Abstract
Considering the importance of BRCA1, BRCA2, CHEK2 and TP53 in the development of hereditary early-onset breast and ovarian cancer and that the genetic susceptibility profile of the Northeast population from Brazil has never been analyzed, this study aimed to verify the frequency of mutations of clinical significance in these genes in high-risk hereditary breast and ovarian cancer (HBOC) syndrome patients from that region. DNA samples from 106 high-risk unrelated patients mostly from Bahia, the biggest state in the Northeast region, were analyzed. These patients underwent full BRCA1 gene sequencing, screening for common founder mutations in the BRCA2, CHEK2 and TP53 genes and genetic ancestry analysis with nine ancestry informative markers. The positive results were confirmed by two sequencing reactions. Three mutations of clinical significance were found: BRCA1 p.R71G (4.71%), 3450del4 (3.77%) and TP53 p.R337H (0.94%). The genetic ancestry analysis showed a high European ancestry contribution (62.2%) as well as considerable African (31.2%) and Amerindian (6.6%) ancestry contributions (r2=0.991); this degree of heterogeneity was also significant in the population structure analysis (r=0.604). This population is highly admixed with a different spectrum of genetic susceptibility, with the Galician founder mutation BRCA1 p.R71G accounting for 50% of all identified mutations in high-risk HBOC patients. TP53 p.R337H was also significantly frequent; thus, the combined screening of BRCA1/2 and TP53 should be offered to high-risk HBOC patients from Northeast Brazil.
Introduction
Breast and ovarian cancers are among the most common cancers in developed and developing countries.1 Approximately 5–10% of familial cases are due to germline mutations in susceptibility genes, such as BRCA1, BRCA2, CHEK2 and TP53. These susceptibility genes have key roles in cell cycle control, apoptosis and DNA repair.2,3
Mutations in the BRCA1 and BRCA2 genes are associated with the hereditary breast and ovarian cancer (HBOC) syndrome. Patients who have HBOC syndrome have a personal and family history of cancer mainly in the following organs: breast, ovarian, prostate and pancreas.2 In contrast, mutations in the CHEK2 and TP53 genes are also associated with breast and ovarian cancers, as well as with other cancers related to the Li–Fraumeni Syndrome and Li–Fraumeni-like syndrome.4,5
Many studies have demonstrated that mutations in these susceptibility genes could result in early-onset carcinoma as well as in a worse prognosis; therefore, it is important to use genetic testing to identify these deleterious mutations, so that risk-reducing and risk-prevention strategies can be applied.6,7
In previously characterized populations, it is easy to verify the genetic susceptibility of high-risk individuals, because some germline mutations have founder effects, such as the BRCA1 c.66_67delAG and c.5266dupC mutations and BRCA2 c.5946_5946delT in the Ashkenazi population; BRCA2 c.156_157insAlu in Portugal; BRCA2 999del5 in Iceland; CHEK2 c.1100 in European populations; and TP53 p.R337H in Southern Brazil.8–13
Thus, knowing that ethnic differences in cancer incidence and mortality are the results of genetic and epidemiological risk factor interactions, it is crucial to characterize a population which will allow for a more accurate risk assessment and proper genetic counseling for the individuals.14,15
The Brazilian population is one of the most heterogeneous in the world. Many populations have contributed to it, mainly Portuguese, African and Amerindian. However, other European populations, such as the Spanish and Italian, have also contributed to its genetic diversity.16 As the pattern of admixture in Brazil varies according to the region17–21 and no study has been performed with the population from the Northeast region of Brazil, our aim was to verify the frequency of mutations of clinical significance in BRCA1, BRCA2, CHEK2 and TP53 in 106 patients at high risk for HBOC from the Northeast region of Brazil.
Materials and methods
Study population
A total of 106 unrelated Brazilian patients at high risk for HBOC who were referred to the Genetics Clinic of the Hospital Universitário Edgar Santos from Universidade Federal da Bahia between 2008 and 2013 were included in the study. The research was approved by the Research Ethics Committee of the institutions that were involved and all of the patients provided informed consent.
During genetic counseling, clinical and epidemiologic data were obtained, and genetic testing was offered to affected individuals who met at least one of the following criteria: (a) two or more family members affected by breast cancer; (b) one family member affected by breast cancer and another affected by ovarian cancer; (c) three or more family members affected by breast or ovarian cancer, with at least one affected with ovarian cancer; (d) breast cancer diagnosed before 45yr of age; (e) ovarian cancer diagnosed before 50yr of age; (f) bilateral breast cancer; (g) male breast cancer at any age; and (h) cancer in multiple organs including breast cancer. The patients’ available relatives were also analyzed.
Genotyping susceptibility genes
DNA was extracted from peripheral blood using the Gentra Puregene Blood kit (QIAGEN, Hilden, Germany) or the AxyPrep Blood genomic DNA Miniprep kit (Axygen Biosciences, Union City, CA, USA). The screening for mutations in BRCA1 was performed in two steps: (a) polymerase chain reaction (PCR) of each exon followed by single-strand conformation polymorphism analysis and electrophoresis of denatured DNA as previously described22 and (b) then all of the PCR fragments that presented a different migration pattern on the gel were analyzed by direct sequencing in an ABI PRISM 3130xl genetic analyzer (Applied BioSystems, Foster City, CA USA) using the BigDye™ terminator sequencing kit (Applied BioSystems) as recommend by the manufacturer. The primers used as well as the PCR cycling conditions are shown in Supplementary Table 1.
