Abstract
The impact of glycopeptide resistance on the molecular regulation of Staphylococcus aureus virulence and attachment to host tissues is poorly documented. We compared stable teicoplanin-resistant methicillin-resistant S. aureus (MRSA) strain 14-4 with its teicoplanin-susceptible MRSA parent, strain MRGR3, which exhibits a high degree of virulence in a rat model of chronic foreign body MRSA infection. The levels of fibronectin-mediated adhesion and surface display of fibronectin-binding proteins were higher in teicoplanin-resistant strain 14-4 than in its teicoplanin-susceptible parent or a teicoplanin-susceptible revertant (strain 14-4rev) that spontaneously emerged during tissue cage infection. Quantitative reverse transcription-PCR (qRT-PCR) showed four- and twofold higher steady-state levels of fnbA and fnbB transcripts, respectively, in strain 14-4 than in its teicoplanin-susceptible counterparts. Analysis of global regulatory activities by qRT-PCR revealed a strong reduction in the steady-state levels of RNAIII and RNAII in the teicoplanin-resistant strain compared to in its teicoplanin-susceptible counterparts. In contrast, sarA mRNA levels were more than fivefold higher in strain 14-4 than in MRGR3 and 14-4rev. Furthermore, the alternative transcription factor sigma B had a higher level of functional activity in the teicoplanin-resistant strain than in its teicoplanin-susceptible counterparts, as evidenced by significant increases in both the sigma B-dependent asp23 mRNA levels and the sarA P3 promoter-derived transcript levels, as assayed by qRT-PCR and Northern blotting, respectively. These data provide further evidence that the emergence of glycopeptide resistance is linked by still poorly understood molecular pathways with significant pleiotropic changes in the expression and regulation of some major virulence genes. These molecular and phenotypic changes may have a profound impact on the bacterial adhesion and colonization properties of such multiresistant organisms.
The recent emergence of multidrug-resistant isolates of methicillin-resistant Staphylococcus aureus (MRSA) exhibiting decreased susceptibilities to glycopeptides (glycopeptide-intermediate S. aureus [GISA]) presents multiple challenges for antimicrobial therapy, antimicrobial susceptibility testing, and hospital infection control (18, 26, 27, 57, 58, 60, 66). In vitro observations indicate a stepwise development of resistance to vancomycin, which initially occurs in very small subpopulations that gradually expand during glycopeptide exposure in vitro (25, 47, 58) and in vivo (27, 40, 42, 48, 57, 58). Compared to vancomycin, teicoplanin can much more easily select first-step mutants or mutants for which MICs are higher and has clearly been associated with clinical treatment failures (22).
Recent genomic studies of gene transcription by microarray analysis support the notion that multiple genes and, likely, multiple metabolic pathways have been altered (30, 31, 39) in S. aureus isolates exhibiting decreased susceptibilities to glycopeptides. These changes in the S. aureus transcriptome likely contribute to the multiple alterations in the morphology and global regulation of cell wall synthesis, composition, and turnover frequently observed in GISA strains (18, 61, 66).
The ability of S. aureus to cause a wide spectrum of infections in humans and animals is attributable to the wide array of extracellular and cell wall-associated virulence determinants that are coordinately expressed in this organism (32, 49). S. aureus strains display a variety of surface protein adhesins collectively known as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs) (46). Several plasma or extracellular matrix host proteins are recognized by MSCRAMMs (46). Fibronectin-binding protein (FnBP) A (FnBPA) and FnBPB, encoded by the fnbA and fnbB genes, respectively, play prominent roles in S. aureus attachment and colonization of host tissues or implanted biomaterials (14, 23, 64). FnBPs also promote endocytic uptake of S. aureus by epithelial and endothelial cell lines and fibroblasts (16, 59).
The regulation of genes coding for extracellular and cell wall-associated virulence determinants in S. aureus involves an intricate network of global regulators (11, 45), in particular, agr and sarA. The agr locus encodes a two-component octapeptide-mediated quorum-sensing system that is involved in the generation of two divergent transcripts, RNAII and RNAIII, from two distinct promoters, P2 and P3, respectively. RNAIII mediates the regulation of numerous target genes (13), in particular, by downregulating several cell wall-associated proteins and upregulating the production of extracellular virulence factors during the postexponential phase (6, 11, 45). The sarA locus is composed of three overlapping transcripts of 0.58, 0.84, and 1.15 kb initiated from the P1, P3, and P2 promoters, respectively, coding for a single 14.5-kDa DNA-binding protein. Like agr, the sarA locus was shown by transcription profiling studies to influence the expression of >100 S. aureus genes (11). The SarA protein can either directly regulate several target genes (7, 29, 67), e.g., hla (alpha-hemolysin gene), spa (protein A gene), fnbA, cna, and sspA, or indirectly influence target gene expression by changing the P2 and/or P3 agr promoter activities (9, 41). In addition, several sar homologues (sarH1, sarR, sarT, sarU, and rot) (34, 36, 37, 51, 56) and two-component global regulons (e.g., arlRS and srrAB) (15, 69) may also regulate major virulence genes either directly or via the global regulators agr or sarA, or both (45).
An important mechanism of cell adaptation to environmental stress that shows some homology between Bacillus subtilis and S. aureus is the alternative transcription factor sigma B (SigB). The sigB operon of S. aureus is composed of four open reading frames (rsbU, rsbV, rsbW, and sigB) that are structurally and functionally homologous to those of B. subtilis (2, 10, 20, 28, 38, 68). The activity of SigB in both species is controlled posttranslationally by a multicomponent signal transduction system that involves proteins RsbU, RsbV, and RsbW. The SigB factor, which is normally sequestered by the anti-sigma factor RsbW, may be released from the SigB-RsbW complex by transduction of external signals mediated by RsbU (a phosphatase) that dephosphorylates RsbV, thus permitting its interaction with RsbW. A number of virulence-associated target genes, such as asp23, coa, clfA, sarH1, and the P3 promoter of sarA, are reported to be transcriptionally regulated by the level of free SigB (28, 43, 44).
