Abstract
Cystic fibrosis (CF), the most common life-threatening autosomal recessive disorder in Caucasians, is caused by mutations in CF transmembrane conductance regulator gene (CFTR). We and others previously identified CFTR mutations in 20 Chinese patients with CF. In this study, eight Chinese patients with a clinical diagnosis of suspected CF were newly collected and screened for CFTR mutations using a combination of conventional Sanger sequencing and multiplex ligation-dependent probe amplification (MLPA) analysis. The CFTR mutations observed in Chinese CF patients, both reported previously and identified in the present study, were also summarized. In the newly collected patients, we identified 10 different CFTR mutations, including p.F508del, the most common CF-causing mutation in Caucasians, and three novel mutations (p.V1212Afs*15; p.L666* and p.A969A). Most notably, the previously reported p.G970D mutation was found in six patients, making it the most frequent CFTR mutation identified in Chinese CF patients thus far. In conclusion, we detected p.F508del for the first time, identified additional novel CFTR mutations and recorded the most frequent CF-causing mutation in Chinese CF patients.
Introduction
Cystic fibrosis (CF, MIM #219700), which is inherited in an autosomal recessive pattern, is the most frequent lethal genetic disorder among Caucasians, with a prevalence of ~1/2,500. The incidence of CF varies dramatically among different races. Only a few reliable estimates of the incidence in Asians are available, one of which indicates an estimated incidence of CF in Asian Pacific Islanders living in Hawaii of 1 in 90,000,1 while the other two reports indicate an estimated incidence of CF in Japan of 1 in 350,000.2,3 CF is a highly variable disorder. The classic phenotypic expression of CF includes recurrent pulmonary disease, an increased sweat chloride concentration ([Cl−]>60 mmol/l), and pancreatic insufficiency (PI). Approximately 60% of all CF patients are diagnosed before 12 months of age and 90% by 10 years of age. However, some adults can present with mild or incomplete phenotypes of CF, without clinical, chemical or histological evidence of pancreatic disease; these cases have been variably referred to as non-classic, pancreatic sufficiency (PS), intermediate, borderline or atypical forms of CF.4
CF is caused by mutations in the CF transmembrane conductance regulator gene (CFTR, MIM #602421), which is expressed by most epithelial cells. CFTR is composed of 27 exons spanning 189 kb of DNA, encoding a protein of 1,480 amino acids. CFTR functions as a chloride channel. A sodium and chloride ion transport defect in the epithelial cells lining the airways of the lungs and in the sweat glands of CF patients is the fundamental defect observed in CF.5 Approximately 2,000 mutations in the CFTR gene have been identified thus far, and this number is continuously updated within the Cystic Fibrosis Genetic Analysis Consortium (CFGAC) database.6 All types of mutations, including missense and nonsense mutations, splicing, small and large rearrangements, frameshift and in-frame deletions/insertions, have been observed in CF patients. One mutation, p.F508del, has been found to occur in ~70% of the CFTR alleles of Caucasian CF patients.7 However, its frequency varies considerably among human populations. Apart from p.F508del, ~20 mutations occur with a frequency >0.1%, together accounting for ~15–20% of the CF alleles of Caucasians. The effects of these mutations on CFTR function have been grouped into six classes: defective synthesis (Class I); defective protein processing and trafficking (Class II); defective regulation or gating (Class III); defective chloride conductance (Class IV); reduced synthesis or trafficking (Class V); and decreased stability (Class VI).8 Complete loss of functional CFTR results in classic CF, and Class I–III mutations are, therefore, generally prone to cause classic CF phenotypes, which are associated with PI, severe lung dysfunction, an increased incidence of malnutrition, and earlier mortality. Mutations that alter, but which do not abolish, the function of CFTR can give rise to mild or non-classic CF, and thus, Class IV–V mutations are usually associated with non-classic CF phenotypes with milder lung disease, longer survival and PS.9
We and others have previously reported 20 CF patients of Chinese origin with CFTR mutations.10 In the present study, we collected data from eight additional Chinese patients with a clinical diagnosis of suspected CF and performed sequencing of CFTR to identify pathogenic mutations and to confirm the diagnosis of CF. We also conducted a systematic literature review11–20 to summarize the mutation spectrum in Chinese CF patients.
