma cell neoplasms35,36,37.
The plasma cell neoplasms that we observe in the Mef−/− Rad50s/s mice appear to originate from post-germinal center memory B cells, which is consistent with our current understanding of plasma cell biology42. Several, recent studies have examined myeloma-initiating cells39. For instance, Matsui et al. reported that CD20+CD27+memory B cells isolated from multiple myeloma patients can give rise to clonogenic, multiple myeloma cell growth in vitro and engraftment in NOD/SCID mice29. The accumulation of post-germinal center, memory B cells likely reflects the earliest steps in the generation of clonogenic plasma cells in the Mef−/− Rad50s/s mice. Together with the chromosomal instability that is seen in the Rad50s/s mouse background, Mef−/− Rad50s/s B cells are clearly predisposed to transform into plasma cell malignancies21,25, likely because they accumulate multiple mutations after CSR.
Our studies of Mef/Elf4, and those of others, have identified both oncogenic and tumor suppressor activities43,44,45,46. Perhaps most relevant here are our studies of the role of Mef in the DNA damage response47. We found that the absence of Mef diminished the cell’s DNA damage response, leading to less activation of p53 and less γH2AX after irradiation. This could allow for a more modest repair of certain types of DNA damage, and also improved cell survival after cellular stress. Mef−/− Rad50s/s mice show a full spectrum of clinical plasma cell disorders, with a time course faster than other mouse models7,8. One possible reason for this, is that the Rad50s/s background is more tolerant to the acquisition of mutations. Another is that both Mef−/− mice30 and Mef−/− Rad50s/s mice have reduced numbers of NK and NK-T cells (data not shown), which may impair the immune response to the transformed plasma cells that are generated within the Mef−/− Rad50s/s mice.
We have summarized the characteristics of the Mef−/− Rad50s/s mouse model of multiple myeloma and plasma cell neoplasms, and compared it with other transgenic myeloma models and human multiple myeloma (Supplementary Table S4). Chang et al. published that chromosomal instability triggered by Rrm2b loss leads to plasma cell neoplasms48. In that model, the malignant cell is provoked by hyperactivation of pro-inflammatory cytokines, including IL-6. IL-6 transgenic mice have been reported to develop extramedullary plasmacytoma49. In contrast, neoplastic plasma cells derived from Mef−/− Rad50s/s mice do not proliferate ex vivo in IL-6 containing media and serum IL-6 levels are not elevated in Mef−/− Rad50s/s mice (data not shown). These findings suggest a lack of IL-6 dependence, and indicate a different mechanism of myelomagenesis, than that seen in the Rrm2b null mice.
Interactions between myeloma cells and various components of the bone marrow microenvironment play essential roles in tumor growth, survival, and drug resistance1. Increased angiogenesis is thought to be important for the proliferation and survival of myeloma cells, as well as for the disease progression50. Published work, using the 5T2MM myeloma mouse model, has demonstrated that multiple myeloma-initiating cells prefer a hypoxic bone marrow microenvironment; nonetheless, hypoxia is apparently lessened during disease progression from MGUS to multiple myeloma51. The micro-vessel density is increased in the Mef−/− Rad50s/s bone marrow of older mice, which likely supports the progressive growth of the neoplastic plasma cells. Future studies can address the importance of these interactions in our model.
The pathogenesis of the plasma cell neoplasm we observed is not linked to the activation of a specific oncogene nor inactivation of a specific tumor suppressor other than the lack of Mef. Its absence is clearly important because it allows Rad50s/s mice to survive long enough so they can develop a plasma cell expansion, but its absence clearly plays an important pathogenic role. We do not see increased plasma cells in the circulation or the bone marrow of the Rad50s/s mice, indicating that the alterations in Rad50 and Mef genes work synergistically to create a cellular environment that promotes plasma cell expansion and transformation. The bone marrow failure induced by Rad50s/s is partially mitigated by p21−/−, p27−/−, and Chk2−/− in addition to Mef−/−25, yet Rad50s/s mice generated on the p21−/−, p27−/−, or Chk2−/− background primarily develop lymphomas (Morales M et al. unpublished data), while the Rad50s/s mice generated on the Mef−/−background develop only plasma cell neoplasms. Thus, while the Rad50 hypermorphic status has strong oncogenic potential in the hematopoietic compartment, both Rad50 hypermorphic status and Mef deletion are needed to develop plasma cell neoplasms. Further, in-depth examination of Mef−/− Rad50s/s plasma cells will allow us to better define myeloma pathogenesis and screen for novel anti-myeloma compounds, or for factors that can delay the onset or progression of plasma cell disorders.