Bacterial exposure to antimicrobial agents represents a most stressful situation for which a fast adaptive response is required for early survival before the eventual emergence of more stable antibiotic resistance determinants. This adaptive response may be mediated by upregulation of efflux pumps and stress-triggered responses involving various repair systems as well as global regulators of virulence and housekeeping genes. Subinhibitory concentrations of antibiotics may downregulate or upregulate specific adhesins (46) or secreted virulence factors (24) of S. aureus, such as FnBPs, collagen-binding protein, or alpha-toxin. It has also been shown that the emergence or acquisition of antibiotic resistance determinants by S. aureus, such as methicillin or glycopeptide resistance, can significantly alter the expression of global regulators and virulence factors, such as agr (52, 53) and alpha-toxin (53), and can downregulate the expression of cell wall-associated surface components, such as clumping factor A and FnBPs (54, 65). In some extreme situations, subinhibitory concentrations of some antibiotics, such as ciprofloxacin, can, paradoxically, increase the levels of expression of FnBPs and potentially provide a survival advantage for quinolone-resistant S. aureus strains (4, 5).
An experimental model of foreign body infections with MRSA which promoted the emergence of subpopulations expressing teicoplanin resistance and which eventually led to the selection of stable highly teicoplanin-resistant strain 14-4 was recently described (62). The presence of the teicoplanin-resistant strain and its teicoplanin-susceptible isogenic parent offered a unique opportunity to evaluate under well-controlled conditions the impact of glycopeptide resistance on the regulation and expression of some major virulence genes, with a particular focus on the FnBPs, which play an important role in bacterial adhesion, colonization, and infection (64).
MATERIALS AND METHODS
Bacterial strains.
MRSA strain MRGR3 was isolated in 1979 from a patient with catheter-related sepsis and was selected for its virulence properties in a rat model of chronic S. aureus tissue cage infection (33, 55). Strain MRGR3 has additional determinants for resistance to penicillin, gentamicin, chloramphenicol, erythromycin, tetracycline, and polymyxin B, as described previously (33, 55). The average MICs of teicoplanin and vancomycin for strain MRGR3 in cation-adjusted Mueller-Hinton broth (MHB; Difco, Detroit, Mich.) are 1 to 2 and 1 μg/ml, respectively.
Strain 14-4 is a stable teicoplanin-resistant derivative of MRGR3 that first emerged during a rat chronic tissue cage infection as a subpopulation growing on agar supplemented with 10 μg of teicoplanin per ml and was further selected by two in vitro passages on teicoplanin-containing agar (62). The average MICs of teicoplanin and vancomycin for strain 14-4 are 16 to 32 and 4 μg/ml, respectively (62). The other determinants of resistance of strain 14-4 to all antibiotics described above are identical to those of its parent, strain MRGR3.
Strain 14-4rev is a teicoplanin-susceptible revertant of strain 14-4, which spontaneously emerged in one rat tissue cage that was experimentally infected with strain 14-4 and essentially replaced the parental teicoplanin-resistant strain (unpublished data). The average MICs of teicoplanin and vancomycin for strain 14-4rev are 1 μg/ml for each drug. The other determinants of resistance of strain 14-4rev to all other antibiotics described above are identical to those of strains 14-4 and MRGR3.
Finally, the genetic relatedness of strains MRGR3, 14-4, and 14-4rev was assessed by standard pulsed-field gel electrophoresis of SmaI-digested chromosomal DNA (data not shown).
Assay of bacterial adhesion to fibronectin.
The attachment properties of the S. aureus strains were measured by an adhesion assay with polymethylmethacrylate coverslips coated in vitro with three different concentrations (0.25, 0.5, and 1 μg/ml) of purified human fibronectin, as described previously (63). Briefly, 107 CFU of washed cultures of late-logarithmic-phase cells, metabolically radiolabeled with [3H]thymidine during 5 h of growth without shaking at 37°C in MHB, were incubated for 1 h at 37°C with the fibronectin-coated coverslips with human albumin-supplemented phosphate-buffered saline as described previously (4). The coverslips were washed, and the radioactivity was determined. Bacterial adhesion data for the different strains whose cell-associated radioactivities and viable counts differed slightly (<20%) were normalized as described previously (63). Relative changes in bacterial adhesion to each fibronectin coating concentration were expressed as the percent increase or decrease in the levels of attachment of strains 14-4 and 14-4rev compared to that of strain MRGR3. Each experiment was performed five times, and the results are expressed as mean percent changes ± standard errors of the means (SEMs). The statistical significance of pairwise differences in the bacterial adhesion of isogenic strains differing in their expression of teicoplanin resistance was evaluated by paired t tests for each fibronectin coating concentration, and data were considered significant when P was <0.05 (50).
Quantification of FnBPs by flow cytometry.
The FnBP-mediated fibronectin binding displayed by the different strains of S. aureus was monitored by flow cytometry, as described previously (17, 63). The specificity of the flow cytometry data was assessed by parallel analysis of control strains DU5883, a mutant of strain 8325-4 which is simultaneously defective in the expression of both FnBPA and FnBPB, and DU5883(pFNBB4), which overexpresses FnBPB (17, 23, 63).
Relative changes in flow cytometric data were expressed as the percent increase or decrease in the levels of fluorescein isothiocyanate (FITC)-fibronectin binding by strains 14-4 and 14-4rev compared to that of strain MRGR3. Each experiment was performed three times, and the results are expressed as the mean percent changes ± SEMs. The statistical significance of pairwise differences in raw data on the FITC-fibronectin binding of isogenic strains differing in their levels of expression of teicoplanin resistance was evaluated by paired t tests for each fibronectin coating concentration, and data were considered significant when P was <0.05 (50).
Total RNA extraction.
MRGR3, 14-4, and 14-4rev cells were cultured for 5 h in MHB as described above and were then harvested and immediately frozen in liquid nitrogen. Bacterial lysis was performed in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]) containing lysostaphin (200 μg/ml) for 5 min at 37°C. RNA was extracted by use of a High Pure RNA isolation kit (Roche Applied Science, Rotkreuz, Switzerland). DNA was removed from the RNA preparations by treatment with DNase I (Roche), and the absence of contaminating DNA was verified by PCR. Purified RNA samples were analyzed by use of the RNA NanoLab chip on the 2100 Bioanalyser (Agilent, Palo Alto, Calif.).