Materials and Methods
Patients and evaluation of clinical data
We collected data from eight CF patients at Peking Union Medical College Hospital (PUMCH). All of the patients were suspected of having CF, showing sweat chloride levels >60 mmol/l. The sweat tests were conducted following the protocol described in detail in our previous study.10 All subjects signed an informed consent form allowing anonymous use of their DNA samples and clinical data for research purposes. The protocol of this study was approved by the Institutional Review Board committee at PUMCH.
DNA extraction and identification of mutations
Genomic DNA was extracted from whole-blood samples. To detect CFTR gene mutations, we subjected all 27 exons and the intronic boundaries to PCR-Sanger sequencing. PCR was performed under the following conditions: initial denaturation at 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s. The list of primer pairs used for PCR was reported previously.10 The Sanger sequencing results were analyzed using CodonCode Aligner Software (CodonCode Aligner Corporation; Centerville, MA, USA). The presence of large CFTR rearrangements was tested in all 27 CFTR exons using a commercial kit (MLPA SALSA kit; MRC-Holland, Amsterdam, The Netherlands) and the generated results were analyzed with Coffalyser software (MRC-Holland, Amsterdam, The Netherlands).
Generation of minigene constructs
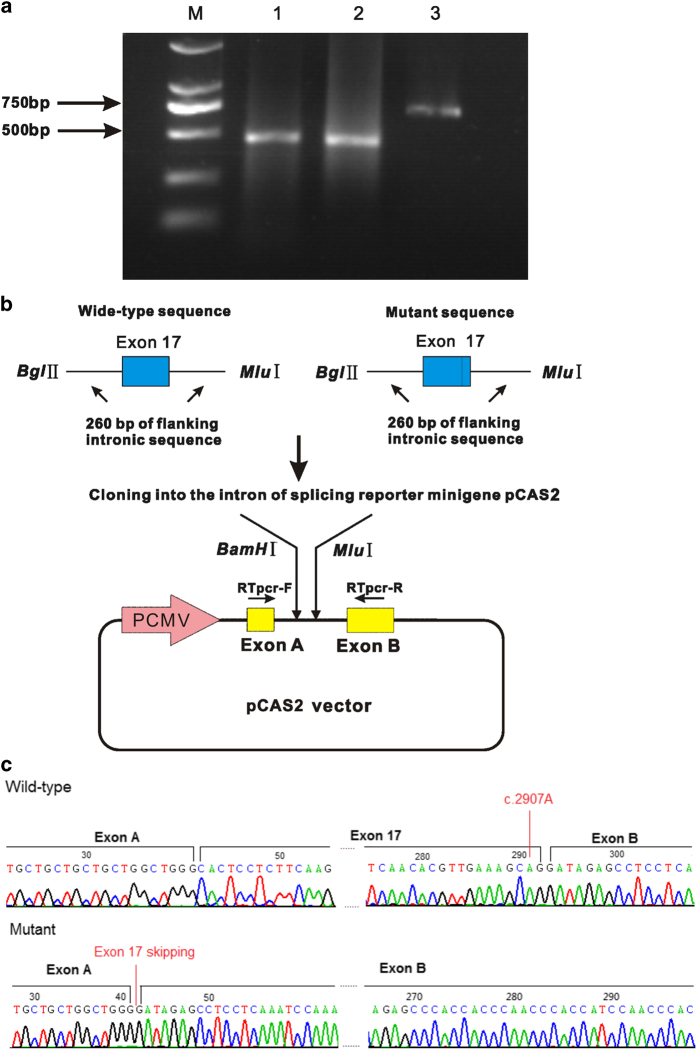
The identified synonymous variant of c.2907A>C (p.A969A) is located in the neighboring region of the splicing donor site of intron 7. To study the effect of this variant on the splicing pattern, a DNA fragment of 951 bp encompassing exon 17 and the flanking intronic sequences of CFTR was directly amplified from the genomic DNA of a CF patient heterozygous for this variant (c.2907A>C; p.A969A). The primers (Minigene-F/R) used for the generation of the minigene constructs are listed in Supplementary Table 1. Each PCR product was digested with the BglII and MluI restriction enzymes and cloned into the pCAS2 vector, which had also been digested with BamHI and MluI21 (Figure 2a). All of the selected clones were sequenced, and the verified clones, referred to as the wild-type (pCFTR-c.2907A) and mutant (pCFTR-c.2907C) clones, were retained for expression experiments.
