Abstract
Interstitial deletion of 6q21–22 has been previously reported in 11 individuals, who presented with intellectual disability, facial dysmorphism, cardiac abnormality, cerebellar hypoplasia and dysplasia of the corpus callosum. Here, we report the first instance of a patient with 6q21–22 deletion presenting with interrupted aortic arch in addition to the previously described clinical signs. Array analysis using Agilent Human genome CGH 180K identified a 13.3-Mb deletion at 6q21–q22.31 (nt. 109885195–123209593).
Common clinical features of 6q21–22 deletion among 11 previously reported patients1–4 and our current patient include intellectual disability, hyper- or hypotelorism, long philtrum or wide nasal bridge, microcephaly and dysplasia of the corpus callosum and cerebellum. Cardiac abnormalities were also detected in 5 of these 12 patients, including ventricular septal defect (VSD), pulmonary atresia, coarctation of aorta (CoA), double outlet right ventricle and patent ductus arteriosus. Interrupted aortic arch (IAA) type B was identified only in our patient (Table 1).
Table 1. Clinical features of 6q21–22 deletion.
| Patient (this study) | Patient 1 (Rosenfeld et al.4) | Patient 2 (Rosenfeld et al.4) | Patient 3 (Rosenfeld et al.4) | Patient 4 (Rosenfeld et al.4) | Patient 5 (Rosenfeld et al.4) | Patient 6 (Rosenfeld et al.4) | Patient 7 (Rosenfeld et al.4) | Patient 8 (Rosenfeld et al.4) | Patient from Chen et al.2 | Patient from Bzduch et al.1 | Patient from Park et al.3 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deletion (Mb or cytogenetic) | 109.9 ~123.2 | 111 ~124 | 113.4 ~121 | 114.3 ~118 | 106.6 ~114.5 | 104.3 ~121 | 113.2 ~115 | 114.2 ~115 | 116 ~129 | 113 ~123.2 | 6q22.2 →q23 | 6q22.2 →23.1 |
| Hyper- or hypo-telorism | Hyper | − | − | − | Hyper | − | Hyper | − | Hypo | Hyper | NA | NA |
| Long philtrum or wide nasal bridge | + | − | + | − | + | + | + | − | + | − | + | + |
| Cardiac abnormality | IAA, VSD | − | − | − | DORV, PA, VSD, ASD | PDA | − | − | − | VSD, PA | CoA (mild) | − |
| Microcephaly | − | + | − | − | + | + | − | + | + | + | + | + |
| Cranial MRI hypoplasia of the cerebellum | + | NA | +mild | − | + | + | − | + | − | NA | NA | NA |
| Dysplasia or hypoplasia of the CC | + | NA | +dysmorphic | +thick | +Partial ACC | + | − | +thick, shorten | − | NA | NA | NA |
| Intellectual disability | + | + | + | + | + | + | mild | + | + | NA | + | + |
Abbreviations: ACC, agenesis of the corpus callosum; ASD, atrial septal defect; CC, corpus callosum; CoA, coarctation of aorta; DORV, double outlet right ventricle; IAA, interrupted aortic arch; MRI, magnetic resonance imaging; NA, not available; PA, pulmonary atresia; PDA, patent ductus arteriosus; VSD, ventricular septal defect.