To control for possible artifacts that may have arisen from the RNA preparation method, a second procedure (24) was used. Briefly, bacteria were recovered, fixed in acetone-ethanol (1:1), and washed in N-Tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)-sucrose buffer. The samples were then incubated on ice for 30 min with lysostaphin (150 μg/ml) in TES-sucrose buffer (20% [wt/vol] sucrose, 20 mM Tris [pH 7.6], 10 mM EDTA, 50 mM NaCl). The RNA was then extracted as described above by use of a High Pure RNA isolation kit. Comparison of the real-time reverse transcription (RT)-PCR data obtained by either method yielded nearly identical results.
Real-time RT-PCR.
mRNA levels were determined by quantitative RT-PCR (qRT-PCR) by use of the one-step reverse transcriptase qPCR Master Mix kit (Eurogentec, Seraing, Belgium), as described previously (63). All primers and probes that were not described previously (63) are listed in Table 1 and were designed with PrimerExpress software (version 1.5; Applied Biosystems) and obtained from Eurogentec or Applied Biosystems.
TABLE 1.
TaqMan primers and probes used in this study
| Gene | Forward primer (sequence) | Reverse primer (sequence) | Probe (sequence [5′→3′]) |
|---|---|---|---|
| asp23 | 299F (GTTAACCACCTTTCATGTCTAAGATAC) | 390R (AAATTAACTTTCTCTGATGAAGTTGTTGA) | 333T (CTTCACGTGCAGCGATACCAGCAATTT3)a |
| hla | 337F (ATGAGTACTTTAACTTATGGATTCAACGG) | 437R (AGTGTATGACCAATCGAAACATTTG3) | 385T (ACAGGAAAAATTGGCGGCCTTATTGGT)a |
| rsbU | 208F (CATAACGATGGCACAATGAGCTT) | 298R (TGCCAAACTTTATCATACTCATTGC) | 234T (CCTTTTCCAATGACATCTGCAACA)a |
| sigB | 4862F (AAATTAGCAGTCATTAACCCATTCG) | 4962R (ACAAAATCCTTCTCTGCTGCC) | 4900T (AAAAGGTGTGATTTATCCGCATCCACCAG)a |
| spA | 1914F (CAGCAAACCATGCAGATGCTAA) | 1992R (ACAGTTGTACCGATGAATGGATTTT) | 1945T (AGCATTACCAGAAACT)b |
The probes were labeled at the 5′ end with 6-carboxyfluorescein and at the 3′ end with 6-carboxytetramethylrhodamine.
The probe was labeled at the 5′ end with 6-carboxyfluorescein and at the 3′ end with a minor groove binder.
RT and PCR were performed with the primers and probes at concentrations of 0.2 and 0.1 μM, respectively. Primers and probes specific for the sigB and hla genes were used at a uniform concentration of 0.1 μM, and those specific for the sarA gene were used at a concentration of 0.05 μM. RT-PCR mixtures containing 4 ng of total RNA were incubated for 30 min at 48°C, followed by incubation for 10 min at 95°C and then 40 cycles of 30 s at 95°C and 60 s at 60°C. Fluorescence emission was detected with an ABI Prism 7700 detector and analyzed with Sequence Detector software (version 1.7; Applied Biosystems). The mRNA levels of the target genes extracted from the different strains were normalized on the basis of their 16S rRNA levels, which were assayed in each round of the qRT-PCR as internal controls, as described previously (63). The statistical significance of strain-specific differences in the normalized threshold cycle (CT) values of each transcript was evaluated by paired t tests, and data were considered significant when P was <0.05 (50).
Northern blotting.
Total RNA (16 μg) was separated in a formaldehyde-agarose gel and blotted onto a nylon membrane (Hybond-N+; Amersham) in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), essentially as described previously (8). A 290-bp sarA fragment generated by PCR amplification with primers 5′-ATGATTGCTTTGAGTTGTTATCAAT-3′ and 5′-ACTCAATAATGATTCGATTTTTTTA-3′ was labeled with [α-32P]dCTP by use of a Prime-it Random Primer Labeling kit (Stratagene) and purified by use of Sephadex G-50 DNA columns (Pharmacia). The blots were prehybridized with Rapid-Hyb buffer (Amersham) for 2 h at 65°C and then hybridized with the labeled sarA probe at 65°C in the same buffer. Washes were done first at room temperature for 20 min with 2× SSC-0.1% sodium dodecyl sulfate (SDS) and second at 65°C for 15 min with 1×SSC-0.1% SDS. Hybridization signals were detected by autoradiography.
agr group typing.
The MRGR3, 14-4, and 14-4rev agr groups were determined by multiplex PCR with primers Pan, agr1, agr2, agr3, and agr4, described by Gilot et al. (21). Strain COL was used as a reference strain for agr group I, and strains Mu50 and N315 were used as reference strains for agr group II. Amplification products were separated by 1.5% agarose gel electrophoresis in the presence of ethidium bromide.
DNA sequencing of sigB and agr loci.
Genomic DNA was isolated by use of a DNeasy Tissue kit (Qiagen). The agr locus (3,500 bp) and the sigB locus (4,100 bp) were amplified by PCR with primers agr1 (5′-CATAGCACTGAGTCCAAGG-3′) and agr2 (5′-GCCAGCTATACAGTGCATTTG-3′) and with primers sigB1 (5′-AAGCTTTTCCGATAGAGTGTGAAG-3′) and sigB2 (5′-AAAATATCCTTCTTTAATTCCTCAGTAAG-3′), respectively. The reactions were performed on an Expand High Fidelity PCR system (Boehringer). Sequencing primers were derived from the published sequence of S. aureus COL from the National Center for Biotechnology Information (NCBI). DNA sequencing was performed with an ABI Prism 377 sequencer (Applied Biosystems). Homology searches and sequence alignments were done by using the BLAST programs provided by NCBI and the Expasy SIM alignment tool http://us.expasy.org/tools/sim-nucl.html).
RESULTS
Increased adhesion of the Teir derivative of MRGR3 on fibronectin-coated surfaces.
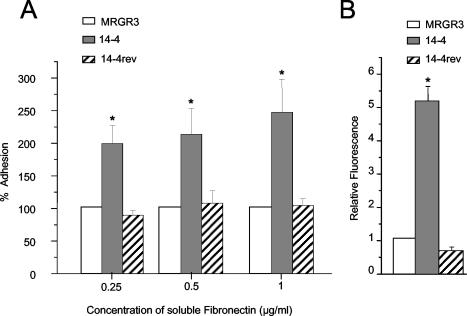
The level of adhesion of teicoplanin-resistant (Teir) strain 14-4 with the three coating concentrations of fibronectin was 2- to 2.5-fold higher (P < 0.05) than that of its isogenic parent, strain MRGR3 (Fig. 1A). Interestingly, the spontaneous Teis revertant strain, strain 14-4rev, that arose during experimental tissue cage infection with strain 14-4 (unpublished data) exhibited bacterial attachment profiles that were nearly identical to those of parent strain MRGR3. These data suggest that the expression of teicoplanin resistance is associated in an unknown way with increased levels of attachment to fibronectin-coated surfaces.
FIG. 1.
(A) Relative changes in levels of adhesion of strains 14-4 and 14-4rev to fibronectin-coated coverslips compared to that for parent strain MRGR3. Values represent the mean ± SEMs of five experiments. (B) Relative changes in binding of soluble FITC-labeled fibronectin by strains 14-4 and 14-4rev compared to that of strain MRGR3. Values represent the means ± SEMs (error bars) of three experiments. *, results significantly different (P < 0.05) from those for MRGR3 and 14-4rev.
To assess whether the increased level of adhesion of Teir strain 14-4 indeed resulted from increased levels of surface display of fibronectin adhesin molecules, FnBP-mediated fibronectin-binding sites were monitored by flow cytometry. Figure 1B shows that strain 14-4 bound fivefold more FITC-labeled fibronectin (P < 0.01) than its parent strain, strain MRGR3, did. In contrast, equivalent amounts of FITC-labeled fibronectin were bound by Teis revertant strain 14-4rev and Teis parent strain MRGR3.
Increased levels of fnbA and fnbB transcripts in Teir strain 14-4.
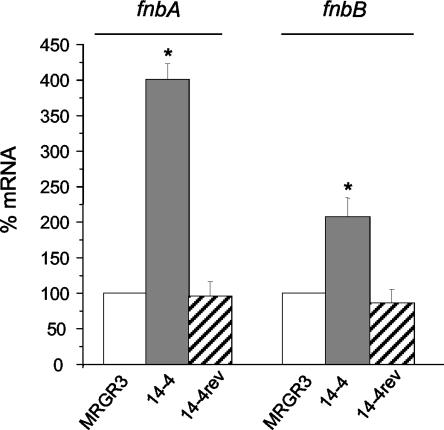
Figure 2 shows that the steady-state fnbA and fnbB mRNA levels in Teir strain 14-4 were significantly (P < 0.01) increased, averaging four- and twofold higher, respectively, over those assayed in parallel in Teis parent strain MRGR3 by qRT-PCR. In contrast, the levels of fnbA and fnbB transcripts in Teis revertant 14-4rev were not significantly different from those in Teis strain MRGR3.
FIG. 2.
Steady-state levels of fnbA and fnbB transcripts of strains 14-4 and 14-4rev compared to those of parent strain MRGR3, determined by real-time RT-PCR and normalized on the basis of adjusted 16S rRNA levels. Values represent the means ± SEMs of five experiments performed in triplicate. *, results significantly different (P < 0.01) from those for MRGR3 and 14-4rev.
Downregulation of agr transcripts in Teir strain 14-4.
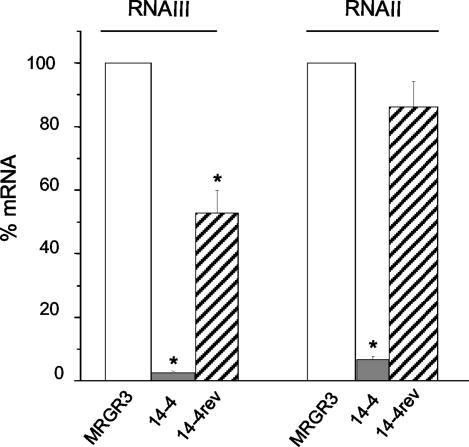
To identify potential global regulators that may contribute in a strain-specific manner to the upregulation of fnb genes in Teir strain 14-4 compared to their regulation in Teis parent strain MRGR3 or Teis revertant 14-4rev, we compared the levels of transcription of agr, sarA, and sigB in the three strains. The agr RNAII and RNAIII levels assayed by real-time RT-PCR were markedly reduced (P < 0.01) by 92 and >98%, respectively, in the Teir strain 14-4 compared to the levels in Teis parent strain MRGR3 (Fig. 3). In contrast, Teis revertant 14-4rev showed no significant decrease in RNAII levels and only a 50% decrease in RNAIII levels compared to those in Teis strain MRGR3. Even if the RNAII and RNAIII levels in Teis revertant 14-4rev were not fully restored to the levels assayed in Teis strain MRGR3, they were at least significantly (P < 0.01) different from those assayed in Teir strain 14-4 (Fig. 3).
FIG. 3.
Steady-state levels of RNAIII and RNAII of strains 14-4 and 14-4rev compared to those of parent strain MRGR3, determined by real-time RT-PCR and normalized on the basis of adjusted 16S rRNA levels. Results are presented as means + SEMs of three experiments performed in triplicate. *, results significantly different (P < 0.01) from those for MRGR3.
Since recent observations have indicated the frequent occurrence of a defective agr function that was associated with agr group II in clinical GISA isolates (52, 53), we also determined the agr group of Teir strain 14-4 and its Teis counterparts by multiplex PCR. All three strains were found to belong to agr group I, as evidenced by the recovery of 441-bp PCR products identical to that of strain COL, used as an agr group I reference strain, as opposed to the 575-bp PCR products for agr group II control strains Mu50 and N315 (data not shown). To confirm the agr group I identification of our strains and to exclude the occurrence of spontaneous mutations that may explain the specific defect in the agr transcripts of the teicoplanin-resistant strain compared to the transcripts of its teicoplanin-susceptible counterparts, we sequenced the entire 3,500-bp agr locus of each strain. No single-nucleotide difference was found between the respective sequences of the agr loci of strains MRGR3, 14-4, and 14-4rev, including the agrD-encoded propeptide of the autoinducing group I peptide (45; data not shown). Thus, no strain-specific mutation could account for the specific downregulation of the agr regulon in Teir strain 14-4 compared to its regulation in the Teis counterparts MRGR3 and 14-4rev.
Upregulation of sarA transcripts in Teir strain 14-4.
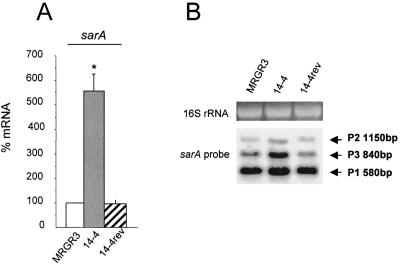
Overall, the steady-state sarA mRNA levels assayed in Teir strain 14-4 by qRT-PCR were more than fivefold higher (P < 0.01) than those recorded in Teis parent strain MRGR3. In contrast, no significant increase in the sarA transcript level of Teis revertant 14-4rev compared to that in MRGR3 was found (Fig. 4A).
FIG. 4.
(A) Steady-state levels of sarA transcripts of strains 14-4 and 14-4rev compared to those of parent strain MRGR3, determined by real-time RT-PCR and normalized on the basis of adjusted 16S rRNA levels. Results are presented as means ± SEMs of three experiments performed in triplicate. *, results significantly different (P < 0.01) from those for MRGR3 and 14-4rev. (B) Northern blot analysis of sarA transcripts in strains MRGR3, 14-4, and 14-4rev. The arrows indicate the three different sarA transcripts.
Since the sarA locus is composed of three overlapping transcripts of 0.58, 0.84, and 1.15 kb, initiated from the P1, P3, and P2 promoters, respectively, we performed Northern blot analysis to evaluate the levels of each individual transcript in Teir strain 14-4 compared to those in Teis strains MRGR3 and 14-4rev. Figure 4B demonstrates a significant increase in the level of the P3-driven transcript, which is specific to strain 14-4, compared to those in strains MRGR3 and 14-4rev. According to previous studies (2, 12, 19, 35), the significant increase in the SigB-dependent sarA P3-derived transcript in strain 14-4 provided the first indirect evidence of a higher level of SigB activity in the Teir strain than in its isogenic Teis parent or revertant.
Transcriptional and functional activities of the SigB global regulon.
Previous studies have documented increased SigB functional activity in S. aureus strains that have acquired teicoplanin resistance (1, 3). We therefore evaluated by qRT-PCR the mRNA levels of asp23, a gene used as a marker for SigB activity. The levels of the SigB-dependent target gene asp23 transcript were 3.5- and 2-fold higher in Teir strain 14-4 than in Teis parent strain MRGR3 and Teis revertant 14-4rev, respectively (Fig. 5A). These data provide further support for increased SigB activity in strain 14-4 than in MRGR3 and 14-4rev, as demonstrated by Northern blotting of the sarA P3-derived transcript (Fig. 4B). To identify any spontaneous strain-specific mutation that could account for the higher level of SigB activity observed in Teir strain 14-4 than in its teicoplanin-susceptible counterparts, we sequenced the entire SigB regions of the three strains, corresponding to positions 2117407 to 2121606 of strain N315 (NC 002745). No single-nucleotide difference between the respective sequences of these three strains was found (data not shown).
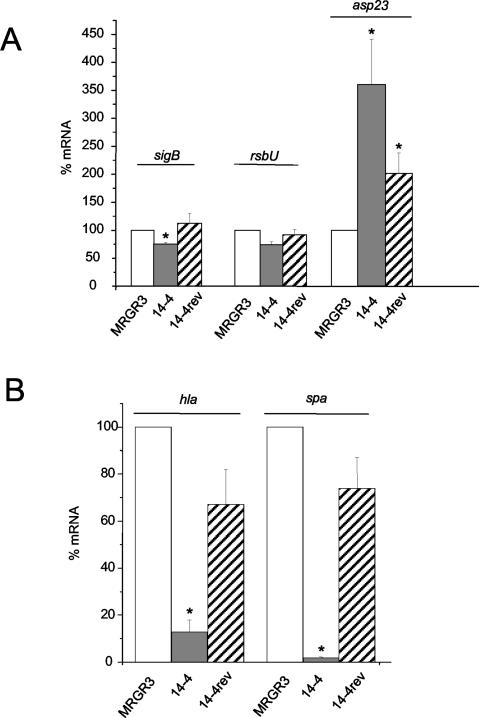
FIG. 5.
Steady-state mRNA levels of sigB, rsbU, and asp23 genes (A) or hla and spA genes (B) of strains 14-4 and 14-4rev compared to those of parent strain MRGR3, determined by real-time RT-PCR and normalized on the basis of adjusted 16S rRNA levels. Values are the mean + SEMs of three experiments performed in triplicate. *, values significantly different (P < 0.01) from those for MRGR3.
The sigB and rsbU transcript levels were also evaluated. Unexpectedly, the steady-state levels of sigB and rsbU mRNA were not significantly elevated in Teir strain 14-4 compared with their levels in its teicoplanin-susceptible counterparts (Fig. 5A), a finding that is thus in conflict with the evidence of significantly increased SigB activities of target gene asp23 and sarA P3-derived transcripts.
Steady-state hla and spa transcript levels in Teir strain 14-4.
Since previous reports indicated that GISA isolates have decreased levels of production of hemolysin (53) and protein A (30), we compared the mRNA levels of Teir strain 14-4 and its isogenic Teir counterparts. Spectacular differences in the hla and spa transcript levels of Teir strain 14-4 and those of its Teis parent strain were found, with the levels in 14-4 being significantly decreased (P < 0.001) compared to those in MRGR3 (Fig. 5B). While the levels of mRNA for hla and spa in strain 14-4 represented only 13 and 1.8% of those assayed in strain MRGR3, respectively, these levels were restored in strain 14-4rev to 67 and 74% of the levels found in MRGR3, respectively.
DISCUSSION
This report provides substantial evidence that the acquisition of teicoplanin resistance may be linked with multiple changes in the expression and regulation of virulence genes. This conclusion was based on the simultaneous testing of isogenic teicoplanin-resistant and teicoplanin-susceptible derivatives of S. aureus that included the teicoplanin-susceptible MRSA parent and a spontaneous teicoplanin-susceptible revertant of teicoplanin-resistant strain 14-4 selected in a tissue cage model of foreign body infection. A major finding of our experimental study was the increased levels of expression and surface display of FnBPs that specifically occurred in Teir strain 14-4 compared to those that occurred in its Teis counterparts. This observation may be of significant clinical relevance, particularly if it could be extended to clinical GISA strains, in view of the prominent roles of FnBPs in S. aureus attachment and colonization of host tissues and implanted biomaterials (64). Since a recent study (53) mentioned that a majority of GISA isolates were recovered from device-associated infections, we can postulate that an increased levels of surface display of FnBPs by such resistant or intermediate organisms might contribute significantly to their attachment to the fibronectin-coated medical devices as an early step of implant colonization and infection (23, 64). In addition, the higher levels of surface display of FnBPs involved in fibronectin-mediated bridging with the host receptor integrin α5β1 may also be expected to promote their endocytic uptake by nonprofessional phagocytes (16, 59).
Two previous studies performed in our laboratory also described increased levels of surface display of FnBPs in S. aureus strains under conditions unrelated to the acquisition of teicoplanin resistance. In one study, the upregulation of FnBPs was due to a specific increase in the levels of fnbB transcription in fluoroquinolone-resistant S. aureus isolates exposed to subinhibitory levels of ciprofloxacin (5). The quinolone-induced overexpression of FnBPs did not require the active contribution of any of the three global regulators agr (5), sarA (C. Bisognano, W. L. Kelley, P. Francois, J. Schrenzel, D. P. Lew, D. C. Hooper, and P. Vaudaux, Abstr. 42nd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-1063, 2002), or sigB [D. Li, A. Renzoni, T. Estoppey, C. Bisognano, P. Francois, W. L. Kelley, D. P. Lew, J. Schrenzel, and P. Vaudaux, Clin. Microbiol. Infect. 9(Suppl. 1), abstr. P-465, 2003], as shown in recent preliminary reports. In a second study (63), upregulation of FnBPs and increased endocytic uptake occurred in hemB mutants of S. aureus expressing small colony variant phenotypes, but they could not be consistently explained by altered activities of their global regulons agr and sarA.
To evaluate the relationship between the three major global regulators and elevated levels of both fnb genes, we assayed the functional activities of the agr, sarA, and sigB regulons in Teir strain 14-4 compared to those in its Teis counterparts. Indeed, the steady-state transcript levels of all three global regulators were significantly altered in strain 14-4 compared to those in MRGR3 and 14-4rev when the levels were assayed by the highly sensitive qRT-PCR technique and, when necessary, by Northern blotting hybridization. First, agr RNAII and RNAIII levels were indeed markedly reduced in the Teir strain compared to those in the Teis strains, in agreement with previous observations obtained with unrelated vancomycin-intermediate S. aureus clinical isolates (52, 53). However, we found that agr downregulation in strain 14-4 occurred in an agr group I background instead of an agr group II background, as previously reported for vancomycin-intermediate S. aureus clinical isolates (52, 53). Furthermore, complete sequencing of the agr group I locus revealed no single-nucleotide difference between the Teir and Teis strains, which indicated that the strain-specific differences in agr functional activities were not intrinsic to the agr regulon per se but likely resulted from additional as yet unknown regulatory elements that may repress RNAII and RNAIII transcription in Teir strain 14-4.
In contrast to agr, the sarA transcript levels assayed by qRT-PCR were significantly elevated in the Teir strain compared to those in the Teis strains. Northern blotting analyses confirmed not only an overall increase in sarA transcript levels but also a strain-specific activation of the sarA P3 promoter-derived transcript in Teir strain 14-4. Since the specific increase in the level of the SigB-dependent sarA P3-derived transcript in strain 14-4 strongly suggested a higher level of strain-specific SigB activity in the Teir strain (2, 12, 19, 35), we also examined the transcriptional activities of the SigB regulon and its specific target gene, asp23. The absence of increased sigB and rsbU transcript levels in the Teir strain compared to those in its Teis counterparts, as assayed by qRT-PCR, was unexpected and was in contrast to the increased functional activity of SigB that was indirectly determined from the significantly higher levels of target gene asp23 and sarA P3-derived transcripts in the former strain. A further criterion for increased SigB functional activity in the Teir strain was the more intense pigmentation of isolated colonies on agar plates compared to those of its Teis counterparts (data not shown). The reasons explaining the discrepant qRT-PCR data for the SigB operon compared with the functional data for SigB activity, as assessed by determination of the target gene transcript levels, have not been elucidated. No spontaneous mutation occurred in the nucleotide sequences of the complete SigB regulons of the Teir and Teis strains that were found to be identical. A trivial methodological error that might have compromised the sensitivity and/or specificity of our qRT-PCR assay for sigB transcripts level seems unlikely, since another study performed in our laboratory by use of an identical assay could demonstrate more than fivefold higher levels of sigB transcripts in a RsbU+-restored mutant of S. aureus compared to those in its RsbU-defective parent [Li et al., Clin. Microbiol. Infect. 9(Suppl. 1), abstr. P-465, 2003].
In conclusion, the data presented here provide further evidence that the emergence of glycopeptide resistance is linked by still poorly understood molecular pathways with significant pleiotropic changes in the levels of expression and regulation of some major virulence genes. These molecular and phenotypic changes may have profound impacts on the bacterial adhesion and colonization properties of such multiresistant organisms. More global approaches, such as transcription profiling studies (13, 30, 39, 51), are required to understand the contributions of recently discovered global regulators and transcription factors to the changes in fitness of glycopeptide-intermediate or -resistant S. aureus strains.
Acknowledgments
This work was supported in part by a research grant from the Swiss National Program on Antibiotic Resistance (grant 4049-063250) and grants 3200-63710.00 (to P.V.) and 632-57950.99 (to J.S.) from the Swiss National Science Foundation. Support was also provided by the Centro Internacional de Fisica, Bogota, Colombia (to A.R.).
We thank Manuela Bento for technical assistance.