Figure 2.
Impact of the c.2907A>C mutation on the CFTR splicing pattern. (a) The wild-type and the mutant exonic sequences of exon 17 of CFTR were PCRamplified from patient genomic DNA, together with ~260 bp of their 5′ and 3′ intronic flanking sequences, using specific primers carrying 5’ tails with BglII and MluI restriction sites. The amplicons are cloned into the pCAS2 reporter vector, which is based on the pcDNA3.1 plasmid and contains a minigene composed of exon A and exon B. (b) A 2% agarose gel showing the different fragments of cDNA obtained using specific primers with total cDNA for each plasmid construct. Lanes 2, 3 and 4 represent three independent transfections of the empty, mutant (c.2907 C) and wild-type (c.2907A) constructs. Lane 1 contains the DL 2000 DNA marker (M). Two fragments are visible: the wild-type fragment, showing normal splicing with exon 17 (717 bp); and the mutant fragment, in which exon 17 is skipped (466 bp). (c) Sequencing traces of the 717 bp fragment (wild-type), with normal splicing, and the 466 bp fragment (mutant), in which exon 17 is skipped.
Cell culture and minigene splicing assay
HEK 293T cells were grown in a 5% CO2 incubator at 37 °C in Dulbecco’s modified Eagle’s medium (Union cell resource center) supplemented with 10% fetal bovine serum (Gibco). HEK 293T cells that had been seeded at 30% confluence in 60 mm dishes 16 h before transfection were transfected with 8 μg of pCFTR-c.2907A, pCFTR-c.2907C and the empty pCAS2 vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Twenty-four hours after transfection, cells were collected and total RNA was extracted using TRIzol (Invitrogen). Then, 1.5 μg of total RNA was reverse transcribed using a Reverse Transcription System (Promega, Madison, WI, USA), according to the manufacturer’s instructions. Following RNA retrotranscription, 400 ng of complementary DNA from the three constructs mentioned above was PCR amplified using the primers (reverse transcriptase–PCR-F/R) shown in Supplementary Table 1. The PCR products were then separated on a 2% agarose gel, and individual bands were excised and sequenced using specific primers (SEQprimer-F/R) (Supplementary Table 1).
Results
Clinical data for eight CF patients in PUMCH
All eight patients were of Chinese Han origin, and none of them had a family history of intermarriage with Caucasians. The concentration of chloride in all eight patients' sweat ranged from 62–140 mmol/l (109.1±25.0 mmol/l). The age at diagnosis ranged from 1 to 22 years (median age, 14 years). The age of symptom onset ranged from newborn to 14 years of age (median age, 1 year). Three of the patients were male (37.5%), and three of the patients (Case C3, C6 and C7) had or might have had a family history of CF. Nearly all patients exhibited diffuse bronchiectasis predominantly affecting the upper lobes of the lung. Three patients (C4, C6 and C8) had allergic bronchopulmonary aspergillosis. Only one patient (C2) showed a positive result of Sudan III staining of the stool and was defined as having PI. Two patients (cases C1 and C3) were suspected of having PI, who manifested with recurrent diarrhea and suspected meconium ileus, respectively, but lacked abnormal Sudan III results for the stool (Table 1). Two patients (cases C7 and C8) exhibited severe growth retardation, with a weight below the third percentile, and their heights were below the third percentile and between the tenth and twenty-fifth percentiles, respectively, according to the same age-standardized growth curve data for Chinese children.22 However, neither patient showed a positive stool Sudan III result in at least two separate tests. The body mass indexes of the two adult patients (cases C1 and C6) were only between 16.1 and 17.2, and these patients exhibited negative stool Sudan III results. The detailed clinical manifestations of the patients are shown in Table 1.