Methods
Mice
The generation of Mef−/− and Rad50s/s mice that were backcrossed to C57BL/6 five times was described previously21,30. All mice were maintained in the Memorial Sloan-Kettering Cancer Center (MSKCC) and University of Miami (UM) Animal Facility, according to IACUC (Institutional Animal Care and Use Committee)-approved protocols, and kept in Thorensten units with filtered germ-free air. All the studies were approved by IACUC of MSKCC and UM and experiments were conducted in accordance with the committee’s approved guidelines.
Pathological and immunohistochemical studies
Peripheral blood was collected from tail veins and analyzed on an automated blood counter, HEMAVET HV950FS (Drew Scientific). Tissue samples were fixed immediately after isolation and processed into paraffin, sectioned and examined histologically using hematoxylin and eosin, Congo red, or immunohistochemical techniques. Immunohistochemical staining was performed using the following anti-mouse antibodies: CD138 (281-2, BD Pharmingen), λ (SouthernBiotech), κ (SouthernBiotech), and c-Myc (Y-69, Abcam). Samples were reviewed by pathologists and diagnosed using uniform criteria.
Transplantation studies
Femoral and tibial bone marrow or splenic nucleated cells from the 400–450-day-old Mef−/− Rad50s/s mice with plasma cell neoplasms were injected intravenously into sub-lethally (4.75 Gy) irradiated 6–8 week-old C57BL/6 SJL (Jackson Lab) recipient mice.
Flow cytometry
The following antibodies were used in combinations: CD138, CD45R/B220, CD20, CD27, IgG1, κ and λ (BD Pharmingen). To detect intracellular κ and λ, we used an Intracellular Staining Kit (Invitrogen). Stained cells were analyzed by flow cytometry using FACScan, FACSCalibur (BD), MoFlo (Cytomation) or LSRII (BD).
Serum protein electrophoresis
Serum protein electrophoresis was performed using a SPIFE 3000 electrophoresis analyzer (Helena Laboratories).
Clonality and VH analysis
In order to perform the clonality analysis, we examined the tumor and the spleen control samples, though we were not able to collect enough numbers of purified tumor cells from tumor block samples because of technical limitations. We used PCR to amplify the variable-joining (VH-JH) region of the immunoglobulin heavy chain locus. For this goal the 5′ primers for VHJ558 (5′-RGCCTGGGRCTTCAGTGAAG-3′ or 5′-AAGGSCACAYTKACTGTAGAC-3′)52,53 (R = A+ G), VHGAM3.8: (5′-GAAGAA GCCTGGAGAGACAGTCAAGAT-3′), VHQ52: (5′ GCCCTCACAGAGCCTGTCCAT-3′), and VH7183: (5′TCCCTGAAACTCTCCTGTGCAGCCTC-3′) were combined with a 3′ primer for J4 (5′-GGAGACGGTGACTGAGGTTCC-3′) were combined with a 3′ primer for J4 (5′- GGAGACGGTGACTGAGGTTCC-3′). Each PCR reaction had a final volume of 25 μl containing 30ng genomic DNA (tumor or spleen), 1 μM of VHJ558, VHGAM3.8, VHQ52, or VH7183 primer and 0.22 μM of J4 primer. All amplifications were performed with AmpliTaq Gold (Applied Biosystems) with a 10 minute initial denaturation step at 95 oC followed by 11 cycles with 30 second denaturation (94 oC), 1 minute annealing (68 oC, −1 oC per cycle), and 1 minute extension (68 oC); and finally 30 cycles with 30 second denaturation (94 oC), 1 minute annealing (57 oC), and 1 minute extension (68 oC). The PCR products were analyzed in 1.5% Agarose gels with ethidium bromide and cloned into the pCR 2.1 vectors (Invitrogen) for DNA sequencing analysis. Primers used to amplify GAPDH gene were FW (5′- CACCTTCAGCTTTCCGGCCACTTAC-3′) and RV (5′- GGAAGCCCATCACCATCTTCCAGGA-3′). Sequences were performed by 3130xL Genetic Analyzer (Applied Biosystems) and analyzed using MacVector software, and the VH, DH, and JH usage and mutations were scored by comparing each sequence (10 sequences per sample) with the germline sequences at the IMGT server (http://www.imgt.org)54. Based on the nucleic acid numbers of the rearranged VH regions, we calculated the percentage of mutations, as described previously8.