The initial screening for the mutations BRCA2 (c.5946_5946delT and c.156_157insAlu), CHEK2 (c.1100delC, c.444+1G>A and p.I157T) and TP53 (p.R337H) was performed by AS-PCR followed by electrophoresis in agarose gels (2%) or PCR/RFLP, which consisted of the following steps: (a) PCR reaction, (b) verification of amplification by electrophoresis in agarose gels (1.5%), (c) digestion with the appropriate restriction enzyme and (d) scoring other electrophoreses in agarose gels (2.5%).
The primers and PCR cycling were based on Struewing et al.,8 Machado et al.,9 Bayram et al.23 and Custodio et al.24 (Supplementary Table 2). All of the RFLP reactions used enzymes reagents of New England Biolabs (New England Biolabs, Beverly, MA, USA) and were performed according to this manufacturer.
Each PCR reaction contained 0.25 μM each primer, 0.5 mM dNTPs, 10 mM Tris-HCl (pH 8.9), 50 mM KCl, 1.5 or 2.0 mM MgCl2, 1 U Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), 25 ng genomic DNA and UltraPure DNase/RNase-Free Distilled Water (Invitrogen) up to 25 μl. Except in the PCR reaction for TP53’s mutation, Go Taq Flexi DNA polymerase was used (Promega, Madison, WI, USA).
The positive results were verified by two sequencing reactions. For the genotyping control, 10% of the negative samples were randomly selected and sequenced. The PCR reactions of the BRCA2 c.156_157insAlu were run with a positive control. The gene reference sequences were NG_005905.2, NG_012772.3, NG_008150.1 and NG_017013.2, which are available in GenBank (NCBI, NIH, USA). The sequences were analyzed using the CLC Bio sequence viewer version 6.8.1 (a QIAGEN Company) and BioEdit version 7.1.9 (Ibis Biosciences, Carlsbad, CA, USA) or FinchTV 1.4.0 (Geospiza, Seattle, WA, USA).
Genetic ancestry analysis
A micro-panel of nine ancestry informative markers was used to estimate the genetic ancestry: APO, AT3-I/D, CKMM, FY-NULL, GC*1S, GC*1F, LPL, PV92 and SB19.3. These ancestry informative markers were genotyped as described above by (a) PCR followed by electrophoresis, (b) PCR/RFLP, or (c) real-time PCR as described by Parra et al.25 and Shriver et al.26
The contents of the PCR reaction were as described above, except for the real-time PCR reactions, which used TaqMan SNP genotyping assays (Applied BioSystems) as recommended by the manufacturer and were monitored in ABI Prism 7700 Sequence Detection Systems (Applied Biosystems).
The restriction enzymes were also from New England Biolabs (New England Biolabs). The primers, enzymes, probes and PCR conditions are described in Supplementary Table 3.
Statistical analysis
The descriptive analyses were performed in Epi Info version 7. GENEPOP 1.227 and ADMIX 9528 were used to verify the allelic frequencies and estimate the genetic ancestry. The population structure was analyzed in STRUCTURE v.2.3.429 with 10.000 for the burn-in period interactions, 10 000 additional interactions and admixture option with the LOCPRIOR model and a K value from 1 to 3. The genotypes of the African (Nigerian), Amerindian and European (German and Spanish) populations were kindly provided by Dr Mark Shriver from Penn State University.
Results
All of the patients were female, and most of the patients had cancer before 50 years of age, with a mean age at diagnosis of 43.06 years (SD=±10.67). The majority of the cases were breast cancer (91.4%), followed by ovarian cancer (4.7%) (Table 1). Among the breast cancer cases, the most common histological type was the invasive ductal carcinoma (IDC) at 69.30% (70/101), and among the ovarian cancer cases, the most common was serous adenocarcinoma, at 77.77% (7/9).
Table 1. Baseline characteristics of the 106 patients analyzed.
| Characteristic | % | N |
|---|---|---|
| Cancer | ||
| Breast | 91.51 | 97 |
| Ovarian | 4.72 | 5 |
| Breast and ovarian | 3.77 | 4 |
|
Age at diagnostic
a
| ||
| <50yrs | 77.36 | 82 |
| ≥50yrs | 22.64 | 24 |
|
Tumor location
| ||
| Unilateral | 69.81 | 74 |
| Bilateral | 4.72 | 5 |
| Missing | 25.47 | 27 |
|
Origin
| ||
| Salvador, BA | 30.19 | 32 |
| Countryside of BA | 52.83 | 56 |
| Other regions of Brazil | 15.09 | 16 |
| Missing | 1.89 | 2 |
|
Self-reported race/color
| ||
| White | 34.91 | 37 |
| Mulatto | 40.57 | 43 |
| Black | 20.75 | 22 |
| Others | 3.77 | 4 |
|
Family history
| ||
| Without family history | 16.98 | 18 |
| Only 1st relatives | 11.32 | 12 |
| 1st and 2nd relatives | 17.92 | 19 |
| 1st and 3rd relatives | 3.77 | 4 |
| Only 2nd relatives | 9.43 | 9 |
| 2nd and 3rd relatives | 12.26 | 13 |
| Only 3rd relatives | 8.49 | 9 |
| 1st, 2nd and 3rd relatives | 19.81 | 21 |
|
Oral contraceptive
b
| ||
| Yes | 64.15 | 68 |
| No | 16.98 | 18 |
| Missing | 18.87 | 20 |
|
Parity
| ||
| Any birth | 23.58 | 25 |
| 1–2 births | 44.34 | 47 |
| 3–4 births | 16.04 | 17 |
| 5 births | 1.89 | 2 |
| Missing | 14.15 | 15 |
|
Smoker
c
| ||
| Yes | 16.04 | 17 |
| No | 64.15 | 68 |
| Missing | 19.81 | 21 |
Age at diagnostic of the first cancer.