Table 1. Clinical data and characteristics of CFTR gene mutations in eight Chinese CF patients examined in the current study.
| Case | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 |
|---|---|---|---|---|---|---|---|---|
| Age at diagnosis | 20 | 15 | 1 | 13 | 15 | 22 | 4 | 13 |
| Gender | M | F | F | M | M | F | F | F |
| Age at onset of symptom | Several months | 12 Years | 4 Months | 1 Year | 14 Years | 12 Years | 3 Months | 12 Years |
| Wt/Ht(kg/m) | 52/1.74 | 42/1.65 | 8/NA | 48/1.6 | 55/1.78 | 45/1.67 | 11/0.93 | 32/1.49 |
| Family history | None | None | Yes | None | Yes | Yes | None | None |
| Sweat test [Cl−] (mmol/l) | 137 | 140 | 108.4 | 95.2 | 106.5 | 101.9 | 122.1 | 62 |
| Gastrointestinal symptoms | Recurrent diarrhea, marasmus | Marasmus | Recurrent diarrhea; Meconium ileus suspected | None | None | Marasmus | Malnutrition | Malnutrition |
| Comorbidity | None | None | None | ABPA | None | ABPA | None | ABPA |
| Pulmonary function test | FEV1 43.6% PRED FEV1/FVC 56.96% | Not available | Not available | FEV1 61.4% PRED FEV1/FVC 74.5% | FEV1 81.7% PRED FEV1/FVC 77.01% | FEV1 30.2% PRED FEV1/FVC 47.98% | Not available | FEV1 72.9% PRED FEV1/FVC 69.48% |
| Diagnosis before CF confirmation | Bronchiectasis | Bronchiectasis | Bartter Syndrome | Bartter Syndrome and bronchiectasis | Bronchiectasis | Bronchiectasis | Bronchiectasis | ABPA |
| PS/PI | PI suspected | PI | PI suspected | PS | PS | PS | PS | PS |
| Sudan III Staining | Negative | Positive | Negative | Negative | Negative | Negative | Negative | Negative |
| Sputum culture | Pseudomonas Aeruginosa | Pseudomonas Aeruginosa | Pseudomonas Aeruginosa and Methicillin Resistant Staphylococus Epidermidis | Pseudomonas Aeruginosa | Pseudomonas Aeruginosa | Escherichia coli (extended-spectrum beta-lactamase+) and Pseudomonas Aeruginosa | Pseudomonas Aeruginosa | Negative |
|
CFTR allele 1 | ||||||||
| Nucleotide change | c.2909G>A | c.2909G>A | c.2909G>A | c.3700A>G | c.3635delTa | c.2909G>A | c.2909G>A | c.2909G>A |
| Amino acid change | p.G970D | p.G970D | p.G970D | p.I1234V | p.V1212Afs*15 | p.G970D | p.G970D | p.G970D |
|
CFTR allele 2 | ||||||||
| Nucleotide change | c.1521_1523delCTT | c.2374C>T | c.2125C>T | c.959–960insA | not detectedb | c.1997T>Ga | c.263T>G | c.2907A>Ca |
| Amino acid change | p.F508del | p.R792* | p.R709* | p.S321Ifs*42 | NA | p.L666* | p.L88* | p.A969A (ΔE17 p.N886Kfs*4) |
Abbreviations: ABPA, allergic bronchopulmonary aspergillosis; CF, cystic fibrosis; CFTR RefSeq, NM_000492.3; F, female; FEV1, forced expiratory volume in 1s; FVC, forced vital capacity; M, male; NA, not applicable; PI, pancreatic insufficiency; pred, predicted; PS, pancreatic sufficiency;
Novel mutation (shown in bold) identified in the present study.
Performed MLPA.