Micro-CT images and measurement of bone mineral density
Mice over 300 days old age were used for this analysis. CT images were obtained using a microCAT-II scanner (ImTek, Knoxville, TN). The image data were processed by an ultra-fast volume reconstruction engine (Real-time Image Reconstruction System), as previously described55.
Array CGH
Genomic DNA was extracted from tissues by DNeasy Tissue & Blood Kit (Qiagen) and the SurePrint G3 Mouse CGH Microarray Kit, 1 × 1M (Agilent Technologies) was used for array CGH analysis. The acquired data were normalized using the MSKCC software and analyzed using the Integrated Genomics Viewer software.
Quantitative PCR
Quantitative PCR was performed by 7500 Fast Real-Time PCR System (Applied Biosystems), using RNA isolated from wild type plasma cells, which were sorted by mouse CD138+ Plasma Cell Isolation Kit (Miltenyi Biotec), and plasma cell tumors derived from Mef−/− Rad50s/s mice. Transcript expression levels were calculated and standardized by the ratio of each transcript vs Hprt. The following Taqman probes (Life Technologies) were used for quantitative PCR: Mm00487804_m1 (Myc).
Exome Sequencing
SureSelect Mouse All Exon Kit (Agilent Technologies) was used for enrichment of the entire mouse exome, and the 5500xl Genetic Analyzer (Applied Biosystems) was used for the sequencing. The BAM files were processed using the GATK toolkit, following the published best practice guidelines. They were first realigned using the InDel realigner and then the base quality values were recalibrated using the BaseQRecalibrator. Variants were then called using the GATK Unified Genotyper. The calls were filtered to remove any mutations scored as LowQual by the Unified Genotyper or with an alternative allele depth <5 reads. The filtered calls were annotated with SNPEff and synonymous mutations were also filtered out from the list. To make the final list from this list, we selected genes with >0.15 of the variant frequency and variants which could not be observed in control samples, and excluded identical variants at an identical base as artifacts.
Statistics
Statistical significance was assayed by Student’s t test (for two groups) and one-way ANOVA with Tukey’s multiple comparison test as a post test (for more than two groups). Survival analysis was performed by Log-rank test. Comparison of slopes of linear regression is performed by ANCOVA; *p < 0.05; **p < 0.01; ***p < 0.005; # < 0.0001; ns, not significant.
Additional Information
How to cite this article: Asai, T. et al. Generation of a novel, multi-stage, progressive, and transplantable model of plasma cell neoplasms. Sci. Rep. 6, 22760; doi: 10.1038/srep22760 (2016).
Supplementary Material
Acknowledgments
The authors would like to thank the staff of the Flow Cytometry, Molecular Cytology and the Genomics and Oncogenomics Core Facilities at the Sylvester Comprehensive Cancer Center and the core facility staffs at MSKCC for their assistance. We also thank the members of the Nimer lab for helpful comments. This work was funded by NIH RO1 grant (DK52208 to S.D.N.), R37GM59413 and RO1-GM56888 (J.H.J.P.), a grant from the Geoffrey Beene Foundation (S.D.N. and T.A.), and support from the Mel Stottlemyre Multiple Myeloma Fund and the Norma and Gordon Smith Cancer Research Foundation.
Footnotes
The authors declare no competing financial interests.
Author Contributions T.A., J.P. and S.D.N. designed the research. T.A., M.A.H., C.L., D.N.L., A.D., K.M., M.F., E.M.C. and R.E.V. performed experiments and analyzed data. T.A. and S.D.N. designed figures and wrote the manuscript.