Oral contraceptive for more than 5 years.
It was considered as smoker the use of tobacco for more than 1 year.
In addition to the personal cancer history, most of these women also had a family history (83.02%, Table 1), and the frequency of the cancers associated with HBOC (breast cancer, ovarian cancer, prostate cancer and pancreatic cancer) was significant among the first (81.82%)-, second (87.83%)- and third (78.71%)-degree relatives.
Regarding other risk factors, such as the use of oral contraceptives, parity and age at menarche, a significant percentage of the study population had used oral contraceptives for more than 5 years (64.15%) or also had children (76.42%), and the mean age at menarche was 12.89 years (SD=±1.63). Another risk factor that was analyzed was the smoking status, but a considerable portion of the subjects were nonsmokers (64.15%) (Table 1).
Knowing that the Brazilian population is composed of at least three ancestral population groups, a trihybrid admixture model was estimated in ADMIX 95, which showed a high European ancestry contribution 62.20% (s.e.=0.034) and considerable African and Amerindian ancestry contributions, 31.20% (s.e=0.011) and 6.60% (s.e.=0.036), respectively. This analysis was statistically significant (r2=0.991), and the study population fit the Hardy–Weinberg Equilibrium (P<0.005).
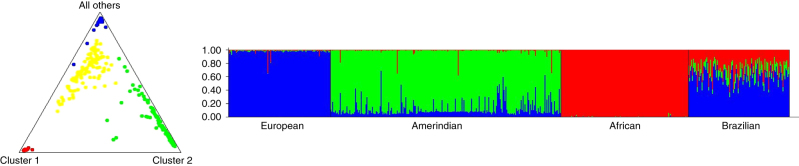
In addition, the population stratification analysis in STRUCTURE was also significant for K=3 (r=0.604); the bar plot in Figure 1 (left) shows the genetic admixture proportions of the Brazilian study population compared with the African, Amerindian and European populations based on the ancestry informative markers panel. The triangle plot in Figure 1 (right) demonstrates the distribution of the subjects according their genetic admixture and population of origin, in which is observed the heterogeneity degree of the study population clustered between the European and African populations. The average distances (expected heterozygosity) between individuals in the same cluster was greater in cluster 3 (0.319), in which the study population was more closely clustered than was that in cluster 1 (0.262) and cluster 2 (0.256), in which the African and Amerindian populations clustered (see triangle plot, Figure 1).The subjects also exhibited a distinct pattern in the assessment of self-reported race/color; most of the subjects self-reported as mulatto (40.57%), followed by white (34.91%) and black (20.75%). Almost all of the subjects were from the State of Bahia (83.02%), but there were also subjects from other regions of Brazil (15.09%) (Table 1).
Figure 1.
Distribution of the subjects according their estimated ancestral background (triangle plot) and population structure for the trihybrid model (bar plot). Note: red dots, African population; green dots, Amerindian subjects; blue, European subjects; yellow dots, Brazilian population (study population).
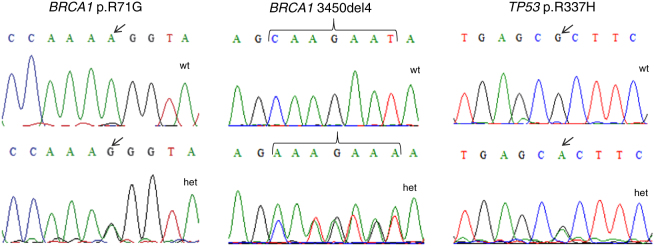
Ten unrelated patients were identified with mutations of clinical significance; the most frequent mutation was BRCA1 c.211A>Grs80357382 (4.71%; 5/10), followed by BRCA1 c.3331_3334delCAAGrs80357777 (3.77%; 4/10) and TP53 p.R337Hrs121912664 (0.94%; 1/10). Six of these patients were from the countryside regions of the State of Bahia and four were from Salvador, the capital of Bahia. The clinic and pathological characteristics of the mutation carriers are described in Table 2, and Figure 2 shows the sequencing fragments of the identified mutations.
Table 2. Characteristics of the patients with BRCA1 and TP53 mutations.
| Family/patient# | Cancer | Age at diagnostica | Histology | Grade | U/B | ER | PgR | HER2 | Self-reported color |
Ancestry (%)b |
Mutation | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Af | Am | Eu | |||||||||||
| 4.1 | Ov/Br | 38 | -/Serous | -/II | U | — | — | — | White | 14.6 | 10.1 | 75.3 | BRCA1 3450del4 |
| 13.1 | Br | 47 | IDC | — | U | — | — | — | White | 29.2 | 9.6 | 61.2 | BRCA1 3450del4 |
| 21.1 | Br | 32 | IDC | — | U | — | — | — | White | 19.8 | 14.4 | 65.8 | BRCA1 3450del4 |
| 34.2 | Br/Ov | 39 | IDC/serous | III/II | U | neg | neg | neg | White | 27.0 | 6.5 | 66.5 | BRCA1 p.R71G |
| 80.2 | Br | 46 | Medular | X | U | neg | neg | neg | Mulatto | 50.7 | 10.5 | 38.8 | BRCA1 p.R71G |
| 97.1 | Br | 53 | Lobular invasive | I | U | pos | pos | neg | White | 17.4 | 6.3 | 76.3 | TP53 p.R337H |
| 98.2 | Br | 38 | IDC | III | U | neg | neg | neg | Mulatto | 21.8 | 11.8 | 66.5 | BRCA1 3450del4 |
| 103.1 | Br | 52 | IDC | II | U | neg | neg | neg | Mulatto | 17.5 | 10.1 | 72.4 | BRCA1 p.R71G |
| 105.1 | Br | 39 | IDC | III | U | neg | pos | neg | Mulatto | 29.3 | 11.2 | 59.5 | BRCA1 p.R71G |
| 106.1 | Br | 46 | IDC | III | U | neg | neg | neg | Black | 25.7 | 10.3 | 64.0 | BRCA1 p.R71G |
Abbreviations: Af, African; Am, Amerindian; B, bilateral; Br, breast cancer; ER, estrogen receptor; Eu, European; HER2, human epidermal growth factor receptor; IDC, invasive ductal carcinoma; Ov, ovarian cancer; PgR, progesterone receptor; U, unilateral; -, data not available.
