Interleukin 1β (IL-1β) is a proinflammatory cytokine remarkable for its broad range of functions (IL-1β plays a central role in innate host defense and host homeostasis) (1). However, IL-1β is also remarkable for the complexity of the physiology surrounding its activation, release, and signaling. In this context, work centered on understanding IL-1β physiology has led to surprise discoveries that have significantly impacted cell biology. The first major impact was the discovery that IL-1β is produced as a leaderless prohormone that requires specific cleavage by an unknown enzyme. Identification of this enzyme, IL-1β-converting enzyme (ICE), foreshadowed the discovery of apoptotic enzymes known as caspases (2). The centrality of caspases to programmed cell death is now accepted (and because of this ICE was renamed caspase-1). Indeed, caspase-1 biology continues to astonish. Research into the activation of caspase-1 has recently led to an intriguing connection to the innate host defense of plants. Intracellular human homologues of plant “guard” molecules (variously described as NOD, NALP, or CATERPILLER proteins) are critical to caspase-1 activation (3–5). However, despite these exciting discoveries, a fundamental question about IL-1β physiology still remains. How does this leaderless molecule pro-IL-1β combine with active caspase-1 and exit from the cytosol of activated monocytes and macrophages? The work of Andrei et al. (6) in a recent issue of PNAS provided a novel synthesis of the prevailing theories and offered a useful new paradigm about how caspase-1 and IL-1β are regulated.
ATP as Inducer of IL-1β Processing and Release
Although endotoxin induces large intracellular stores of pro-IL-1β, the release of processed IL-1β is relatively inefficient (7). However, exogenous ATP induces rapid processing and massive release of endotoxin-induced IL-1β (8–10). Therefore, ATP has become a standard second stimulus used to telescope IL-1β processing and release down to ≈30 min. ATP works by activating P2X7 receptors (11), which open plasma membrane pores promoting the rapid efflux of intracellular K+ (9, 12). Culturing monocytes in the presence of elevated K+ prevents the K+ efflux and thereby blocks ATP-induced processing and release of IL-1β, confirming the important role that intracellular K+ plays in regulating IL-1β processing and release (9).
Models of IL-1 Release
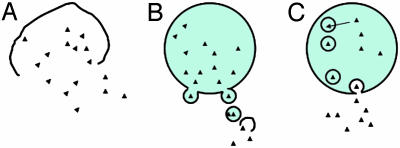
How this ATP-induced event triggers the release remains controversial. Three models that have been previously suggested are outlined in Fig. 1. The first and oldest model proposes that the ATP simply triggers cell death and cell lysis (Fig. 1 A) (8). The cell rupture idea has long been favored by many investigators as it is known that injurious stimuli that perturb membrane function are particularly adept at inducing IL-1β release from mononuclear phagocytes (13–15). Silicates that are known macrophage cytotoxic agents are excellent inducers of IL-1β release, which may in part explain the fibrotic potential of inhaled silica (15, 16). Although it is true that ATP stimulation eventually leads to cell death and lysis, studies that follow the kinetics of IL-1β release and lactic dehydrogenase release show that the IL-1β release precedes pore formation-induced cell lysis (17). So there is a clear link to cell death, but it appears that the process is more orchestrated than can be explained by simple lysis.
Fig. 1.
Three proposed pathways to explain IL-1β release from macrophages. (A) Lysis (as with cytolytic cell death). (B) Blebbing (as with the ATP induced extrusion of microvesicles containing IL-1β.(C) Exocytosis (as with the PLA2-mediated exocytosis of lysosomal vesicles, see ref. 6).
A second proposed mechanism for the IL-1β escape is via the release of microvesicles in a process of membrane blebbing (Fig. 1B). ATP induces the formation of tight-necked blebs in monocytes (12, 18). In this model, it has been shown that human THP-1 cells produce buds that shed as microvesicles as early as 2–5 s after ATP. These vesicles contain bioactive IL-1β, and eventually these blebs rupture, releasing IL-1β (18). This model provides a mechanism that allows IL-1β to get through the plasma membrane without having to invoke a terminal lysis event or membrane crossing event. However, it has been demonstrated that these vesicles are released in the absence of prior endotoxin priming (18). This finding is notable because priming is necessary for the ATP induction of processing and release (17). The microvesicle concept has been further challenged by the recent observation that these blebs depend on the activation of Rho-effector kinases and that inhibitors of these kinases can prevent blebbing but not IL-1β processing and release (12). Together, these issues raise the possibility that the microvesicles may represent a parallel pathway that is only partly responsible for IL-1β release. This is where the synthesis provided by the work of Andrei et al. (6) provides clear new answers to the puzzle.
The third pathway, originally described by Bakouche et al. (19, 20), involves the exocytosis of secretory lysosomes (Fig. 1C). Andrei et al. (6) expand on this third pathway by describing a novel framework that brings the experimental data together into a cohesive model. They provide explanations for how pro-IL-1β and caspase-1 get together and how the two seemingly divergent phospholipase pathways, Ca2+-independent phospholipase A2 and Ca2+-dependent phospholipase A2, work in concert. By using subcellular fractionation and immunoelectron microscopy, Andrei et al. demonstrate that pro-IL-1β is transported into preterminal endocytic vesicles. There IL-1β is detected in the same compartment as the acid hydrolase cathepsin D, the lysosomal marker Lamp-1, and caspase-1. This finding is important because placing pro-IL-1β and caspase-1 together in the same protected compartment provides a mechanism whereby pro-IL-1β can be safely processed by active caspase-1 without the caspase-1 inducing apoptosis. Of note here is that, in contrast to active caspase-3 that is readily detectable in cells undergoing apoptosis, active caspase-1 has not been convincingly demonstrated within the cytosol of activated monocytes even when actively releasing processed IL-1β (21). Therefore, this model provides a solution to a number of questions about IL-1β processing and release. It explains how caspase-1 can be activated but not induce programmed cell death, why IL-1β and caspase-1 secretion are synchronous, and why active caspase-1 has been difficult to detect during active processing and release.
Role of Phospholipases
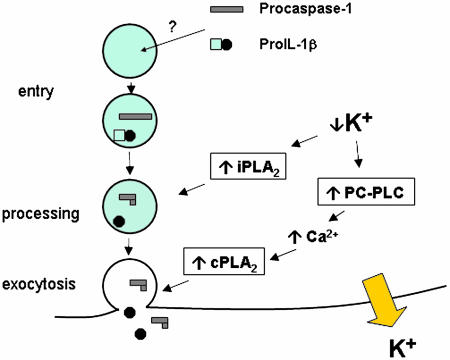
Finally, an event that requires penetration and modification of membranes requires the work of phospholipases. These enzymes help in modifying lipid surfaces and triggering downstream signaling events critical to cell function. In this context, ATP-induced K+ release from cells triggers IL-1β processing. Walev et al. (22) recently documented that K+ depletion mediates IL-1β processing via activation of Ca2+-independent phospholipase A2 (iPLA2). Specific inhibition of iPLA2 prevents IL-1β processing, but it does not prevent IL-1β release. ATP also increases the concentrations of intracellular Ca2+, but intracellular Ca2+ levels activate Ca2+-dependent PLA2 (cPLA2), which has been shown to inhibit IL-1β processing (21). Thus, it is controversial how ATP-induced intracellular Ca2+ changes can be associated with IL-1β release. The new model (Fig. 2) converges the two competing PLA2 models into a coherent one. The link is the intracellular loss of K+. Andrei et al. (6) show that loss of intracellular K+ activates phosphatidylcholine-specific phospholipase C (PC-PLC), which in turn is responsible for the increase in cytosolic Ca2+. This results in activation of cPLA2 and lysosomal exocytosis of IL-1β, along with caspase-1 and cathepsin D. Importantly, although specific inhibition of iPLA2 blocks the processing of both pro-IL-1β and procaspase-1, it does not impair their release. It has been known that IL-1β release does not require prior processing (17, 23). Thus, it appears that iPLA2 activates processing, whereas cPLA2 activates endosomal release. Both phospholipases depend on loss of intracellular K+ stores. Low K+ triggers PC-PLC-induced increases in intracellular Ca2+, which activates cPLA2 and lysosomal exocytosis. Concurrently, low K+ also directly triggers iPLA2 in a PC-PLC/Ca2+-independent manner. Thus, the compartmentalization in exosomes and the separate function of the phospholipases solve many of the issues in understanding the links between IL-1β processing and IL-1β release.
Fig. 2.
Steps in exosome pathway. Procaspase-1 and pro-IL-1β enter the endosome by a pathway yet to be fully characterized. Loss of intracellular K+ (which can be induced by exogenous ATP) triggers activation of the calcium-independent phosholipase A2 (iPLA2). Endosomal caspase-1 is then activated by the iPLA2 to cleave pro-IL-1β to its functional form. The low intracellular K+ levels also activate phosphatidylcholine phospholipase C (PC-PLC), which induces increases in intracellular calcium (Ca2+). Increased cytosolic Ca2+ activates the calcium-dependent phospholipase A2 (cPLA2) and exocytosis.
Of course, many questions remain. Future studies will need to address how the recently described CATERPILLER proteins function in the context of exosomal caspase activation. Are CATERPILLERs also secreted into the exosomes and do they play a role in regulating exosome trafficking? And, finally, if the exosome pathway proves to be the primary secretory route for IL-1β, one major unsolved question remains. How does IL-1β cross into these secretory vesicles? If the past discoveries are any predictor, expect to be surprised.
Acknowledgments
This work was supported by National Institutes of Health Grant HL40871.
See companion article on page 9745 in issue 26 of volume 101.
References
- 1.Dinarello, C. A. (2000) Chest 118, 503–508. [DOI] [PubMed] [Google Scholar]
- 2.Yuan, J., Shaham, S., Ledoux, S., Ellis, H. M. & Horvitz, H. R. (1993) Cell 75, 641–652. [DOI] [PubMed] [Google Scholar]
- 3.Tschopp, J., Martinon, F. & Burns, K. (2003) Nat. Rev. Mol. Cell. Biol. 4, 95–104. [DOI] [PubMed] [Google Scholar]
- 4.Inohara, N. & Nunez, G. (2003) Nat. Rev. Immunol 3, 371–382. [DOI] [PubMed] [Google Scholar]
- 5.Dangl, J. L. & Jones, J. D. (2001) Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- 6.Andrei, C., Margiocco, P., Poggi, A., Lotti, L. V., Torrisi, M. R. & Rubartelli, A. (2004) Proc. Natl. Acad. Sci. USA 101, 9745–9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wewers, M. D., Rennard, S. I., Hance, A. J., Bitterman, P. B. & Crystal, R. G. (1984) J. Clin. Invest. 74, 2208–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogquist, K. A., Nett, M. A., Unanue, E. R. & Chaplin, D. D. (1991) Proc. Natl. Acad. Sci. USA 88, 8485–8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perregaux, D. & Gabel, C. A. (1994) J. Biol. Chem. 269, 15195–15203. [PubMed] [Google Scholar]
- 10.Griffiths, R. J., Stam, E. J., Downs, J. T. & Otterness, I. G. (1995) J. Immunol. 154, 2821–2828. [PubMed] [Google Scholar]
- 11.Dubyak, G. R. & El-Moatassim, C. (1993) Am. J. Physiol. 265, C577–C606. [DOI] [PubMed] [Google Scholar]
- 12.Verhoef, P. A., Estacion, M., Schilling, W. & Dubyak, G. R. (2003) J. Immunol. 170, 5728–5738. [DOI] [PubMed] [Google Scholar]
- 13.Gery, I., Davies, P., Derr, J., Krett, N. & Barranger, J. A. (1981) Cell. Immunol. 64, 293–303. [DOI] [PubMed] [Google Scholar]
- 14.Vogel, S. N., English, K. E. & O'Brien, A. D. (1982) Infect. Immun. 38, 681–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt, J. A., Oliver, C. N., Lepe-Zuniga, J. L., Green, I. & Gery, I. (1984) J. Clin. Invest. 73, 1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava, K. D., Rom, W. N., Jagirdar, J., Yie, T. A., Gordon, T. & Tchou-Wong, K. M. (2002) Am. J. Respir. Crit. Care Med. 165, 527–533. [DOI] [PubMed] [Google Scholar]
- 17.Mehta, V. B., Hart, J. & Wewers, M. D. (2001) J. Biol. Chem. 276, 3820–3826. [DOI] [PubMed] [Google Scholar]
- 18.Mackenzie, A., Wilson, H. L., Kiss-Toth, E., Dower, S. K., North, R. A. & Surprenant, A. (2001) Immunity 15, 825–835. [DOI] [PubMed] [Google Scholar]
- 19.Bakouche, O., Brown, D. C. & Lachman, L. B. (1987) J. Immunol. 138, 4249–4255. [PubMed] [Google Scholar]
- 20.Andrei, C., Dazzi, C., Lotti, L., Torrisi, M. R., Chimini, G. & Rubartelli, A. (1999) Mol. Biol. Cell 10, 1463–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahy, R. J., Doseff, A. I. & Wewers, M. D. (1999) J. Immunol. 163, 1755–1762. [PubMed] [Google Scholar]
- 22.Walev, I., Klein, J., Husmann, M., Valeva, A., Strauch, S., Wirtz, H., Weichel, O. & Bhakdi, S. (2000) J. Immunol. 164, 5120–5124. [DOI] [PubMed] [Google Scholar]
- 23.Perregaux, D. G. & Gabel, C. A. (1998) J. Immunol. 160, 2469–2477. [PubMed] [Google Scholar]