References
- Palumbo A. & Anderson K. Multiple myeloma. N Engl J Med 364, 1046–60 (2011). [DOI] [PubMed] [Google Scholar]
- Bird J. M. et al. Guidelines for the diagnosis and management of multiple myeloma 2011. Br J Haematol. 154, 32–75 (2011). [DOI] [PubMed] [Google Scholar]
- Mitsiades C. S., Anderson K. C. & Carrasco D. R. Mouse models of human myeloma. Hematol Oncol Clin North Am. 21, 1051–69, viii (2007). [DOI] [PubMed] [Google Scholar]
- Yaccoby S., Barlogie B. & Epstein J. Primary myeloma cells growing in SCID-hu mice: a model for studying the biology and treatment of myeloma and its manifestations. Blood 92, 2908–13 (1998). [PubMed] [Google Scholar]
- Pilarski L. M. et al. Myeloma progenitors in the blood of patients with aggressive or minimal disease: engraftment and self-renewal of primary human myeloma in the bone marrow of NOD SCID mice. Blood 95, 1056–65 (2000). [PubMed] [Google Scholar]
- Asosingh K., Radl J., Van Riet I., Van Camp B. & Vanderkerken K. The 5TMM series: a useful in vivo mouse model of human multiple myeloma. Hematol J 1, 351–6 (2000). [DOI] [PubMed] [Google Scholar]
- Carrasco D. R. et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 11, 349–60 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesi M. et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell 13, 167–80 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Duenas C. et al. A novel molecular mechanism involved in multiple myeloma development revealed by targeting MafB to haematopoietic progenitors. EMBO J 31, 3704–17 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl W. M. & Bergsagel P. L. Molecular pathogenesis of multiple myeloma and its premalignant precursor. J Clin Invest 122, 3456–63 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corneo B. et al. Rag mutations reveal robust alternative end joining. Nature 449, 483–6 (2007). [DOI] [PubMed] [Google Scholar]
- Yan C. T. et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–82 (2007). [DOI] [PubMed] [Google Scholar]
- Stracker T. H. & Petrini J. H. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 12, 90–103 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinkelmann M. et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol 16, 808–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki M., Fujii M. M. & Maizels N. The MRE11-RAD50-NBS1 complex accelerates somatic hypermutation and gene conversion of immunoglobulin variable regions. Nat Immunol 6, 730–6 (2005). [DOI] [PubMed] [Google Scholar]
- Rollinson S., Kesby H. & Morgan G. J. Haplotypic variation in MRE11, RAD50 and NBS1 and risk of non-Hodgkin’s lymphoma. Leuk Lymphoma 47, 2567–83 (2006). [DOI] [PubMed] [Google Scholar]
- Schuetz J. M. et al. Genetic variation in the NBS1, MRE11, RAD50 and BLM genes and susceptibility to non-Hodgkin lymphoma. BMC Med Genet 10, 117 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Betti C., Singh S., Toor A. & Vaughan A. Impaired NHEJ function in multiple myeloma. Mutat Res 660, 66–73 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M. A. et al. Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–72 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roddam P. L. et al. Non-homologous end-joining gene profiling reveals distinct expression patterns associated with lymphoma and multiple myeloma. Br J Haematol 149, 258–62 (2010). [DOI] [PubMed] [Google Scholar]
- Bender C. F. et al. Cancer predisposition and hematopoietic failure in Rad50(S/S) mice. Genes Dev 16, 2237–51 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales M. et al. The Rad50S allele promotes ATM-dependent DNA damage responses and suppresses ATM deficiency: implications for the Mre11 complex as a DNA damage sensor. Genes Dev 19, 3043–54 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y., Sun X., Uchida H., Zhang J. & Nimer S. MEF, a novel transcription factor with an Elf-1 like DNA binding domain but distinct transcriptional activating properties. Oncogene 13, 1721–9 (1996). [PubMed] [Google Scholar]
- Lacorazza H. D. et al. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell 9, 175–87 (2006). [DOI] [PubMed] [Google Scholar]
- Morales M. et al. DNA damage signaling in hematopoietic cells: a role for Mre11 complex repair of topoisomerase lesions. Cancer Res 68, 2186–93 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman J. R. et al. The Ig(kappa) enhancer influences the ratio of Ig(kappa) versus Ig(lambda) B lymphocytes. Immunity 5, 241–52 (1996). [DOI] [PubMed] [Google Scholar]
- Terpos E. & Dimopoulos M. A. Myeloma bone disease: pathophysiology and management. Ann Oncol 16, 1223–31 (2005). [DOI] [PubMed] [Google Scholar]
- Chesi M. et al. Drug response in a genetically engineered mouse model of multiple myeloma is predictive of clinical efficacy. Blood 120, 376–85 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui W. et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res 68, 190–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacorazza H. D. et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity 17, 437–49 (2002). [DOI] [PubMed] [Google Scholar]
- Chng W. J. et al. Clinical and biological implications of MYC activation: a common difference between MGUS and newly diagnosed multiple myeloma. Leukemia 25, 1026–35 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Largo C. et al. Identification of overexpressed genes in frequently gained/amplified chromosome regions in multiple myeloma. Haematologica 91, 184–91 (2006). [PubMed] [Google Scholar]
- Bergsagel P. L. & Kuehl W. M. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol 23, 6333–8 (2005). [DOI] [PubMed] [Google Scholar]
- Bergsagel P. L. et al. Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood 106, 296–303 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan F. et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood 99, 1745–57 (2002). [DOI] [PubMed] [Google Scholar]
- Zhan F. et al. Gene-expression signature of benign monoclonal gammopathy evident in multiple myeloma is linked to good prognosis. Blood 109, 1692–700 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnelli L. et al. A SNP microarray and FISH-based procedure to detect allelic imbalances in multiple myeloma: an integrated genomics approach reveals a wide gene dosage effect. Genes Chromosomes Cancer 48, 603–14 (2009). [DOI] [PubMed] [Google Scholar]
- Kyle R. A. et al. Prevalence of monoclonal gammopathy of undetermined significance. N Engl J Med 354, 1362–9 (2006). [DOI] [PubMed] [Google Scholar]
- Hajek R., Okubote S. A. & Svachova H. Myeloma stem cell concepts, heterogeneity and plasticity of multiple myeloma. Br J Haematol 163, 551–64 (2013). [DOI] [PubMed] [Google Scholar]
- Muramatsu M. et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102, 553–63 (2000). [DOI] [PubMed] [Google Scholar]
- Ramiro A. R. et al. AID is required for c-myc/IgH chromosome translocations in vivo. Cell 118, 431–8 (2004). [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams L. J. & McHeyzer-Williams M. G. Antigen-specific memory B cell development. Annu Rev Immunol 23, 487–513 (2005). [DOI] [PubMed] [Google Scholar]
- Seki Y. et al. The ETS transcription factor MEF is a candidate tumor suppressor gene on the X chromosome. Cancer Res 62, 6579–86 (2002). [PubMed] [Google Scholar]
- Yao J. J. et al. Tumor promoting properties of the ETS protein MEF in ovarian cancer. Oncogene 26, 4032–7 (2007). [DOI] [PubMed] [Google Scholar]
- Sashida G. et al. ELF4/MEF activates MDM2 expression and blocks oncogene-induced p16 activation to promote transformation. Mol Cell Biol 29, 3687–99 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzoli E. et al. MEF promotes stemness in the pathogenesis of gliomas. Cell Stem Cell 11, 836–44 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashida G. et al. The mef/elf4 transcription factor fine tunes the DNA damage response. Cancer Res 71, 4857–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L., Guo R., Huang Q. & Yen Y. Chromosomal instability triggered by Rrm2b loss leads to IL-6 secretion and plasmacytic neoplasms. Cell Rep 3, 1389–97 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalchuk A. L. et al. IL-6 transgenic mouse model for extraosseous plasmacytoma. Proc Natl Acad Sci USA 99, 1509–14 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca A. & Ribatti D. Bone marrow angiogenesis in multiple myeloma. Leukemia 20, 193–9 (2006). [DOI] [PubMed] [Google Scholar]
- Asosingh K. et al. Role of the hypoxic bone marrow microenvironment in 5T2MM murine myeloma tumor progression. Haematologica 90, 810–7 (2005). [PubMed] [Google Scholar]
- Andersen S. et al. Monoclonal B-cell hyperplasia and leukocyte imbalance precede development of B-cell malignancies in uracil-DNA glycosylase deficient mice. DNA Repair (Amst) 4, 1432–41 (2005). [DOI] [PubMed] [Google Scholar]
- Keenan R. A. et al. Censoring of autoreactive B cell development by the pre-B cell receptor. Science 321, 696–9 (2008). [DOI] [PubMed] [Google Scholar]
- Brochet X., Lefranc M. P. & Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36, W503–8 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. F., Zanzonico P., Ling C. C. & O’Donoghue J. Visualization of experimental lung and bone metastases in live nude mice by X-ray micro-computed tomography. Technol Cancer Res Treat 5, 147–55 (2006). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.