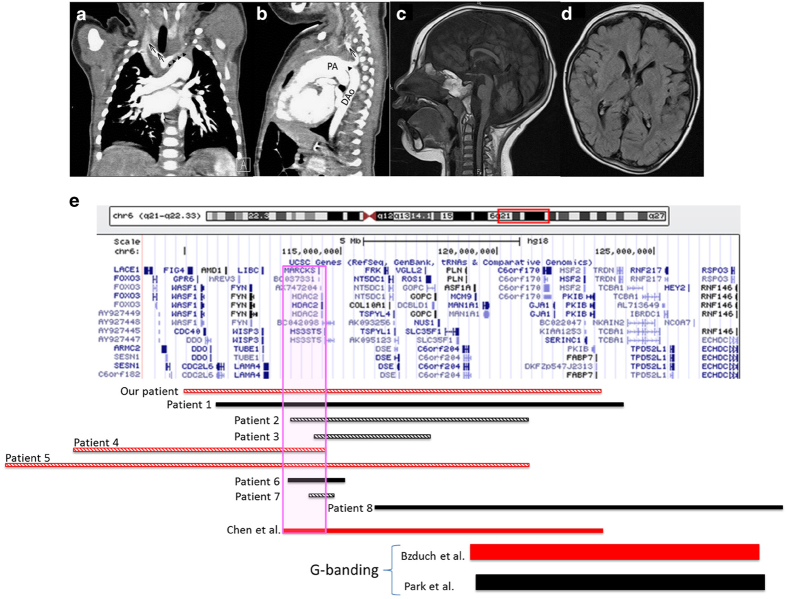
The patient was a 5-year-old boy and the first-born son of unrelated and healthy Japanese parents without notable family history. The patient was delivered at 38 weeks gestation with a birth weight of 3,075 g (+0.60 standard deviation (s.d.)), height of 48.5 cm (+0.20 s.d.) and head circumference of 33.4 cm (+0.30 s.d.). The Apgar score was 9 at 5 min. He failed to thrive, and his body weight was 3,560 g at 1 month of age. A systolic murmur was detected, and echocardiogram and thoracic contrast-enhanced computer tomography revealed IAA and a VSD (Figure 1a,b). IAA repair and VSD patch closure under cardiopulmonary bypass was performed at 1 month of age. At 4 years of age, his height, weight and occipitofrontal circumference were 103.5 cm (+0.65 s.d.), 16.4 kg (+0.30 s.d.) and 48.2 cm (−1.25 s.d.), respectively. He could sit and walk with assistance, and had an intellectual disability (development quotient of 20) without meaningful words. Additional presenting phenotypes included a prominent forehead, hypotelorism, short upturned nose, long philtrum, small mandible, high-arched palate, low-set ears, sparse hair and internal strabismus. Cranial magnetic resonance imaging revealed dysgenesis of the cerebral cortex and the corpus callosum, and mild hypoplasia of the brainstem and cerebellum (Figure 1c,d). G-banding and fluorescence in situ hybridization (FISH) analysis of the 22q11.2 region were normal. For genetic analysis, blood was obtained from the patient with written informed consent from the mother for testing. This test was approved by the bioethics committee for human gene analysis at Jichi Medical University. Parental samples were not made available for analysis. Array comparative genomic hybridization (aCGH) analysis was performed using SurePrint G3 Human CGH Microarray Kit, 4×180K (Agilent Technologies, Santa Clara, CA, USA) and a 13.3-Mb deletion at 6q21–q22.31 (nt. 109885195–123209593) was detected.
Figure 1.
Thoracic contrast-enhanced computer tomography at 1 month of age (a, b), cranial magnetic resonance imaging findings at 2 years of age (c, d) and comparison of the deleted positions (e). (a) Interruption of the aortic arch proximal to the left subclavian artery is shown. Arrows indicate the right brachiocephalic artery. Triangles indicate the left common carotid artery. (b) The pulmonary artery inserts into the descending aorta and subclavian artery through the arterial duct. Arrow indicates the right brachiocephalic artery. Triangle indicates the arterial duct. PA, pulmonary artery; DAo, descending aorta. (c) Sagittal and T1-weighted image, showing mild hypoplasia in the corpus callosum, brainstem and cerebellum. (d) Axial and T1-weighted image, showing mild atrophy of the cerebral cortex. (e) Bars indicate the deletion of individual patients. Red bars indicate cardiac abnormalities. Black bars indicate no cardiac abnormalities. Slanting lines indicate brain abnormalities. The main genes existing in this area were listed as modified University of California Santa Cruz (UCSC) data. Genes surrounding the pink box were included in the common deleted region in those with cardiac abnormalities except for the patient reported by Bzduch et al.1 Patients 1–8 were reported by Rosenfeld et al.4
Concerning cardiac abnormalities in 6q21–22 deletion syndrome, all patients shared the deleted region from 113 to 114.5 Mb including the myristoylated alanine-rich protein kinase c substrate (MARCKS) and histone deacetylase 2 (HDAC2) genes, among others (Figure 1e). MARCKS is an actin and calmodulin-binding protein, and MARCKS-null mice experience embryonic lethality because of a neural defect.5 No cardiac phenotype has been reported in MARKS-null mice, although MARCKS is expressed in the endothelial cells where it regulates migration.6 HDAC2 regulates cardiac morphogenesis and growth with uncontrolled proliferation of ventricular cardiomyocytes, obliteration of the right ventricle and perinatal lethality occurring in HDAC2-null mice. Abnormal HDAC2 expression is associated with hypoplastic right heart syndrome, right ventricular dysplasia and transposition of the great arteries.7 The DECIPHER database describes Patient 2,498, who harbors a deletion within 6q21–22.1 (nt. 11321360–115432628). This region includes MARCKS and HDAC2. Abnormalities of the vascular system are described for Patient 2,498; however, detailed information was not available.