CFTR mutations identified in the present study
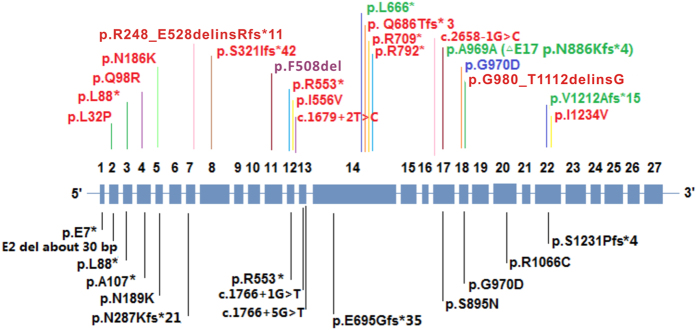
All eight newly identified patients underwent screening for mutations in CFTR coding regions and flanking sequences using direct Sanger sequencing or multiplex ligation-dependent probe amplification (MLPA). All of the patients were compound heterozygotes for CFTR mutations, except for case C5, in whom only one mutation was detected. In total, we identified 10 different mutations, including 3 novel mutations (c.3635delT/p.V1212Afs*15; c.1997T>G/p.L666*; c.2907A>C/p.A969A); c.1521_1523delCTT/p.F508del was found in one case in a heterozygous state; and c.2909G>A/p.G970D was detected in six cases (Table 1, Figure 1). The seven known mutations detected are uncommon CFTR mutations according to both the Human Gene Mutation Database (HGMD)23 and CFGAC, with all of these mutations occurring in fewer than six reports, except for p.F508del.
Figure 1.
CFTR mutation spectrum in Chinese patients with CF. Upper panel: CFTR mutations identified in our current (eight CFs) and previous (seven CFs) (#14–20, as in Table 2) samples of Chinese CF patients collected at PUMCH. The three novel mutations identified in the current study are highlighted in green; the most common mutation found in Chinese CF patients thus far is highlighted in blue; the most common mutation found in Caucasian CF patients is highlighted in purple. Lower panel: CFTR mutations identified in previous studies (#1–13 as in Table 2) in Chinese patients with CF. The CFTR gene is shown with rescaled exons and minimized introns, and is not drawn to scale. CF, cystic fibrosis; PUMCH, Peking Union Medical College Hospital.
Impact of the c.2909A>C mutation on CFTR mRNA splicing
Following transient transfection of the wild-type (pCFTR-c.2907A) and mutant (pCFTR-c.2907C) plasmids into HEK293 cells, mRNA analysis was performed via reverse transcriptase–PCR, and direct mRNA sequencing was carried out using specific primers (Supplementary Table 1). Electrophoresis on a 2% agarose gel showed two fragments: a normally spliced fragment with exon 17 and an aberrantly spliced fragment with exon 17 skipping (Figure 2b). The sequencing traces presented in Figure 2c showed two different fragments: the 717-bp fragment represents normal splicing with the complete exon 17; and the 466-bp fragment represents abnormal splicing, skipping exon 17.
Discussion
CF is a highly allelically heterogeneous genetic disorder, for which ~2,000 putative disease-causing mutations have been reported to the CFGAC. p.F508del is the most commonly identified CF mutation, occurring in ~70% of the CFTR genes of CF patients. However, its frequency varies considerably among human populations. The highest frequency observed in Northern Europeans corresponds to 75–88% of CF alleles, while the frequency in Southern Europeans is 50–60%.7 In non-Caucasian CF patients, individuals of African descent living in the United States carry the p.F508del allele at a frequency of 48%, probably due to admixture.24 Because of a misconception that CF is prevalent only in Caucasians, only a few studies on CFTR mutations have been carried out in Chinese CF patients, and p.F508del has never been previously reported in this group. In the present study, we detected the p.F508del mutation for the first time in one of our patients. The p.F508del mutation is a relatively ancient mutation, arising more than 52,000 years ago in Europe.25 Almost all F508del alleles occur within a (TG)10-T9 haplotype, which is quite rare in Asian populations. We also examined the (TG)m-Tn haplotype in intron 9 of CFTR in our patients (Table 1) and observed that in patient C1, F508del was present in the (TG)10-T9 haplotype inherited from his mother. In fact, this is not the first time that p.F508del in CFTR has been detected in a Chinese population; it has previously been reported that p.F508del in CFTR occurs in CFTR-related disorders such as CBAVD (congenital bilateral absence of vas deferens) in the Chinese population with a very low frequency.26
In the present study, three novel mutations were identified: c.3635delT, p.V1212Afs*15; c.1997T>G, p.L666* and c.2907A>C and p.A969A/ΔE17 p.N886Kfs*4, which are all predicted to lead to truncated proteins. These mutations are highly likely to have pathogenic potential, although there is currently no evidence from functional testing to support this possibility.