Age at diagnostic of the first cancer.
Estimated genetic ancestry.
Figure 2.
Sequencing fragments of carriers and noncarriers for the BRCA1 p.R71G, BRCA1 3450del4 and TP53 p.R337H mutations. Note: Het, heterozygous type; Wt, wild type.
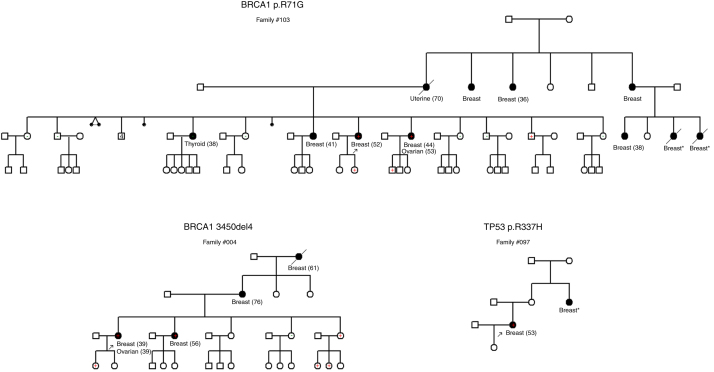
All of the mutation carriers had a family history of breast and ovarian cancer involving first-, second- and/or third-degree relatives (80.00%), except for two carriers who had no suggestive family history of any cancer (patients #21.1 and #106.1, Table 2). The pedigrees of three subjects who are carriers for each mutation are shown in the Figure 3, which presents the segregation of these mutations among first- and second-degree relatives, except for the TP53 p.R337H mutation carrier (Patient #97.1) for whom no family members were available for testing.
Figure 3.
Pedigree of families harboring the BRCA1 p.R71G, BRCA1 3450del4 and TP53 p.R337H mutations. Note: positive (+) symbol, mutation carrier; negative (−) symbol, noncarrier; *age unknown. The numbers between parentheses represent the age at diagnosis in years.
No patient with the Galician founder mutation (BRCA1 c.211A>G) reported a Spanish ancestry contribution in their families, whereas among the patients with the BRCA1 c.3331_3334delCAAG (3450del4), three reported a Portuguese and/or French ancestry contribution and the fourth patient reported an Italian ancestry contribution. The TP53 p.R337H mutation’s carrier did not report her ancestry contribution. Table 2 presents the estimated genetic ancestry proportion for each mutation carrier, which was similarly estimated for the study population as a whole as described above.
Discussion
Here, 106 unrelated high-risk subjects for HBOC from Northeast Brazil, a population that until now was uncharacterized, were analyzed. In this population, two mutations of clinical significance were found in BRCA1, 3450del4 and p.R71G (8.49%) and one in TP53, p.R337H (0.94%). These mutations were associated with aggressive clinic-pathological characteristics; most of the subjects had IDC and triple-negative tumors, and a family history of cancer involving first-, second- and/or third-degree relatives (Table 2 and Figure 3). Only two mutation carriers did not have a family history of any cancer, but both of these carriers had early-onset breast cancer (patients #21.1 and #106.1, Table 2).
The 3450del4 mutation has been described in many populations worldwide.30–34 In South America, this mutation has been described among patients with HBOC from other regions of Brazil33,34 and among patients with ovarian cancer from Colombia.31 In fact, in Colombia, 3450del4 was the only mutation of clinical significance, suggesting a founder effect. Here, this mutation also had a significant frequency among the patients from Northeast Brazil, with a frequency higher (3.77%) than that previously observed in the others regions of Brazil, at 0.16%33 and 2.12%.34
The other mutation that was found in this study, BRCA1 p.R71G, which accounted for 50% of the mutations identified, is a Galician founder mutation that was first described by Vega et al.35 Based on the mtDNA analysis, the Galician population is very homogeneous, with traits of a Culdesac population with a striking similarity to the Basque population owing to its geographic location and cultural barriers (dialect).36 Others studies corroborate the restriction of this mutation in Galician or Hispanic descendants in Portugal,37 USA38,39 and Asturias (Northern Spain).30
Although no Brazilian patient with the p.R71G reported a Spanish ancestry contribution, these patients presented considerable European ancestry contributions (Table 2), which was statistically significant (r=0.604), and historical data corroborate the hypothesis that the Northeast region of Brazil has a high Spanish ancestry contribution, especially Galician, because the State of Bahia received more Galician immigrants than others states. In Salvador, the capital of Bahia, from 1883 to 1950, approximately 3–6 generations ago (~20 years per generation), 94.3% of the Spanish immigrants were from Galicia, of which 90.8% were from Pontevedra Province.40,41 It is also important to emphasize that the likelihood of a de novo mutation occurring in BRCA1 and BRCA2 genes is very low.42
The third mutation that was found, TP53 p.R337H (0.94%), is a low-penetrance mutation that has a founder effect in the Southern region of Brazil and is associated with breast cancer cases that are related to Li–Fraumeni-like syndrome syndrome.4,11 It seems that p.R337H originated in the Portuguese population because the haplotypes of the Portuguese (A3) and the Brazilian (A1) carriers differ by only one SNP, rs9894946:C>T.11
The frequency of the TP53 p.R337H mutation in Southeast and South Brazil varies from 0.51 to 8.6%,43–45 and in the Northeast population, this mutation occurred at a significant frequency (0.94%). However, here the mutation carrier had a family history that was suggestive of HBOC and not Li–Fraumeni Syndrome or Li–Fraumeni-like syndrome (Figure 2) based on the NCCN Clinical Practice Guidelines in Oncology v.1.201046 and criteria that were proposed by Li–Fraumeni, Birch, Eeeles and Chompret.47
However, this association was also recently found by Cury et al.;48 therefore, this finding demonstrated that in Brazil, the TP53 p.R337H mutation is important for patients at high risk for HBOC as well as Li–Fraumeni Syndrome and Li–Fraumeni-like syndrome. Consequently, in agreement with it has been recently proposed, the combined screening of BRCA1, BRCA2 and TP53 should be offered to high-risk as well as early-onset breast cancer patients from Brazil.44,49
Interestingly, in this study we did not observe the BRCA1 mutation that is the most frequent among the Brazilians: the BRCA1 c.5266dupC (5382insC), which accounted for 56% of all of the identified mutations in patients from Southeast Brazil.50 The highest frequency that was ever found in Brazil was 5% by Ewald et al.51 among high-risk patients for HBOC from Porto Alegre and Rio de Janeiro. Other studies in different regions of Brazil also found BRCA1 c.5266dupC with a significant frequency,33,34,52 except among Ashkenazi women from Porto Alegre.53
BRCA1 c.5266dupC was first described as an Ashkenazi founder mutation, but an analysis of the origin of this mutation in the European population demonstrated that c.5266dupC originated from a common ancestor in Northern Europe long before becoming an Ashkenazi founder mutation.54 Because the Brazilian population has an ancestry contribution of many European populations, this mutation is likely to be found.
BRCA1 c.66_67delAG and BRCA2 6174delT, also considered Ashkenazi founder mutations,8 were found in other regions of Brazil34,53 as well as in Spanish and Latin America populations,26,35 and interestingly were not present in the population of Northeast Brazil; this result could also be because of the pattern of admixture of the Northeast population or even to the sampling size. However, the study population fit the Hardy–Weinberg Equilibrium (P<0.005).
The other mutation of BRCA2, c.156_157insAlu, a Portuguese founder mutation,9,10 was also not found in this population, although historical data demonstrated that most of the European immigrants who arrived in Brazil were Portuguese, and it was the Portuguese that settled the country (1500), with a great immigrant flux between 1850 and 1970 that was two times greater than that of the Spanish population. However, when considering the number of immigrants by state, in 1950, Bahia State registered 2509 Spanish immigrants and 1531 Portuguese immigrants. In 1970, the number of Spanish immigrants was also higher than that of the Portuguese, at 3225 and 1586, respectively.16,55
From the data presented here and the available historical data, it can be inferred that even though Salvador (the capital of the State of Bahia) was the first capital of Brazil (from 1549 to 1763), the presence of the Portuguese population in Bahia was smaller than that of the Spanish population, most likely due to the transference of the Portuguese court to Rio de Janeiro, the second capital of Brazil.16
In the estimated period of the first occurrence of BRCA2 c.156_157insAlu in Portugal, 120–130 generations ago (~20 years per generation),9 the Portuguese immigrants were well established in Brazil, but because of economic and political motives, their presence was greater in Southeast Brazil, mainly in Rio de Janeiro and São Paulo. In Rio de Janeiro, BRCA2 c.156_157insAlu was found in three unrelated patients with breast cancer.56
Therefore, due to the bottleneck effect result of the Portuguese population movements, (a) immigration from Portugal to Brazil (settlement, 1500), (b) emigration from the Northeast to the Southeast regions of Brazil (the establishment of Rio de Janeiro as the capital of Brazil in 1973) and (c) others fluxes did not allow the fixation of BRCA2 c.156_157insAlu in the population of Bahia.
The most frequent CHEK2 germline mutations worldwide, c.1100delC, c.444+1G>A and p.I157T,5,57 were also not found in any of the analyzed patients. These data are interesting because mutations of low-penetrance genes are more likely to be found than are those of high-penetrance genes.
Nevertheless, it has to be considered that the presence of these mutations could also be related to the pattern of the admixture of the population. Other studies reported results similar to those presented here,22,52 and in Brazil, only one study found c.1100delC, but the affected individual had a family history matching the clinical criteria for hereditary breast and colorectal cancer.58
In this study, the profile of a population that until now uncharacterized was analyzed; most of the subjects were from the largest state of the Northeast region (83.02%), but some were from other regions of Brazil (15.09%). In fact, it is important to consider that every population at some level is heterogeneous, some more than others, and that these variations can interfere with the level of genetic susceptibility.
Although the ancestry informative markers panel was relatively small, it could be observed that the study population was highly heterogeneous, presenting high levels of European ancestry contribution (62.20%), as well as significant African (31.20%) and Amerindian (6.60%) ancestry contributions. This high European ancestry contribution could be owing to fact that most of the subjects self-reported as white or mulatto (75.48%) and/or were from the countryside regions (52.83%), where there are more people with Caucasian traits than in Salvador, the capital of Bahia.17,18,59
However, this degree of heterogeneity was confirmed in the population stratification analysis that was also statistically significant (r=0.604). In the bar plot, the degree of the genetic admixture of the study population was higher than that of the other population groups analyzed. In the triangle plot, the distribution of the study population was between the European and African populations. However, most Brazilian subjects clustered with the European populations, in which the degree of heterozygosity between individuals was higher (0.319) compared with the clusters of the African (0.262) and Amerindian (0.256) populations.
In Figure 1 (triangle plot), some individuals from the Amerindian population were not near its cluster (cluster 2) or seemed closer to the European cluster (cluster 3); this effect could be related to the size of the panel. Nonetheless, we must consider that there is genetic variation among and within populations; with this panel, a higher degree of heterogeneity of the study population was demonstrated.
In conclusion, in this highly admixed population, three founder mutations, BRCA1 3450del4 and p.R71G and TP53 p.R337H, were found and can be added to the mutation panels that are used for high-risk HBOC patients and for early-onset breast cancer patients from this region. However, more importantly, these findings indicate that the combined screening of BRCA1/2 and TP53 is important and should be offered to high-risk patients. In addition, it is notable that this population seems to have a high Spanish ancestry contribution, considering that BRCA1 p.R71G was found in a total of five unrelated patients. However, further genetic ancestry analysis could demonstrate whether the haplotype of these Brazilian patients is similar to that of the Galician patients.
Funding
This study was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (Process #408858/2006-0) and Secretaria de Saúde do Estado da Bahia (Contract #028/2011).
Acknowledgments
We thank all of the patients and the institutions, NOB, Complexo HUPES/UFBA, HSA/OSID and CICAN, for their collaboration. We also thank Dr Fátima Vaz (Instituto Português de Oncologia, de Francisco Gentil) for kindly helping us develop the genotyping method for the BRCA2 c.156_157insAlu mutation and Dr Mark Shriver (Penn State University) for kindly providing the genotypes of the African, Amerindian and European populations.
The authors declare no conflict of interest.
Footnotes
Supplemental Information for this article can be found on the Human Genome Variation website (http://www.nature.com/hgv)
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global Cancer Statistics. CA Cancer J Clin 2011; 61: 69–90. [DOI] [PubMed] [Google Scholar]
- Smith EC. An overview of hereditary breast and ovarian cancer syndrome. J Midwifery Womens Health 2012; 57: 577–584. [DOI] [PubMed] [Google Scholar]
- Apoustolou P, Fostira F. Hereditary breast cancer: the era of new susceptibility genes. Biomed Res Int 2013; 2013: 74318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achatz MIW, Olivier M, Le Calvez F, Martel-Planche G, Lopes A, Rossi BM et al. The TP53 mutation, R337H, is associated with Li–Fraumeni and Li–Fraumeni-like syndromes in Brazilian families. Cancer Lett 2007; 245: 96–102. [DOI] [PubMed] [Google Scholar]
- Allinen M M, Huusko P, Mäntyniemi S, Launonen V, Winqvist R. Mutation analysis of the CHEK2 gene in families with hereditary breast cancer. Br J Cancer 2001; 85: 209–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh T, Casadei S, Coats KH, Swisher E, Stray SM, Higgins J et al. Spectrum of mutations in BRCA1, BRCA2, CHEK2, e TP53 in families at high risk of breast cancer. J Am Med Assoc 2006; 295: 1378–1388. [DOI] [PubMed] [Google Scholar]
- Bordeleau L, Panchal S, Goodwin P. Prognosis of BRCA-associated breast cancer: a summary of evidence. Breast Cancer Res Treat 2010; 119: 13–24. [DOI] [PubMed] [Google Scholar]
- Struewing JP, Hartge P, Wacholder S, Baker SM, Berlin M, McAdams M et al. The risk of cancer associated with specific mutations of BRCA1 and BRCA2 among Ashkenazi Jews. N Eng J Med 1997; 336: 1401–1408. [DOI] [PubMed] [Google Scholar]
- Machado PM, Brandão RD, Cavaco BM, Eugénio J, Bento S, Nave M et al. Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin Oncol 2007; 25: 2027–2034. [DOI] [PubMed] [Google Scholar]
- Peixoto A, Santos C, Rocha P, Pinheiro M, Príncipe S, Pereira D et al. The c.156_157insAlu BRCA2 rearrangement accounts for more than one-fourth of deleterious BRCA mutations in northern/central Portugal. Breast Cancer Res Treat 2009; 114: 31–38. [DOI] [PubMed] [Google Scholar]
- Garritano S, Gemignani F, Palmero EI, Olivier M, Martel-Planche G, Le Calvez-Kelm F et al. Detailed haplotype analysis at the TP53 locus in p.R337H mutation carriers in the population of Southern Brazil: evidence for a founder effect. Hum Mutat 2010; 31: 143–150. [DOI] [PubMed] [Google Scholar]
- Thorlacius S, Olafsdottir G, Tryggvadottir L, Neuhausen S, Jonasson JG, Tavtigian SV et al. A single BRCA2 mutation in male and female breast cancer families from Iceland with varied cancer phenotypes. Nat Genet 1996; 13: 117–119. [DOI] [PubMed] [Google Scholar]
- Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R et al. Low-penetrance susceptibility to breast cancer due to CHEK2*1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 2002; 31: 55–59. [DOI] [PubMed] [Google Scholar]
- Gilliland FD. Ethnic differences in cancer incidence: a marker for inherited susceptibility? Environ Health Perspect 1997; 105: 897–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhausen SL. Ethnic differences in cancer risk resulting from genetic variation. Cancer 1999; 86: 2575–2582. [DOI] [PubMed] [Google Scholar]
- Instituto Brasileiro de Geografia e Estatística. Brasil 500 anos de povoamento, Centro de Documentação e Disseminação de Informações IBGE: Rio de Janeiro: Brasil, 2007. [Google Scholar]
- Felix GES, Abe Sandes K, Bonfim TM, Bendicho MT, Cisneiros P, Guedes R et al. Ancestry informative markers and complete blood count parameters in Brazilian blood donors. Rev Bras Hematol Hemoter 2010; 32: 282–285. [Google Scholar]
- Abe-Sandes K, Bomfim TF, Machado TMB, Abe-Sandes C, Acosta AX et al. Ancestralidade genômica, nível socioeconômico vulnerabilidade ao HIV-aids na Bahia, Brasil. Saúde Soc São Paulo 2010; 19: 75–84. [Google Scholar]
- Luizon MR, Mendes-Junior CT, De Oliveira SF, Simões AL. Ancestry informative markers in Amerindians from Brazilian Amazon. Am J Hum Biol 2008; 20: 86–90. [DOI] [PubMed] [Google Scholar]
- Parra FC, Amado RC, Lambertucci JR, Rocha J, Antunes CM, Pena SD. Color and genomic ancestry in Brazilians. Proc Natl Acad Sci 2003; 100: 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos NP, Ribeiro-Rodrigues EM, Ribeiro-Dos-Santos AK, Pereira R, Gusmão L, Amorim A et al. Assessing individual interethnic admixture and population substructure using a 48-insertion-deletion (INSEL) ancestry-informative marker (AIM) panel. Hum Mutat 2010; 31: 184–190. [DOI] [PubMed] [Google Scholar]
- Friedman L, Ostermeyer EA, Szabo CI, Dowd P, Lynch ED, Rowell SE, King Marie-Claire. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet 1994; 8: 399–404. [DOI] [PubMed] [Google Scholar]
- Bayran S, Topaktaş M, Akkız H, Bekar A, Akgöllü E. CHEK2 1100delC, IVS2+1G>A and I157T mutations are not present in colorectal cancer cases from Turkish population. Cancer Epidemiol 2012; 36: 453–457. [DOI] [PubMed] [Google Scholar]
- Custodio G, Taques GR, Figueiredo BC, Gugelmin ES, Oliveira Figueiredo MM, Watanabe F et al. Increased incidence of choroid plexus carcinoma due to the germline TP53 R337H mutation in Southern Brazil. PLoS ONE 2011; 6: e18015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, Martinson J, Batzer MA, Cooper R et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet 1998; 63: 1839–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C et al. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet 2003; 112: 387–399. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 1995; 86: 248–249. [Google Scholar]
- Chakraborty R. Gene identity in racial hybrids and estimation of admixture rates. In: Ahuja YR, Neel JV (eds). Genetic Differentiation in Human and other Animal Populations. Indian Anthropological Association: Delhi 1985: 171–180. [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Eco Notes 2007; 7: 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blay P, Santamaría I, Pitiot AS, Luque M, Alvarado MG, Lastra A et al. Mutational analysis of BRCA1 and BRCA2 in hereditary breast and ovarian cancer families from Asturias (Northern Spain). BMC Cancer 2003; 13: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez AO, Pardo GG, Royer R, Larson G, Weitzel JN et al. BRCA1 and BRCA2 mutations among ovarian cancer patients from Colombia. Gynecol Oncol 2012; 124: 236–243. [DOI] [PubMed] [Google Scholar]
- Blesa JR, García JA, Ochoa E. Frequency of germ-line BRCA1 mutations among Spanish families from a Mediterranean area. Hum Mut 2000; 15: 381–382. [DOI] [PubMed] [Google Scholar]
- Lourenço JJ, Vargas FR, Bines J, Santos EM, Lasmar CAP, Costa CH et al. BRCA1 mutations in Brazilian patients. Genet Mol Biol 2004; 27: 500–504. [Google Scholar]
- Esteves VF, Thuler LC, Amêndola LC, Koifman RJ, Koifman S, Frankel PP et al. Prevalence of BRCA1 and BRCA2 gene mutations in families with medium and high risk of breast and ovarian cancer in Brazil. Braz J Med Biol Res 2009; 42: 453–457. [DOI] [PubMed] [Google Scholar]
- Vega A, Campos B, Bressac-De-Paillerets B, Bond PM, Janin N, Douglas FS et al. The R71G BRCA1 is a founder Spanish mutation and leads to aberrant splicing of the transcript. Hum Mutat 2001; 17: 520–521. [DOI] [PubMed] [Google Scholar]
- Salas A, Comas D, Lareu MV, Bertranpetit J, Carracedo A. mtDNA analysis of the Galician population: a genetic edge of the European variation. Eur J Hum Genet 1998; 6: 365–375. [DOI] [PubMed] [Google Scholar]
- Santos C, Peixoto A, Rocha P, Vega A, Soares MJ, Cerveira N et al. Haplotype and quantitative transcript analyses of Portuguese breast/ovarian cancer families with the BRCA1 R71G founder mutation of Galician origin. Fam Cancer 2009; 8: 203–208. [DOI] [PubMed] [Google Scholar]
- John EM, Miron A, Gong G, Phipps AI, Felberg A, Li FP, West DW, Whittemore AS. Prevalence of pathogenic BRCA1 mutation carriers in 5 US racial/ethnic groups. J Am Med Assoc 2007; 298: 2869–2876. [DOI] [PubMed] [Google Scholar]
- Weitzel JN, Lagos V, Blazer KR, Nelson R, Ricker C, Herzog J et al. Prevalence of BRCA mutations and founder effect in high-risk hispanic families. Cancer Epidemiol Biomark Prev 2005; 14: 1666–1671. [DOI] [PubMed] [Google Scholar]
- Bacelar JA. Galegos no Paraíso Racial. Centro Editorial e Didático da UFBA. IANAMÁ: Salvador, Brasil, 1994. [Google Scholar]
- Leira LP. Galegos na Bahia de todos os Santos Galicia en El Mundo Edicións. Editora Graciela Alba Burgos: Spain 2002: 10–22. [Google Scholar]
- Zhang L, Fleischut MH, Kohut K, Spencer S, Wong K, Stadler ZK et al. Assessment of the prevalence of de novo mutations in the BRCA1 and BRCA2 genes. Clin Genet 2011; 80: 97–98. [DOI] [PubMed] [Google Scholar]
- Ashton-Prolla P, Osorio CABT, Khoeler-Santos P, Graudenz MS, Palmero EI, Fernandes G et al. Contribution of TP53 p.R337H mutation to breast cancer prevalence in Brazil. J Clin Oncol 2012; (suppl 30): (abstr 1522).
- Assumpção JG, Seidinger AL, Mastellaro MJ, Ribeiro RC, Zambetti GP, Ganti R et al. Association of the germline TP53 R337H mutation with breast cancer in southern Brazil. BMC Cancer 2008; 11: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MCB, Kotsopoulos J, de Almeida GL, Costa MM, Vieira R, Filho Fde A et al. The R337H mutation in TP53 and breast cancer in Brazil. Hered Cancer Clin Pract 2012; 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCCN - National Comprehensive Cancer Network: Clinical Practice Guidelines in Oncology™. Genetic/Familial High-Risk Assessment: Breast and Ovarian, Hereditary Breast and/or Ovarian Cancer (HBOC-1) V.1; 2010. www.nccn.org.
- Online Mendelian Inheritance in Man, OMIM®Johns Hopkins University, Baltimore, MD. MIM Number: #151623. http://omim.org/. Accessed 4 January 2013.
- Cury NM, Ferraz VE1, Silva WA Jr. TP53 p.R337H prevalence in a series of Brazilian hereditary breast cancer families. Hered Cancer Clin Pract 2014; 12: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCuaig JM1, Armel SR, Novokmet A, Ginsburg OM, Demsky R, Narod AS et al. Routine TP53 testing for breast cancer under age 30: ready for prime time? Fam Cancer 2012; 11: 607–613. [DOI] [PubMed] [Google Scholar]
- Gomes MC, Costa MM, Borojevic R, Monteiro AN, Vieira R, Koifman S, Koifman RJ et al. Prevalence of BRCA1 and BRCA2 mutations in breast cancer patients from Brazil. Breast Cancer Res Treat 2007; 103: 349–353. [DOI] [PubMed] [Google Scholar]
- Ewald IP, Izetti P, Vargas FR, Moreira MA, Moreira AS, Moreira-Filho CA et al. Prevalence of the BRCA1 founder mutation c.5266dupin Brazilian individuals at-risk for the hereditary breast and ovarian cancer syndrome. Hered Cancer Clin Pract 2011; 9: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro DM, Folgueira MAAK, Lisboa BCG, Olivieri EHR, Krepischi ACV, Carvalho AF et al. Comprehensive analysis of BRCA1, BRCA2 and TP53 germline mutation and tumor characterization: a portrait of early-onset breast cancer in Brazil. PLoS ONE 2013; 8: e57581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillenburg CV, Bandeira IC, Tubino TV, Rossato LG, Dias ES, Bittelbrunn AC et al. Prevalence of 185delAG and 5382insC mutations in BRCA1, and 6174delT in BRCA2 in women of Ashkenazi Jewish origin in southern Brazil. Genet Mol Biol 2012; 35: 599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel N, Feng BJ, Foretova L, Stoppa-Lyonnet D, Narod SA, Imyanitov E et al. On the origin and diffusion of BRCA1 c.5266dupC (5382insC) in European populations. Eur J Hum Genet 2011; 19: 300–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MSF. O papel da migração internacional na evolução da população brasileira (1872 a 1972). Rev de Saúde Pública 1974; 3: 49–90. [PubMed] [Google Scholar]
- Moreira MAM, Bobrovnitchaia IG, Lima MA, Santos AC, Ramos JP, Souza KR et al. Portuguese c.156_157insAlu BRCA2 founder mutation: gastrointestinal and tongue neoplasias may be part of the phenotype. Fam Cancer 2012; 11: 657–660. [DOI] [PubMed] [Google Scholar]
- Cybulski C, Górski B, Huzarski T, Masojć B, Mierzejewski M, Debniak T et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet 2004; 75: 1131–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abud J, Prolla JC, Rossi C, Palmero EI, Vargas FR, Nunes LM et al. CHEK2 1100DELC germline mutation: a frequency study in hereditary breast and colon cancer Brazilian families. Arq Gastroenterol 2012; 49: 273–278. [DOI] [PubMed] [Google Scholar]
- Azevêdo E, Fortuna CM, Silva KM, Sousa MG, Machado MA, Lima AM et al. Spread and diversity of human populations in Bahia, Brazil. Hum Biol 1982; 54: 329–341. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.