REFERENCES
- 1.Bischoff, M., and B. Berger-Bächi. 2001. Teicoplanin stress-selected mutations increasing sigma(B) activity in Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1714-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischoff, M., J. M. Entenza, and P. Giachino. 2001. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J. Bacteriol. 183:5171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff, M., M. Roos, J. Putnik, A. Wada, P. Glanzmann, P. Giachino, P. Vaudaux, and B. Berger-Bächi. 2001. Involvement of multiple genetic loci in Staphylococcus aureus teicoplanin resistance. FEMS Microbiol. Lett. 194:77-82. [DOI] [PubMed] [Google Scholar]
- 4.Bisognano, C., P. Vaudaux, D. P. Lew, E. Y. W. Ng, and D. C. Hooper. 1997. Increased expression of fibronectin-binding proteins by fluoroquinolone-resistant Staphylococcus aureus exposed to subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 41:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bisognano, C., P. Vaudaux, P. Rohner, D. P. Lew, and D. C. Hooper. 2000. Induction of fibronectin-binding proteins and increased adhesion of quinolone-resistant Staphylococcus aureus by subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 44:1428-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blevins, J. S., A. F. Gillaspy, T. M. Rechtin, B. K. Hurlburt, and M. S. Smeltzer. 1999. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol. Microbiol. 33:317-326. [DOI] [PubMed] [Google Scholar]
- 8.Brown, T., and K. Mackey. 1997. Analysis of RNA by Northern and slot blot hybridization, p. 4.9.1-4.9.16. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 9.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung, A. L., Y. T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, A. L., S. J. Projan, and H. Gresham. 2002. The genomic aspect of virulence, sepsis, and resistance to killing mechanisms in Staphylococcus aureus. Curr. Infect. Dis. Rep. 4:400-410. [DOI] [PubMed] [Google Scholar]
- 12.Deora, R., T. Tseng, and T. K. Misra. 1997. Alternative transcription factor σSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 179:6355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer, B., P. Vaudaux, M. Magnin, Y. El Mestikawy, R. A. Proctor, D. P. Lew, and H. Vasey. 1996. Novel animal model for studying the molecular mechanisms of bacterial adhesion to bone-implanted metallic devices: role of fibronectin in Staphylococcus aureus adhesion. J. Orthoped. Res. 14:914-920. [DOI] [PubMed] [Google Scholar]
- 15.Fournier, B., A. Klier, and G. Rapoport. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247-261. [DOI] [PubMed] [Google Scholar]
- 16.Fowler, T., E. R. Wann, D. Joh, S. A. Johansson, T. J. Foster, and M. Höök. 2000. Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell β1 integrins. Eur. J. Cell Biol. 79:672-679. [DOI] [PubMed] [Google Scholar]
- 17.Francois, P., J. Schrenzel, C. Stoerman-Chopard, H. Favre, M. Herrmann, T. J. Foster, D. P. Lew, and P. Vaudaux. 2000. Identification of plasma proteins adsorbed on hemodialysis tubing that promote Staphylococcus aureus adhesion. J. Lab. Clin. Med. 135:32-42. [DOI] [PubMed] [Google Scholar]
- 18.Geisel, R., F. J. Schmitz, A. C. Fluit, and H. Labischinski. 2001. Emergence, mechanism, and clinical implications of reduced glycopeptide susceptibility in Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 20:685-697. [DOI] [PubMed] [Google Scholar]
- 19.Gertz, S., S. Engelmann, R. Schmid, A. K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giachino, P., S. Engelmann, and M. Bischoff. 2001. σB activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilot, P., G. Lina, T. Cochard, and B. Poutrel. 2002. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J. Clin. Microbiol. 40:4060-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldstein, F. W., and M. D. Kitzis. 2003. Vancomycin-resistant Staphylococcus aureus: no apocalypse now. Clin. Microbiol. Infect. 9:761-765. [DOI] [PubMed] [Google Scholar]
- 23.Greene, C., D. McDevitt, P. François, P. Vaudaux, D. P. Lew, and T. J. Foster. 1995. Adhesion properties of mutants of Staphylococcus aureus defective in fibronectin binding proteins and studies on the expression of fnb genes. Mol. Microbiol. 17:1143-1152. [DOI] [PubMed] [Google Scholar]
- 24.Herbert, S., P. Barry, and R. P. Novick. 2001. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus. Infect. Immun. 69:2996-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiramatsu, K., N. Aritaka, H. Hanaki, S. Kawasaki, Y. Hosoda, S. Hori, Y. Fukuchi, and I. Kobayashi. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670-1673. [DOI] [PubMed] [Google Scholar]
- 26.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 27.Hiramatsu, K., K. Okuma, X. X. Ma, M. Yamamoto, S. Hori, and M. Kapi. 2002. New trends in Staphylococcus aureus infections: glycopeptide resistance in hospital and methicillin resistance in the community. Curr. Opin. Infect. Dis. 15:407-413. [DOI] [PubMed] [Google Scholar]
- 28.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. Sigma(B) modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor SarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 31.Kuroda, M., K. Kuwahara-Arai, and K. Hiramatsu. 2000. Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. Biochem. Biophys. Res. Commun. 269:485-490. [DOI] [PubMed] [Google Scholar]
- 32.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 33.Lucet, J. C., M. Herrmann, P. Rohner, R. Auckenthaler, F. A. Waldvogel, and D. P. Lew. 1990. Treatment of experimental foreign body infection caused by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 34:2312-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manna, A., and A. L. Cheung. 2001. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect. Immun. 69:885-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manna, A. C., and A. L. Cheung. 2003. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect. Immun. 71:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyazaki, E., J. M. Chen, C. Ko, and W. R. Bishai. 1999. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J. Bacteriol. 181:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mongodin, E., J. Finan, M. W. Climo, A. Rosato, S. Gill, and G. L. Archer. 2003. Microarray transcription analysis of clinical Staphylococcus aureus isolates resistant to vancomycin. J. Bacteriol. 185:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore, M. R., F. Perdreau-Remington, and H. F. Chambers. 2003. Vancomycin treatment failure associated with heterogeneous vancomycin-intermediate Staphylococcus aureus in a patient with endocarditis and in the rabbit model of endocarditis. Antimicrob. Agents Chemother. 47:1262-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morfeldt, E., K. Tegmark, and S. Arvidson. 1996. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol. Microbiol. 21:1227-1237. [DOI] [PubMed] [Google Scholar]
- 42.Naimi, T. S., D. Anderson, C. O'Boyle, D. J. Boxrud, S. K. Johnson, F. C. Tenover, and R. Lynfield. 2003. Vancomycin-intermediate Staphylococcus aureus with phenotypic susceptibility to methicillin in a patient with recurrent bacteremia. Clin. Infect. Dis. 36:1609-1612. [DOI] [PubMed] [Google Scholar]
- 43.Nair, S. P., M. Bischoff, M. M. Senn, and B. Berger-Bächi. 2003. The sigma B regulon influences internalization of Staphylococcus aureus by osteoblasts. Infect. Immun. 71:4167-4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicholas, R. O., T. Li, D. McDevitt, A. Marra, S. Sucoloski, P. L. Demarsh, and D. R. Gentry. 1999. Isolation and characterization of a sigB deletion mutant of Staphylococcus aureus. Infect. Immun. 67:3667-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novick, R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429-1449. [DOI] [PubMed] [Google Scholar]
- 46.Patti, J. M., B. L. Allen, M. J. McGavin, and M. Höök. 1994. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu. Rev. Microbiol. 48:585-617. [DOI] [PubMed] [Google Scholar]
- 47.Pfeltz, R. F., V. K. Singh, J. L. Schmidt, M. A. Batten, C. S. Baranyk, M. J. Nadakavukaren, R. K. Jayaswal, and B. J. Wilkinson. 2000. Characterization of passage-selected vancomycin-resistant Staphylococcus aureus strains of diverse parental backgrounds. Antimicrob. Agents Chemother. 44:294-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ploy, M. C., C. Grelaud, C. Martin, L. de Lumley, and F. Denis. 1998. First clinical isolate of vancomycin-intermediate Staphylococcus aureus in a French hospital. Lancet 351:1212. [DOI] [PubMed] [Google Scholar]
- 49.Projan, S. J., and R. P. Novick. 1997. The molecular basis of pathogenicity, p. 55-81. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone, New York, N.Y.
- 50.Rosner, B. 1990. Analysis of variance, p. 474-526. In M. R. Payne, S. Hankinson, and S. London (ed.), Fundamentals of biostatistics. PWS-KENT Publishing Company, Belmont, Calif.
- 51.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakoulas, G., G. M. Eliopoulos, R. C. J. Moellering, R. P. Novick, L. Venkataraman, C. Wennersten, P. C. Degirolami, M. J. Schwaber, and H. S. Gold. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187:929-938. [DOI] [PubMed] [Google Scholar]
- 53.Sakoulas, G., G. M. Eliopoulos, R. C. J. Moellering, C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savolainen, K., L. Paulin, B. Westerlund-Wikström, T. J. Foster, T. K. Korhonen, and P. Kuusela. 2001. Expression of pls, a gene closely associated with the mecA gene of methicillin-resistant Staphylococcus aureus, prevents bacterial adhesion in vitro. Infect. Immun. 69:3013-3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaad, H. J., C. Chuard, P. Vaudaux, F. A. Waldvogel, and D. P. Lew. 1994. Teicoplanin alone or combined with rifampin compared with vancomycin for prophylaxis and treatment of experimental foreign body infection by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 38:1703-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt, K. A., A. C. Manna, S. Gill, and A. L. Cheung. 2001. SarT, a repressor of α-hemolysin in Staphylococcus aureus. Infect. Immun. 69:4749-4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sieradzki, K., T. Leski, J. Dick, L. Borio, and A. Tomasz. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J. Clin. Microbiol. 41:1687-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sieradzki, K., R. B. Roberts, S. W. Haber, and A. Tomasz. 1999. The development of vancomycin resistance in a patient with methicillin-resistant Staphylococcus aureus infection. N. Engl. J. Med. 340:517-523. [DOI] [PubMed] [Google Scholar]
- 59.Sinha, B., P. P. Francois, O. Nüsse, M. Foti, O. M. Hartford, P. Vaudaux, T. J. Foster, D. P. Lew, M. Herrmann, and K. H. Krause. 1999. Fibronectin-binding protein acts as Staphylococcus aureus invasin via fibronectin bridging to integrin alpha5beta1. Cell. Microbiol. 1:101-118. [DOI] [PubMed] [Google Scholar]
- 60.Smith, T. L., M. L. Pearson, K. R. Wilcox, C. Cruz, M. V. Lancaster, B. Robinson-Dunn, F. C. Tenover, M. J. Zervos, J. D. Band, E. White, and W. R. Jarvis. 1999. Emergence of vancomycin resistance in Staphylococcus aureus. N. Engl. J. Med. 340:493-501. [DOI] [PubMed] [Google Scholar]
- 61.Tenover, F. C., J. W. Biddle, and M. V. Lancaster. 2001. Increasing resistance to vancomycin and other glycopeptides in Staphylococcus aureus. Emerg. Infect. Dis. 7:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaudaux, P., P. Francois, B. Berger-Bächi, and D. Lew. 2001. In vivo emergence of subpopulations expressing teicoplanin or vancomycin-resistant phenotypes in a glycopeptide-susceptible, methicillin-resistant strain of Staphylococcus aureus. J. Antimicrob. Chemother. 47:163-170. [DOI] [PubMed] [Google Scholar]
- 63.Vaudaux, P., P. Francois, C. Bisognano, W. L. Kelley, D. P. Lew, J. Schrenzel, R. A. Proctor, P. J. McNamara, G. Peters, and C. Von Eiff. 2002. Increased expression of clumping factor and fibronectin-binding proteins by hemB mutants of Staphylococcus aureus expressing small colony variant phenotypes. Infect. Immun. 70:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vaudaux, P., P. Francois, D. P. Lew, and F. A. Waldvogel. 2000. Host factors predisposing to and influencing therapy of foreign body infections, p. 1-26. In F. A. Waldvogel and A. L. Bisno (ed.), Infections associated with indwelling medical devices. American Society for Microbiology, Washington, D.C.
- 65.Vaudaux, P., V. Monzillo, P. Francois, D. P. Lew, T. J. Foster, and B. Berger-Bächi. 1998. Introduction of the mec element (methicillin resistance) into Staphylococcus aureus alters in vitro functional activities of fibrinogen and fibronectin adhesins. Antimicrob. Agents Chemother. 42:564-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walsh, T. R., and R. A. Howe. 2002. The prevalence and mechanisms of vancomycin resistance in Staphylococcus aureus. Annu. Rev. Microbiol. 56:657-675. [DOI] [PubMed] [Google Scholar]
- 67.Wolz, C., P. Pöhlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. Van Wamel, and A. L. Cheung. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
- 68.Wu, S. W., H. De Lencastre, and A. Tomasz. 1996. Sigma-B, a putative operon encoding alternate sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J. Bacteriol. 178:6036-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yarwood, J. M., J. K. McCormick, and P. M. Schlievert. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]