In the present study, six CF patients were found to carry the p.G970D mutation, which exhibited the highest frequency of 37.5% (6/16) in our Chinese CF patients. Previously, p.G970D had been reported in Chinese CF patients in three independent studies (Table 2), at a frequency of 7.5% (3/40). Considering these results together, p.G970D is the most frequent mutation that has been identified in Chinese CF patients, with a frequency of 16.1% (9/56) (Tables 1 and 2). p.G970D was first identified by Wagner JA in 199915 and the same patient was later revisited by Wine JJ in 2001 in an ethnically heterogeneous population.27 This patient who was assessed in these two studies was of Chinese origin. Therefore, to our knowledge, p.G970D has only been identified in Chinese CF patients thus far. The recurrence of p.G970D is likely to be due to a founder effect. We explored this possibility by analyzing the haplotypes created by a set of SNPs on chromosomes bearing p.G970 or p.D970. Within an interval of ~150 Kb, harboring p.G970D in the center, we detected an identical haplotype carrying p.D970 in six trios from the current study. Therefore, we cannot exclude the possibility of a founder effect, in spite of the fact that the six families all came from different regions of China (data not shown).
Table 2. Characteristics of CFTR gene mutations in Chinese CF patients in the available published literature.
| Case |
CFTR allele 1 |
CFTR allele 2 |
||
|---|---|---|---|---|
| Nucleotide change | Amino acid change | Nucleotide change | Amino acid change | |
| 1 (ref. 9) | E2 del about 30 bp | NA | NA | NA |
| 2 (ref. 10) | c.1766+5G>T | NA | NA | NA |
| 3 (ref. 11) | c.1766+5G>T | NA | c.1766+5G>T | NA |
| 4 (ref. 12) | c.1766+1G>T | NA | NA | NA |
| 5 (ref. 13) | c.319–326delGCTTCCTA | p.A107X | c. 2909G>A | p.G970D |
| 6 (ref. 14) | c.1766+5G>T | NA | c.2083dupG | p.E695GfsX35 |
| c.2684G>A | p.S895N | |||
| 7 (ref. 14) | c.1766+5G>T | NA | c.2083dupG | p.E695GfsX35 |
| c.2684G>A | p.S895N | |||
| 8 (ref. 15) | c.19G>T | p.E7X | c. 860dupA | p.N287KfsX21 |
| 9 (ref. 15) | c.1766+5G>T | NA | c.2083dupG | p.E695GfsX35 |
| c.2684G>A | p.S895N | |||
| 10 (ref. 16) | c.1657C>T | p.R553X | c.1657C>T | p.R553X |
| 11 (ref. 17) | c.567C>A | p.N189K | c.3691delT | p.S1231PfsX4 |
| 12 (ref. 18) | c.263T>G | p.L88X | c.2909G>A | p.G970D |
| 13 (ref. 18) | c.3196C>T | p.R1066C | c.3196C>T | p.R1066C |
| 14 (ref. 8) | c.293A>G | p.Q98R | c.293A>G | p.Q98R |
| 15 (ref. 8) | c.95T>C | p.L32P | c.1657C>T | p.R553X |
| 16 (ref. 8) | c.293A>G | p.Q98R | c.558C>G | p.N186K |
| 17 (ref. 8) | c.2052 dupA | p. Q686TfsX3 | ΔE18-E20 (c.2909−?_3367+?del) | p.G980_T1112 delinsG |
| 18 (ref. 8) | c.2909G>A | p.G970D | ΔE7-E11 (c.744−?_1584+?del) | p.R248_E528 delinsRfsX11 |
| 19 (ref. 8) | c.1666A>G | p.I556V | Not detected | NA |
| 20 (ref. 8) | c.1679+2T>C | NA | c.2658-1G>C | NA |
Abbreviations: CF, cystic fibrosis; NA, not applicable.
p.G970D, located within the third cytoplasmic loop of CFTR, impairs chloride conductance. This leads to partial function of CFTR,15 allowing us to group it within the category of Class IV mutations, which are generally associated with mild CF phenotypes (e.g., PS).8 In a total of nine cases carrying p.G970D (Tables 1 and 2), each of the patients also carried a nonsense, frameshift or deletion mutation, all of which belong to the Class I or II mutations, generally causing severe CF phenotypes (e.g., PI).8 Considering that Class IV mutations are phenotypically dominant when they occur in combination with class I–III mutations,9 we expected these nine p.G970D-carriers to present a mild CF phenotype (i.e., PS). However, three of the nine p.G970D-carriers (Tables 1 and 2) were suspected of having PI.15,20 This inconsistency could be explained by the fact that general tendencies8 are only expected to apply to patient populations, and not to individuals.28
In addition to two mutations (p.F508del, p.R553*) included in the Caucasian screening panel, as shown in Tables 1 and 2, the limited numbers of studies in Chinese CF patients have indicated that all of the other mutations are only found in Asian populations, with certain mutations predominantly being detected in Chinese patients.23 Together, these studies demonstrate that Chinese populations exhibit a unique CF mutation spectrum, consistent with our previous statement.10 For most of the mutations listed in Tables 1 and 2, we did not find any concrete data that would allow these mutations to be classified based on their biological consequences after carefully checking the literature and the CFTR2 database.29 We then roughly estimated that 16 of 28 CF Chinese patients might carry one Class I or II (classic) mutation in combination with one Class III–V (mild) mutation according to the rules for categorizing CFTR mutations.8 As suggested by Ferec,9 these patients would be expected to present with PS more commonly than PI. The fact that 5/16 of the patients presented with PI again emphasized that general dispositions should not be used to make individual prognostic predictions. As a matter of fact, the potential for a mutation to produce clinical disease depends on multiple factors, including the type of mutation, the molecular mechanism at the cellular level and many others. Moreover, it is well-known that the presence of modifier genes in CF may benefit or harm its clinical outcome,30 which could explain the variability of the CF phenotype observed in CF patients with similar genotypes.
Classic CF is determined as a childhood disease characterized by chronic lung disease, sinusitis, nasal polyposis, PI that causes malnutrition, chronic diarrhea, meconium ileus, rectal prolapse and elevated [Na+]s and [Cl−]s in sweat, while atypical CF is a milder form of CF disorder. Individuals with atypical CF might only exhibit dysfunction of one system, which may be much milder compared with the classic type. In a previous study,10 we suspected Chinese CF patients of exhibiting the atypical rather than the classic form of CF because only a small proportion of the patients presented with gastrointestinal symptoms; more than half of our patients exhibited allergic bronchopulmonary aspergillosis, which is far beyond the prevalence reported for Caucasians.10 Our phenotypic speculation regarding Chinese CF patients was also based on the known correlation between the CFTR genotype and the CF phenotype, which indicates that patients with atypical CF usually carry one severe mutation and one less common mutation.31 However, among all the 15 patients diagnosed at our center (including our previous reports10), 6 of the 15 CF patients (40%) were suspected of having PI, and more than half of our patients (8/15) exhibited growth retardation or marasmus. Allergic bronchopulmonary aspergillosis was slightly less common in the present group (3/8) compared with our previous study (4/7).10 Taken together, such results suggest that the phenotypic prediction made for Chinese CF patients in our previous study might be not correct, or at least it is too early to make this prediction. The atypical clinical manifestations of Chinese patients may be due to selection bias, or to low awareness of the disease in China, or to a lack of sensitive testing methods for pancreatic exocrine insufficiency. In the future, as we collect more CF patients in China, we might be more confident in summarizing the phenotypic spectrum of Chinese CF patients.
In conclusion, we have identified several CFTR mutations, some of which are novel (p.V1212Afs*15; p.L666* and p.A969A), while one is frequent (p.G970D) and one has been detected for the first time (p.F508del) in Chinese patients with CF. These results indicate that, at the molecular genetic level, Chinese CF patients exhibit a unique mutation spectrum and complex genotype–phenotype correlations, similar to other ethnic populations. Sequencing of the entire CFTR gene followed by MLPA analysis, rather than using the targeted sequencing-based screening panel for mutations commonly found in Caucasian populations, is recommended for genetic analysis in Chinese patients with CF. Owing to the small sample size, these results are preliminary and must be confirmed in a larger study, possibly requiring a multicenter collaboration.
Acknowledgments
We thank Min Peng, Xue-feng Sun and Jiangna Han for their support in this study. We thank Zhanjun Zhang for his assistance with the sweat test. This work was supported by the Beijing Municipal Science and Technology Commission (z151100003915078 to XZ), the National Natural Science Foundation of China (81300001 to XT) and Funding for Peking Union Medical College Hospital Middle-aged and Young Researchers (I449000 to XT).
The authors declare no conflict of interest.
Footnotes
Supplementary Information for this article can be found on the Human Genome Variation website (http://www.nature.com/hgv).
References
- Wright SW , Morton NE . Genetic studies on cystic fibrosis in Hawaii. Am J Hum Genet 1968; 20: 157–169. [PMC free article] [PubMed] [Google Scholar]
- Imaizumi Y . Incidence and mortality rates of cystic fibrosis in Japan, 1969-1992. Am J Med Genet 1995; 58: 161–168. [DOI] [PubMed] [Google Scholar]
- Yamashiro Y , Shimizu T , Oguchi S , Shioya T , Nagata S , Ohtsuka Y . The estimated incidence of cystic fibrosis in Japan. J Pediatr Gastroenterol Nutr 1997; 24: 544–547. [DOI] [PubMed] [Google Scholar]
- Bombieri C , Claustres M , De Boeck K , Derichs N , Dodge J , Girodon E et al. Recommendations for the classification of diseases as CFTR-related disorders. J Cyst Fibros 2011; 10: S86–S102. [DOI] [PubMed] [Google Scholar]
- Knowles M , Stutts M , Spock A , Fischer N , Gatzy J , Boucher R . Abnormal ion permeation through cystic fibrosis respiratory epithelium. Science 1983; 221: 1067–1070. [DOI] [PubMed] [Google Scholar]
- Cystic Fibrosis Mutation Database. Available at http://www.genet.sickkids.on.ca/cftr.
- Bobadilla JL , Macek M Jr , Fine JP , Farrell PM . Cystic fibrosis: a worldwide analysis of CFTR mutations--correlation with incidence data and application to screening. Hum Mutat 2002; 19: 575–606. [DOI] [PubMed] [Google Scholar]
- Bombieri C , Seia M , Castellani C . Genotypes and phenotypes in cystic fibrosis and cystic fibrosis transmembrane regulator-related disorders. Semin Resp Crit Care Med 2015; 36: 180–193. [DOI] [PubMed] [Google Scholar]
- Ferec C , Cutting GR . Assessing the Disease-Liability of Mutations in CFTR. Cold Spring Harb Perspect Med 2012; 2: a009480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y , Wang L , Tian X , Xu KF , Xu W , Li X et al. Characterization of gene mutations and phenotypes of cystic fibrosis in Chinese patients. Respirology 2015; 20: 312–318. [DOI] [PubMed] [Google Scholar]
- Baihua C , Sizhong Z , Yuan Y . The first case of cf in mainland China identified by DNA analysis [J]. Chin J Med Cenet 1995; 12: 229–232. [Google Scholar]
- Wang M-C , Shu S-G , Chang S-M , Ho WL , Chi C-S . Cystic fibrosis in two Chinese infants in Taiwan. Zhonghua Minguo Xiao Er Ke Yi Xue Hui Za Zhi 1992; 34: 314–321. [PubMed] [Google Scholar]
- Zielenski J , Markiewicz D , Lin SP , Huang FY , Yang Feng TL , Tsui LC . Skipping of exon 12 as a consequence of a point mutation (1898+5G→T) in the cystic fibrosis transmembrane conductance regulator gene found in a consanguineous Chinese family. Clin Genet 1995; 47: 125–132. [DOI] [PubMed] [Google Scholar]
- Crawford J , Labrinidis A , Carey WF , Nelson PV , Harvey JS , Morris CP . A splicing mutation (1898+1G→T) in the CFTR gene causing cystic fibrosis. