Abstract
Poly(ADP-ribose) polymerase-1 (PARP-1) has a vital role in the progression of the inflammatory response, and its inhibition confers protection in various models of inflammatory disorders. Therefore, we investigated the effect of promoter and exon variations of the PARP-1 gene on the risk for the inflammatory disease Hashimoto’s thyroiditis (HT). This case–control association study was comprised of 141 HT patients and 150 controls from a group of women in a Turkish population. Two polymorphisms in the promoter region of the PARP-1 gene, rs2793378 and rs7527192, were analyzed using the PCR-RFLP method. In addition, single nucleotide polymorphism (SNP) rs1136410, which is located at codon 762, was analyzed using bidirectional sequencing. The combined genotype and haplotype analyses of these polymorphisms were performed using SPSS 18 and Haploview 4.2. The results were statistically analyzed by calculating the odds ratios and 95% confidence interval using Pearson’s χ2-test and Fisher’s exact test (two-sided). Although we had a number of significant results, the associations became nonsignificant following a Bonferroni correction for multiple comparisons. Nonetheless, a protective factor against HT was still observed for the heterozygous genotype (TC) of SNP rs1136410 (P=0.001), even following Bonferroni correction, and according to the rs2793378–rs7527192 combined analysis, the occurrence of the TT/GA combined genotype was significantly higher in the controls (P=0.007). These results prove that the heterozygosity of SNP rs1136410 provides a protective effect against HT disease in a group of women in a Turkish population.
Introduction
Hashimoto’s thyroiditis (HT), which is characterized by the production of autoantibodies against the thyroid gland, is a localized autoimmune disease. HT has been found to have a 55% concordance rate in monozygotic twins.1 Repplinger et al.2 reported that there is a high prevalence for the increased occurrence of HT in women, which is seven times greater than in men. Different genetic and environmental factors are involved in the pathogenesis of this disease.3 It is well established that the chronic inflammation that occurs during the course of HT causes a significant increase in the levels of cytokines and other inflammatory mediators, which ultimately leads to the destruction of the thyroid gland.4 More precisely, the interactions of multiple genes and environmental factors can increase susceptibility to HT and the severity of the disease.5
Poly(ADP-ribose) polymerase-1 (PARP-1) is thought to be one of the candidate genes responsible for the pathogenesis of inflammatory situations. PARP-1 is thought to have an important role in the initiation of the DNA repair pathway, although high levels of its activation are also associated with increased apoptosis. Recently, it was shown that the development and progression of inflammatory diseases were impeded in PARP-1 knockout animal models that used PARP-1 inhibitors.6,7 Results from several studies have shown that the inactivation of PARP-1 protects against endotoxic shock, colitis, lung inflammation and other diseases, such as traumatic brain injury, cerebral and myocardial ischemia, stroke, rheumatoid arthritis and diabetes mellitus-associated heart disease.8–15 Although, the exact role of PARP-1 in the inflammatory cascade is not yet known, all of the available studies on PARP-1 suggest that this gene is a strong candidate and genetic factor for the development of HT.
The PARP-1 gene (OMIM 173870), which localizes to chromosome one (1q41-43), has been shown to accommodate allelic polymorphisms that have been linked to susceptibility for various diseases, some of which are inflammatory disorders.16 In addition, multiple binding sites for transcription factors have been identified in the PARP-1 gene promoter region; therefore, genetic variants within this region may affect PARP-1 expression.17 Several variations (C410T (rs2793378), Poly(A)n, C1362T and G1672A (rs7527192)) have been identified in the promoter region, and there are several studies that have investigated the relationship between these promoter polymorphisms and the pathogenesis of many diseases.16,17 In addition, the active-site polymorphism T2444C (rs1136410) (also known as Val762Ala), which causes a T to C, Val to Ala change in exon 17, was identified by Cottet et al.18 According to Wang and colleagues, the T2444C polymorphism reduces the enzymatic activity of PARP-1 by ~40%.16,19,20
In this study, we questioned whether two promoter polymorphisms (rs2793378 and rs7527192) and an active-site polymorphism (rs1136410) in the PARP-1 gene could predispose individuals to HT disease in a female Turkish population.
Patients and methods
Study subjects
One hundred and forty-one women who were diagnosed with HT in the Endocrine and Metabolic Diseases outpatient clinic at Cerrahpasa Medical School, Istanbul University, were included in this study. The control group consisted of one hundred and fifty healthy women with a similar mean age. The exclusion criteria for participation in the study included pregnancy, smoking, use of alcohol or prescription drugs and having an autoimmune disease that might affect the results of the study.
Although Turks are ethnically heterogeneous, all of the cases and controls were carefully chosen from the European side of Istanbul to ensure that all of the individuals were of Turkish origin. All of the study subjects were unrelated and provided signed informed consent prior to the sample and data collection. The study protocol was approved by the Institutional Ethical Committee of Cerrahpasa Medical School, Istanbul University.
Genotyping of the PARP-1 polymorphisms
The genomic DNA from the study subjects was isolated using an ethanol precipitation method. The concentration and purity of the DNA was measured on a NanoDrop Spectrophotometers (NanoDrop Technologies Inc., Wilmington, DE, USA) and determined using a 260/280 nm optic density ratio. The PARP-1 C410T and G1672A genotypes were determined using a polymerase chain reaction (PCR) based restriction fragment length polymorphism (RFLP) method. The primers used to amplify the polymorphic sites were F: 5′-TCCAGTGGCACTATCAT-3′ and R: 5′-GTTGTGAGACATAGGCCGAATC-3′ for rs2793378, and F: 5′-GCGAGACCCTGTCCCTAA-3′ and R: 5′-TCCCCCTTTTATTTTTGAGACTG-3′ for rs7527192.16
The amplification products yielded a 298-base pair (bp) fragment for rs2793378 and a 187 bp fragment for rs7527192. Ten microliters of the amplification product of rs2793378 were digested with one unit of Hpy31 (Dde I) (New England Biolabs, Beverly, MA, USA), incubated for 12 h at 37 °C, and subjected to electrophoresis for 30 min at 120 V on a 3% agarose gel. The homozygosity of the common allele (represented by the CC genotype) was denoted via 145 and 109 bp bands, while the homozygosity of the variant allele (represented by the TT genotype) was represented by 127 and 109 bp bands. Ten microliters of the amplification product of rs7527192 were digested with one unit of Bsh1236I (BstUI) (MBI, Fermentas, Vilnius, Lithuania), incubated for 12 h at 37 °C, and separated on a 3% agarose gel. The homozygosity of the common allele (represented by the GG genotype) was denoted via 152 and 35 bp bands, while the homozygosity of the variant allele (represented by the AA genotype) was represented by a single 187 bp band, as this polymorphism destroys a restriction site for Bsh1236I.20
For rs1136410, the PCR conditions were set at 95 °C for 4 min for the denaturation, followed by 35 cycles of 94 °C for 30 s, 62 °C for 45 s and 72 °C for 45 s for the annealing step, followed by a final extension at 72 °C for 10 min. The primers that were used are as follows: forward primer 5′-TGAGGAAGGCCTGACCCTGT-3′ and reverse primer 5′-TGACCAGCAGGAGGGTTTGC-3′. The sequential alterations for rs1136410 were determined using bidirectional sequencing. The amplicon sequences were evaluated using CLC workbench 3.6.1 (Denmark).
Haplotype analysis
Haplotypes were generated from the genotype data. The linkage disequilibrium (LD) and haplotype analyses were performed using Haploview 4.2, which uses the expectation maximization (EM) algorithm.
Data processing and analysis
The statistical analyses were performed using the Statistical Package for Social Sciences Statistical Software release 18 (SPSS Windows Version 18, SPSS, Inc., Chicago, IL, USA). The Hardy–Weinberg equilibrium (HWE) was determined in order to identify the compatibility between the patient and control groups using χ2-tests. The relationships between different parameters were evaluated using Spearman’s correlation analysis. The comparison of the frequencies and ratios between the groups was evaluated using χ2- and Fisher’s exact tests. The quantitative data containing normally distributed parameters between the groups were evaluated using Student’s t-test, while Mann–Whitney U-test was used for the remaining samples. The associations between the rs2793378, rs7527192 and rs1136410 genotypes and HT were analyzed by calculating the odds ratios (ORs) and their 95% confidence intervals (95% CIs) using the χ2-test, accepting the homozygous common genotypes as the reference category for each polymorphism. As this study was based on a small sample size and there were several multiple comparisons, the results could have arisen from a type I error. The confidence in the results will generally be weaker if it is conducted as part of a multiple comparison analysis, rather than a single comparison analysis. Therefore, to address the multiple comparison problem, a Bonferroni correction was applied to the comparisons. The Bonferroni correction is a conservative method that is free of independency and distribution assumptions. The Bonferroni correction set the significance level at α (alpha)/n (number of comparisons). The specific multiple comparisons that were used for each Bonferroni-corrected P value are stated below each table.
Results
Study population
A total of 141 Hashimoto thyroiditis patients, aged between 39 and 55 years old (mean, 48.9 year), and 150 control subjects, aged between 37 and 54 years old (mean, 46.6 year), were recruited in this study. There was no significant difference between the ages of the cases and the controls.
The mean levels of thyroid stimulating hormone (TSH), AntiTPO and AntiTG antibody in the HT patients in this study were found to be 8.3±6.8 μIU/ml, 806.7±447.3 IU/ml and 382.7±207.4 IU/ml, respectively. The TSH, AntiTg and AntiTPO antibody levels of the control subjects were 2.8±1.2 μIU/ml, 65.4±59.7 IU/ml and 27±15.8 IU/ml, respectively. The levels of the TSH, AntiTPO and AntiTG antibodies of the HT patients were statistically higher than the control group, as was expected (P=0.0305, 0.0296, 0.0242, respectively).
The 141 HT patients and 150 control subjects were tested for rs2793378, rs7527192 and rs1136410. There were no statistically significant differences between the patients and controls in terms of their mean age (41.1±11.7 years, P=0.352), gender (all female, P=0.881) and ethnicity (Turkish), indicating that the frequency matching between the groups was adequate.
Distribution of the PARP-1 SNPs in HT
The distribution of the rs2793378, rs7527192 and rs1136410 genotypes in the control group were within HWE. The distribution of the genotypes and the allele frequencies of all of the studied polymorphisms are shown in Table 1.
Table 1. Distribution of three SNPs in PARP-1 gene in Hashimoto thyroiditis.
| Genotype/allele | Cases, n (%) | Controls, n (%) | OR (95% CI) | P-value |
|---|---|---|---|---|
| rs1136410 | 141 | 150 | ||
| TT (Val/Val)a | 98 (70) | 77 (51) | 1 | |
| TC (Val/Ala) | 31 (21) | 61 (41) | 2.504 (1.48–4.23) | 0.001 |
| CC (Ala/Ala) | 12 (9) | 12 (8) | 1.273 (0.54–2.99) | 0.579 |
| T (Val) allele frequency | 227 (81) | 215 (72) | 1 | |
| C (Ala) allele frequency | 55 (19) | 85 (28) | 1.632 (1.1–2.403) | 0.013 |
| rs2793378 | 141 | 150 | ||
| CC | 62 (44) | 60 (40) | 1 | |
| CT | 62 (44) | 65 (43) | 0.92 (0.56–1.52) | 0.752 |
| TT | 17 (12) | 25 (17) | 0.66 (0.32–1.34) | 0.247 |
| C allele frequency | 186 (66) | 185 (62) | 1 | |
| T allele frequency | 96 (34) | 115 (38) | 0.84 (0.47–1.50) | 0.556 |
| rs7527192 | 141 | 150 | ||
| GG | 75 (53) | 60 (40) | 1 | |
| GA | 62 (44) | 81 (54) | 1.633 (1.017–2.623) | 0.042 |
| AA | 4 (3) | 9 (6) | 2.813 (0.826–9.581) | 0.087 |
| G allele frequency | 212 (75) | 201 (67) | 1 | |
| A allele frequency | 70 (25) | 99 (33) | 1.492 (1.039–2.142) | 0.03 |
Abbreviations:
CI, confidence intervals: OR, odds ratios.
The corresponding amino acids of each genotypes of rs1136410 polymorphism were given in parentheses.
The differences resulting from the genotypes were determined by proceeding χ2-test. P value after Bonferroni correction; P=5.6×10−3 (calculated as 0.05/9). P value in bold is still maintained significance after Bonferroni correction.
We found a significant association between PARP-1 rs1136410 and rs7527192 and HT. The frequency of the heterozygous TC genotype of rs1136410 was significantly greater in the controls compared to the patients (OR: 2.504, 95% CI: 1.48–4.23, P=0.001). Moreover, the frequency of the C allele of rs1136410 was found to be statistically higher in the healthy group (OR: 1.632, 95% CI: 1.1–2.403, P=0.013), and rs1136410 maintained its significance following Bonferroni correction. The frequency of the heterozygous GA genotype of rs7527192 was significantly higher in the controls (OR: 1.633, 95% CI: 1.017–2.623, P=0.042) than in the patients. Additionally, the frequency of the A allele of rs7527192 was found to be statistically higher in the healthy group (OR: 1.492, 95% CI: 1.039–2.142, P=0.03); although, neither the GA genotype nor the A allele of rs7527192 maintained significance following Bonferroni correction. In addition, the C allele of rs1136410 showed no significance following Bonferroni correction, and PARP-1 rs2793378 showed no protective or risk effect in HT disease. All of the results are shown in Table 1.
Haplotype analysis of the PARP-1 SNPs
We also investigated whether the three polymorphisms were in linkage disequilibrium, and any common haplotypes associated with diseases and rare haplotypes (with a frequency <5%) were excluded from the association analyses. The most common haplotypes of the three polymorphisms, calculated by Haploview 4.2, are summarized in Table 2.
Table 2. The distribution of PARP-1 haplotypes.
|
PARP-1 haplotypes (SNPs)
|
Frequency
|
x2 | P-value | ||||
|---|---|---|---|---|---|---|---|
| rs2793378 | rs7527192 | rs1136410 | Total | Cases | Controls | ||
| T | G | C | 0.488 | 0.521 | 0.406 | 2.899 | 0.0887 |
| T | A | T | 0.128 | 0.132 | 0.115 | 0.145 | 0.7036 |
| C | G | T | 0.116 | 0.121 | 0.102 | 0.182 | 0.6694 |
| T | A | C | 0.096 | 0.091 | 0.106 | 0.142 | 0.7068 |
| C | G | C | 0.085 | 0.061 | 0.146 | 5.01 | 0.0252 |
| T | G | T | 0.078 | 0.065 | 1.109 | 1.465 | 0.2262 |
Abbreviation: SNP, single-nucleotide polymorphism.
P value after Bonferroni correction; P=8.3×10−3 (calculated as 0.05/6).
The haplotypes were generated using the three PARP-1 intragenic SNPs (rs2793378, rs7527192 and rs1136410) among the HT cases and controls, and six different haplotypes were generated (with a frequency >5%).
The prevalence of the CGC haplotype was significantly higher in the controls (ratio of the cases to the controls: 0.061:0.146, P=0.0252, x2=5.01), as shown in Table 2. The first marker of the CGC haplotype, the C allele, is wild type for rs2793378. The second marker of the CGC haplotype, the G allele, is wild type for rs7527192. The third marker, the C allele, is mutant for rs1136410. The difference was based on the third marker, the C allele of rs1136410, which serves as a protective factor. However, because this study was based on a small sample size, we applied a Bonferroni correction to decrease the type I error. Following Bonferroni correction, the haplotype analysis did not show any significance. Furthermore, two of the SNPs in PARP-1 (rs1136410 and rs7527192) were in strong LD (Figure 1).
Figure 1.
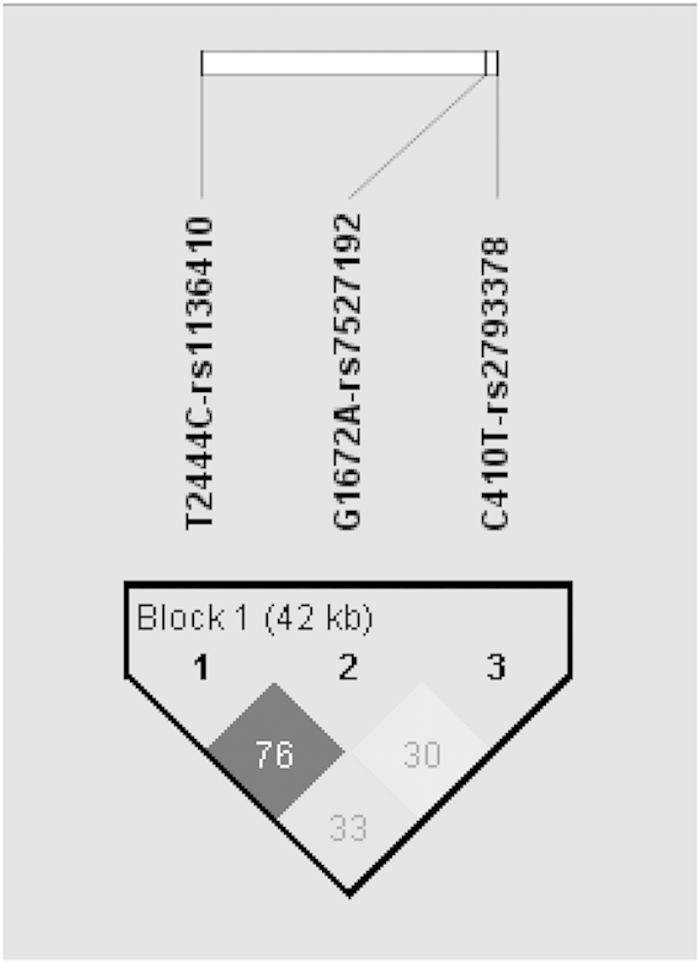
Pairwise linkage disequilibrium plot for examined PARP-1 SNPs. The higher r2-value shows the darker color. Chromosome position of PARP-1 rs2793378 site is 226597341, rs7527192 site is 226596079 and rs1136410 site is 226555302.
Combined genotype analysis of the PARP-1 SNPs
Tables 3, 4, 5 summarize the association studies among the combined genotypes of the three SNPs and the overall risk for HT. The PARP-1 rs2793378 and rs7527192 SNPs showed statistical significance among the combined genotypes in the HT patients and controls. The combined CC/GA, CT/GG and TT/GA genotype frequencies were significantly higher in the control group versus the HT group when compared with the combined CC/GG genotype (OR: 2.956, 95% CI: 1.114–7.840, P=0.027; OR: 2.66, 95% CI: 1.13–6.259, P=0.023; OR: 4.644, 95% CI: 1.455–14.822, P=0.007, respectively). Following Bonferroni correction, only the combined TT/GA genotype remained significant and was found to be a significant protective factor against HT disease (P=0.007), as shown in Table 3.
Table 3. The distribution of rs2793378- rs7527192 combined genotypes.
| rs2793378- rs7527192 combined genotypes | Cases, n | Controls, n | OR (95% CI) | P-value |
|---|---|---|---|---|
| CC/GG | 38 | 15 | 1 | |
| CC/GA | 12 | 14 | 2.956 (1.114–7.840) | 0.027 |
| CT/GG | 20 | 21 | 2.66 (1.13–6.259) | 0.023 |
| CT/GA | 32 | 17 | 1.346 (0.582–3.113) | 0.487 |
| TT/GG | 6 | 3 | 1.267 (0.28–5.73) | 0.711 |
| TT/GA | 6 | 11 | 4.644 (1.455–14.822) | 0.007 |
Abbreviations: CI, confidence intervals; OR, odds ratios.
CC/AA, CT/AA, and TT/AA combined genotypes were excluded in the table as none observed in the subjects or the size was not enough for the statistical analyses. P value after Bonferroni correction; P=8.3×10−3 (calculated as 0.05/6). P value in bold is still maintained significance after Bonferroni correction.
Table 4. The distribution of rs7527192–rs1136410 combined genotypes.
| rs7527192–rs1136410 combined genotypes | Cases n | Controls n | OR (95% CI) | P-value |
|---|---|---|---|---|
| GG/TT | 29 | 10 | 1 | |
| GG/TC | 13 | 9 | 2.008 (0.66–6.111) | 0.216 |
| GG/CC | 7 | 2 | 0.829 (0.147–4.665) | 1 |
| GA/TT | 30 | 13 | 1.257 (0.477–3.314) | 0.644 |
| GA/TC | 4 | 2 | 1.45 (0.23–9.16) | 0.65 |
Abbreviations: CI, confidence intervals; OR, odds ratios.
GA/CC, AA/TC, AA/TT and AA/CC combined genotypes were excluded in the table as none observed in the subjects or the size was not enough for the statistical analyses. P value after Bonferroni correction; P=0.01 (calculated as 0.05/5).
Table 5. The distribution of rs2793378–rs1136410 combined genotypes.
| rs2793378–rs1136410 Combined Genotypes | Cases n | Controls n | OR (95% CI) | P-value |
|---|---|---|---|---|
| CC/TT | 34 | 10 | 1 | |
| CC/TC | 3 | 1 | 1.133 (0.106–12.129) | 0.918 |
| CC/CC | 3 | 2 | 2.267 (0.331–15.509) | 0.584 |
| CT/TT | 21 | 8 | 1.295 (0.441–3.803) | 0.637 |
| CT/TC | 12 | 7 | 0.58 (0.19–1.78) | 0.340 |
| TT/TT | 2 | 2 | 1.983 (0.616–6.382) | 0.247 |
| TT/TC | 3 | 1 | 1.133 (0.106–12.129) | 0.918 |
Abbreviations: CI, confidence intervals; OR, odds ratios.
CT/CC and TT/CC combined genotypes were excluded in the table as none observed in the subjects or the size was not enough for the statistical analysis. P value after Bonferroni correction; P=7.1×10−3 (calculated as 0.05/7).
Aside from the abovementioned results, there were no other significant differences between the multiple comparisons of rs7527192-rs1136410, and rs2793378-rs1136410 in the HT patients and control groups (Tables 4 and 5).
Discussion
Based on genome-wide association studies (GWAS) of thyroid disorders, it is clear that genetic factors are thought to be considerable determinants of thyroid function and autoimmunity. Recently, an intronic variant of VAV3 (vav 3 guanine nucleotide exchange factor, rs12126655) was found to be associated with TSH concentrations, and another variant (rs2071403) located 75 bp proximal to the translation start site of TPO was found to be significantly associated with plasma anti-TPO antibody positivity. Another intronic variant of HLA-DPB2 (rs733208) was associated with anti-TPO antibody positivity as well.21 Eriksson et al.22 showed evidence of an association between hypothyroidism and autoimmune thyroid diseases with SNPs in the HLA class I and II regions (rs2517532 and rs2516049, respectively), and near the CAPZB (capping protein (actin filament) muscle Z-line, beta, rs12091047), PDE8B (phosphodiesterase 8B, rs4704397) and CTLA-4 (toxic T lymphocyte antigen-4, rs231779) genes. Based on candidate gene studies, it is known that CTLA-4 is widely associated with many other autoimmune diseases, including autoimmune hypothyroidism.
In addition, variants near FOXE1, a thyroid transcription factor, were identified as genetic risk factors for primary hypothyroidism.23 In a GWAS of FoxE1, it was suggested that FoxE1 and PARP-1 participate in the human NIS repressor complex, which is a trans-active transcriptional repressor found in thyroid cancer cells that contains PARP-1 and functions via its poly(ADP-ribosyl)ation activity.24,25 Therefore, PARP-1 is a strong and suitable candidate gene for identifying associations between genetic factors in thyroid autoimmunity.
Several studies strongly support that PARP-1 plays a considerable role in autoimmune inflammatory disorders.17,26,27 PARP-1 inhibitors or PARP-1 deletions are being used in many therapeutic approaches that are based on blocking tissue or systemic inflammation.28,29 Recently, alternative pathways have been suggested involving the capability of PARP-1 to regulate the transcription of inflammation-linked genes. Several recent publications indicate that the nuclear protein PARP-1 regulates the transcription and synthesis of inflammatory mediators.30 In addition, the relationship between PARP-1 activity and inflammation has been widely investigated, and the link with inflammation was firmly established via the pharmacological inhibition of PARP-1 activity, which allowed the attenuation of the inflammatory response.6,30 The inhibition of PARP-1 activity is protective in a wide range of inflammatory conditions, such as cardiovascular disease, diabetes, rheumatoid arthritis and stroke.13,31 Therefore, the PARP-1 gene may be a strong candidate based on the observations made in knockout mice, and it provides promising therapeutic approaches to inflammatory diseases using PARP-1 inhibitors.
Alternatively, PARP-1 protein concentration is controlled by the direct regulation of promoter activity. Therefore, polymorphisms within the promoter region may also affect PARP-1 activity. Multiple transcription sites, as well as 3 sets of CCAAT/TATA boxes, have been identified in the promoter region, which provide a binding site for YY1 transcription factors.20 In addition, four sequence variations have been identified in this region: rs2793378, Pol(A)n, C1362T and rs7527192.16,20 According to Infante et al.,32 the C410T polymorphism that is found in the heterozygous state increased the risk for Parkinson’s disease, while rs7527192 heterozygosity significantly delayed the onset of the disease. In this study, we did not find any significance in regards to rs2793378.
Recently, rs2793378, rs7527192, and rs1136410 have been investigated in many inflammatory autoimmune diseases, including RA,16 AR,33 asthma,34 nephritis and arthritis in SLE patients35 and Alzheimer's disease,36,37 as described above. In our study, the heterozygous GA genotype of PARP-1 rs7527192 was found to have a protective effect on HT disease in the Turkish population. This is comparable to the results of Infante et al., which showed that certain promoter variations (rs2793378 and rs7527192) are associated with protection against Parkinson’s disease.24 We also previously determined that the heterozygous genotype of rs7527192 was significantly associated with susceptibility to allergic rhinitis (AR) in the Turkish population. Moreover, in this study, according to the multiple comparisons of the combined genotypes of rs2793378–rs7527192, the frequency of the TT/GA combined genotype was significantly higher in the controls versus the HT group when compared with CC/GG combined genotype. Following Bonferroni correction, only the TT/GA genotype continued to show significance as a protective factor. When the mutant form of rs2793378 and the heterozygous form of rs7527192 were combined, a significant protective effect was observed in the HT patients.
The PARP-1 promoter haplotypes have also been shown to be related to susceptibility to some chronic inflammatory diseases, such as AR, RA and celiac disease.16,33,38 The protective effect of a haplotype of the PARP-1 gene polymorphisms for Alzheimer's disease was also detected.37 The rs2793378, rs7527192 and rs1136410 haplotypes were not associated with an increased risk for AR.33 According to Pascual et al.,16 PARP-1 haplotypes (−410, −1327, CA microsatellite repeat) play a role in susceptibility to RA in the Spanish population, whereas the rs7527192 variant did not appear to be part of this haplotype. Although the CGC haplotype was found to have a protective effect based on the C (Ala) allele of rs1136410, it did not survive Bonferroni correction.
rs1136410 is present in approximately 5–33% of the general population.19 We previously reported that the T (Val) allele of rs1136410 is associated with an increased risk for asthma.34 Furthermore, rs1136410 was significantly associated with coronary artery disease, which is also an inflammatory process.39 Hur et al.35 demonstrated that the nonsynonymous variant +40329T→C(V762A) was also significantly associated with an increased risk for arthritis in Korean SLE patients. However, according to Onaran et al.,40 rs1136410 was not associated with a risk for rheumatoid arthritis. The Val762 variants of PARP-1 rs1136410 have also been found to increase the risk of diabetic polyneuropathy in Russian type 1 diabetic patients.41 Additionally, we found that the heterozygous TC (Val/Ala) genotype of rs1136410 still had a protective effect on susceptibility to HT in the Turkish population even following Bonferroni correction.
PARP-1 and nuclear factor kappa B (NF-κB) have both been suggested to play a considerable role in inflammatory diseases. It has been shown that PARP-1 can act as a coactivator of NF-κB in terms of inflammatory processes in the cell.42 These findings provided us with new insights into the pathogenesis of different inflammatory disorders, such as HT. We previously reported that the frequency of the deletion allele of NF-κB1 rs28362491 that accompanies high interleukin-6 (IL-6) levels was significant in an HT patient group.43 This study is a follow-up to our previous research.
Moreover, it should be kept in mind that the small sample size, which may have influenced the statistical power of our analyses, may be considered to be a limitation of this study. Although a study with low statistical power has a reduced chance of detecting a true effect, case–control studies with small sample sizes are still widely used, and they can be used to assess previously identified candidate regions under selection and more precisely determine the selection targets. Nonetheless, a larger sample size or replication study will provide reassurance for further studies.
We carried out single and combined genotype and haplotype analyses to evaluate the SNPs rs2793378, rs7527192 and rs1136410 in a population-based case–control study. To the best of our knowledge, there are no published studies exploring the associations between these PARP-1 variants and HT disease. We found that the heterozygous genotype of rs1136410 had a significantly decreased risk for HT disease in a group of women from the Turkish population. Molecular heterosis may be present in 50% of all of the gene associations,44 and it has also been reported for variations in the PARP-1 gene.32,34 The biological meaning of the 'protective' effect of the heterozygous genotype of rs1136410 toward HT cannot be easily explained by the existing observations regarding the PARP-1 SNPs. However, it has been demonstrated that the modulation of cytokine production by various stimulators is more strongly affected by the heterozygous variants of several SNPs in various genes than by the homozygous variant. For example, the heterozygous genotypes of four SNPs (rs7527192, rs2793378, rs28362491 and rs696) were recently found to be related to elevated IL-6, IL-1β and tumor necrosis factor alpha (TNF-α) cytokine production in Graves’ disease in a Turkish population.45 In addition, the 4257GA heterozygous genotype was found to be associated with higher IL-13 levels in asthmatics.46 A relationship between the heterozygous -137 G/C genotype in the IL-18 gene with elevated IL-18 serum levels and bladder cancer risk was detected in a North Indian population.47 Moreover, the T cells from heterozygous TC-carrier rheumatoid arthritis patients produced higher TNF-α, IL-17, and interferon-γ when compared with patients carrying the non-TC haplotype.48 All of these studies show meaningful associations between the heterozygous genotype and increasing cytokine production.
PARP-1 is required for the activation of several transcription factors via the inflammation of several mediators. It has been speculated that a different PARP-1 binding and activation mechanism because of the heterozygous genotype of rs1136410 may affect the expression of the genes involved in the inflammatory response, perhaps via an altered inflammatory pathway. Therefore, this polymorphism may confer a protective effect against HT. However, this hypothesis still requires further investigation.
In summary, it is possible that the interactions of variants of the PARP-1 gene have key roles in organizing the various changes that occur during the inflammatory response in HT disease. However, replication studies or additional functional studies with a larger sample size are required to confirm the putative protective role of these PARP-1 gene variants. More precisely, larger trials and replication studies that include different ethnic groups are highly recommended to properly define the relationship between the PARP-1 polymorphisms (especially rs1136410 TC) and the development of HT, as well as the prognosis of the disease.
Acknowledgments
This study was supported by a grant from the Scientific Research Projects Coordination Unit of Istanbul University (Project no. 4055).
The authors declare no conflict of interest.
References
- Vaidya B, Kendall-Taylor P, Pearce SHS. The genetics of autoimmune thyroid disease. J Clin Endocrinol Metab 2002; 87: 5385–5397. [DOI] [PubMed] [Google Scholar]
- Repplinger D, Bargren A, Zhang Y-W, Adler J, Haymart M, Chen H. Is Hashimoto's thyroiditis a risk factor for papillary thyroid cancer?. J Surg Res 2008; 150: 49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistiakov DA. Immunogenetics of Hashimoto's thyroiditis. J Autoimmune Dis 2005; 2: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajjan RA, Watson PF, Weetman AP. Cytokines and thyroid function. Adv Neuroimmunol 1996; 6: 359–386. [DOI] [PubMed] [Google Scholar]
- Paknys G, Kondrotas AJ, Kevelaitis E. Risk factors and pathogenesis of Hashimoto's thyroiditis. Medicina (Kaunas) 2009; 45: 574–583. [PubMed] [Google Scholar]
- Virág L, Bai P, Bak I, Pacher P, Mabley JG, Liaudet L et al. Effects of poly(ADP-ribose) polymerase inhibition on inflammatory cell migration in a murine model of asthma. Med Sci Monit 2004; 10: 77–83. [PubMed] [Google Scholar]
- Virág L, Szabó C. The therapeutic potential of poly(ADP-Ribose) polymerase inhibitors. Pharmacol Rev 2002; 54: 375–429. [DOI] [PubMed] [Google Scholar]
- Oliver FJ, Me´nissier-deMurcia J, Nacci C, Decker P, Andriantsitohaina R, Muller S et al. Resistance to endotoxic shock as a consequence of defective NF-kappaB activation in poly (ADP-ribose) polymerase-1 deficient mice. EMBO J 1999; 18: 4446–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strosznajder R, Gadamski R, Walski M. Inhibition of poly(ADPribose) polymerase activity protects hippocampal cells against morphological and ultrastructural alteration evoked by ischemia-reperfusion injury. Folia Neuropathol 2005; 43: 156–165. [PubMed] [Google Scholar]
- Strosznajder RP, Jesko H, Zambrzycka A. Poly(ADP-ribose) polymerase: the nuclear target in signal transduction and its role in brain ischemia-reperfusion injury. Mol Neurobiol 2005; 31: 149–167. [DOI] [PubMed] [Google Scholar]
- Endres M, Wang ZQ, Namura S, Waeber C, Moskowitz MA. Ischemic brain injury is mediated by the activation of poly(ADP-ribose) polymerase. J Cereb Blood Flow Metab 1997; 17: 1143–1151. [DOI] [PubMed] [Google Scholar]
- Boulares AH, Zoltoski AJ, Sherif ZA, Jolly P, Massaro D, Smulson ME. Gene knockout or pharmacological inhibition of poly(ADP-ribose) polymerase-1 prevents lung inflammation in a murine model of asthma. Am J Respir Cell Mol Biol 2009; 28: 322–329. [DOI] [PubMed] [Google Scholar]
- Peralta-Leal A, Rodríguez-Vargas JM, Aguilar-Quesada R, Rodríguez MI, Linares JL, de Almodóvar MR et al. PARP inhibitors: new partners in the therapy of cancer and inflammatory diseases. Free Radic Biol Med 2009; 47: 13–26. [DOI] [PubMed] [Google Scholar]
- Choi SK, Galán M, Kassan M, Partyka M, Trebak M, Matrougui K. Poly(ADP-ribose) polymerase 1 inhibition improves coronary arteriole function in type 2 diabetes mellitus. Hypertension 2012; 59: 1060–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komjáti K, Besson VC, Szabó C. Poly (adp-ribose) polymerase inhibitors as potential therapeutic agents in stroke and neurotrauma. Curr Drug Targets CNS Neurol Disord 2005; 4: 179–194. [DOI] [PubMed] [Google Scholar]
- Pascual M, López-Nevot MA, Cáliz R, Ferrer MA, Balsa A, Pascual-Salcedo D et al. A poly(ADP-ribose) polymerase haplotype spanning the promoter region confers susceptibility to rheumatoid arthritis. Arthritis Rheum 2003; 48: 638–641. [DOI] [PubMed] [Google Scholar]
- Masutani M, Nakagama H, Sugimura T. Poly(ADP-ribosyl)ation in relation to cancer and autoimmune disease. Cell Mol Life Sci 2005; 62: 769–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottet F, Blanché H, Verasdonck P, Le Gall I, Schächter F, Bürkle A et al. New polymorphisms in the human poly(ADP-ribose) polymerase-1 coding sequence: lack of association with longevity or with increased cellular poly(ADP-ribosyl)ation capacity. J Mol Med (Berl) 2000; 78: 431–440. [DOI] [PubMed] [Google Scholar]
- Wang XG, Wang ZQ, Tong WM, Shen Y. PARP1 Val762Ala polymorphism reduces enzymatic activity. Biochem Biophys Res Commun 2007; 354: 122–126. [DOI] [PubMed] [Google Scholar]
- Kato N, Morita H, Sugiyama T, Kurihara H, Tsubaki S, Nabika T. Evaluation of the poly(ADP-ribose) polymerase gene in human stroke. Atherosclerosis 2000; 148: 345–352. [DOI] [PubMed] [Google Scholar]
- Kwak SH, Park YJ, Go MJ, Lee KE, Kim SJ, Choi HS et al. A genome-wide association study on thyroid function and anti-thyroid peroxidase antibodies in Koreans. Hum Mol Genet 2014; 23: 4433–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson N, Tung JY, Kiefer AK, Hinds DA, Francke U, Mountain JL et al. Novel associations for hypothyroidism include known autoimmune risk loci. PLoS ONE 2012; 7: e34442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 2011; 89: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ain KB. Human sodium-iodide symporter (hNIS) gene expression is inhibited by a trans-active transcriptional repressor, NIS-repressor, containing PARP-1 in thyroid cancer cells. Endocr Relat Cancer 2010; 17: 383–398. [DOI] [PubMed] [Google Scholar]
- Fernández LP, López-Márquez A, Martínez ÁM, Gómez-López G, Santisteban P. New insights into FoxE1 functions: identification of direct FoxE1 targets in thyroid cells. PLoS ONE 2013; 8: e62849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj V, Soundarapandian MM, Chechneva O, Williams AJ, Sidorov MK, Soulika AM. PARP-1 deficiency increases the severity of disease in a mouse model of multiple sclerosis. J Biol Chem 2009; 284: 26070–26084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog NR, Dinnall JA, Gallucci S, Madaio MP, Caricchio R. Poly(ADP-ribose) polymerase-1 regulates the progression of autoimmune nephritis in males by inducing necrotic cell death and modulating inflammation. J Immunol 2009; 182: 7297–7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesel R, Kurpisz M, Kroger H. Modulation of inflammatory arthritis by inhibition of poly(ADP ribose) polymerase. Inflammation 1995; 19: 379–387. [DOI] [PubMed] [Google Scholar]
- Szabo C, Virag L, Cuzzocrea S, Scott GS, Hake P, O’Connor MP et al. Protection against peroxynitrite-induced fibroblast injury and arthritis development by inhibition of poly(ADP-ribose) synthase. Proc Natl Acad Sci USA 1998; 95: 3867–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Rey E, Martínez-Romero R, O'Valle F, Aguilar-Quesada R, Conde C, Delgado M et al. Therapeutic effect of a poly(ADP-Ribose) polymerase-1 inhibitor on experimental arthritis by downregulating inflammation and Th1 Response. PLoS ONE 2007; 2: e1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti V, Donà F, Tillhon M, Scovassi AI. PARP inhibitors: new tools to protect from inflammation. Biochem Pharmacol 2010; 80: 1869–1877. [DOI] [PubMed] [Google Scholar]
- Infante J, Sánchez-Juan P, Mateo I, Rodríguez-Rodríguez E, Sánchez-Quintana C, Llorca J et al. Poly (ADP-ribose) polymerase-1 (PARP-1) genetic variants are protective against Parkinson's disease. J Neurol Sci 2007; 256: 68–70. [DOI] [PubMed] [Google Scholar]
- Ozaydin A, Akbas F, Aksoy F, Yildirim YS, Demirhan H, Karakurt F et al. Investigation of poly (adp-ribose) polymerase-1 genetic variants as a possible risk for allergic rhinitis. Genet Test Mol Biomarkers 2014; 18: 57–61. [DOI] [PubMed] [Google Scholar]
- Tezcan G, Gurel CB, Tutluoglu B, Onaran IKanigur-Sultuybek G. The Ala allele at Val762Ala polymorphism in poly(ADP-ribose) polymerase-1 (PARP-1) gene is associated with a decreased risk of asthma in a Turkish population. J Asthma 2009; 46: 371–374. [DOI] [PubMed] [Google Scholar]
- Hur JW, Sung YK, Shin HD, Park BL, Cheong HS, Bae SC. Poly(ADP-ribose) polymerase (PARP) polymorphisms associated with nephritis and arthritis in systemic lupus erythematosus. Rheumatology (Oxford) 2006; 45: 711–717. [DOI] [PubMed] [Google Scholar]
- Infante J, Llorca J, Mateo I, Rodríguez-Rodríguez E, Sánchez-Quintana C, Sánchez-Juan P et al. Interaction between poly(ADP-ribose) polymerase 1 and interleukin 1A genes is associated with Alzheimer's disease risk. Dement Geriatr Cogn Disord 2007; 23: 215–218. [DOI] [PubMed] [Google Scholar]
- Liu HP, Lin WY, Wu BT, Liu SH, Wang WF, Tsai CH et al. Evaluation of the poly(ADP-ribose) polymerase-1 gene variants in Alzheimer's disease. J Clin Lab Anal 2010; 24: 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda B, Koeleman BP, López-Nevot MA, Ortega E, Maldonado J, López M et al. Poly (ADP-ribose) polymerase-1 haplotypes are associated with coeliac disease. Int J Immunogenet 2005; 32: 245–248. [DOI] [PubMed] [Google Scholar]
- Narne P, Ponnaluri KC, Singh S, Siraj M, Ishaq M. Relationship between NADPH oxidase p22phox C242T, PARP-1 Val762Ala polymorphisms, angiographically verified coronary artery disease and myocardial infarction in South Indian patients with type 2 diabetes mellitus. Thromb Res 2012; 130: 259–265. [DOI] [PubMed] [Google Scholar]
- Onaran I, Tezcan G, Özgönenel L, Çetin E, Özdemir AT, Kanıgür-Sultuybek G. The Val762Ala polymorphism in the poly(ADP-ribose) polymerase-1 gene is not associated with susceptibility in Turkish rheumatoid arthritis patients. Rheumatol Int 2009; 29: 797–800. [DOI] [PubMed] [Google Scholar]
- Nikitin AG, Chudakova DA, Strokov IA, Bursa TR, Chistiakov DA, Nosikov VV. Leu54Phe and Val762Ala polymorphisms in the poly(ADP-ribose)polymerase-1 gene are associated with diabetic polyneuropathy in Russian type 1 diabetic patients. Diabetes Res Clin Pract 2008; 79: 446–452. [DOI] [PubMed] [Google Scholar]
- Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci 2002; 59: 1534–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc A, Batar B, Celik O, Onaran I, Tasan E, Kanigur-Sultuybek G. Polymorphism of the NFKB1 affects the serum inflammatory levels of IL-6 in Hashimoto thyroiditis in a Turkish population. Immunobiology 2014; 219: 531–536. [DOI] [PubMed] [Google Scholar]
- Comings DE, MacMurray JP. Molecular heterosis: a review. Mol Genet Metab 2000; 71: 19–31. [DOI] [PubMed] [Google Scholar]
- Niyazoglu M, Baykara O, Koc A, Aydoğdu P, Onaran I, Dellal FD et al. Association of PARP-1, NF-κB, NF-κBIA and IL-6, IL-1β and TNF-α with Graves disease and Graves ophthalmopathy. Gene 2014; 547: 226–232. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan AK, Raj VL, Tan LK, Liam CK. Single nucleotide polymorphism in the promoter of the human interleukin-13 gene is associated with asthma in Malaysian adults. Biomed Res Int 2013; 2013: 981012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal PK, Singh V, Srivastava P, Mittal RD. Association of IL-12, IL-18 variants and serum IL-18 with bladder cancer susceptibility in North Indian population. Gene 2013; 519: 128–134. [DOI] [PubMed] [Google Scholar]
- Snir O, Hesselberg E, Amoudruz P, Klareskog L, Zarea-Ganji I, Catrina AI et al. Genetic variation in the serotonin receptor gene affects immune responses in rheumatoid arthritis. Genes Immun 2013; 14: 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]