The patient presenting with CoA reported by Bzduch et al.1 was examined using G-banding, but not aCGH, and so the exact position of the deletion was not defined . However, both the CoA patient and our current patient shared deletion of 6q22.2 to 12.3 Mb and this region includes gap junction protein alpha-1 (GJA1). GJA1 knockout mice die in utero because of swelling and blockage of the right ventricular outflow track from the heart. However, IAA was not reported.8
We report a patient with 6q21–22 deletion presenting with IAA. Previous patient reports and knockout mouse analyses indicate that 6q21–22 deletion leads to right cardiac anomalies. Our findings and those reported by Bzduch et al.1 suggest that IAA/CoA is also a possible phenotype of 6q21–22 deletion. Understanding the genotype–phenotype relationship and identification of the gene deletion(s) responsible for IAA/CoA will require study of additional patients with 6q21–22 deletion.
Acknowledgments
This work was supported by JSPS KAKENHI grant number 26893252.
Footnotes
The authors declare no conflict of interest.
References
- Bzduch V , Lukacova M . Interstitial deletion of the long arm of chromosome 6(q22.2q23) in a boy with phenotypic features of Williams syndrome. Clin Genet 1989; 35: 230–231. [DOI] [PubMed] [Google Scholar]
- Chen CP , Wang TH , Lin SP , Chern SR , Chen MR , Lee CC et al. 24Mb deletion of 6q22.1–>q23.2 in an infant with pulmonary atresia, ventricular septal defect, microcephaly, developmental delay and facial dysmorphism. Eur J Med Genet 2006; 49: 516–519. [DOI] [PubMed] [Google Scholar]
- Park JP , Graham JM Jr , Berg SZ , Wurster-Hill DH . A de novo interstitial deletion of chromosome 6 (q22.2q23.1). Clin Genet 1988; 33: 65–68. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JA , Amrom D , Andermann E , Andermann F , Veilleux M , Curry C et al. Genotype–phenotype correlation in interstitial 6q deletions: a report of 12 new cases. Neurogenetics 2012; 13: 31–47. [DOI] [PubMed] [Google Scholar]
- Stumpo DJ , Bock CB , Tuttle JS , Blackshear PJ . MARCKS deficiency in mice leads to abnormal brain development and perinatal death. Proc Natl Acad Sci USA 1995; 92: 944–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monahan TS , Andersen ND , Martin MC , Malek JY , Shrikhande GV , Pradhan L et al. MARCKS silencing differentially affects human vascular smooth muscle and endothelial cell phenotypes to inhibit neointimal hyperplasia in saphenous vein. FASEB J 2009; 23: 557–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery RL , Davis CA , Potthoff MJ , Haberland M , Fielitz J , Qi X et al. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 2007; 21: 1790–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume AG , de Sousa PA , Kulkarni S , Langille BL , Zhu D , Davies TC et al. Cardiac malformation in neonatal mice lacking connexin43. Science 1995; 267: 1831–1834. [DOI] [PubMed] [Google Scholar]
Data Citations
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.600. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.602. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.604. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.606. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.608. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.610. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.612. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.614. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.616. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.618. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.620. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.622. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.624. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.626. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.628. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.630. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.632. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.634. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.636. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.638. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.640. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.642. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.644. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.646. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.648. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.650. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.652. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.654. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.656. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.658. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.660. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.662. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.664. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.666. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.600. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.602. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.604. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.606. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.608. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.610. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.612. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.614. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.616. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.618. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.620. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.622. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.624. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.626. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.628. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.630. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.632. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.634. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.636. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.638. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.640. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.642. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.644. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.646. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.648. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.650. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.652. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.654. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.656. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.658. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.660. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.662. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.664. [DOI]
- Yamagata Takanori.HGV Database. 2015. 10.6084/m9.figshare.hgv.666. [DOI]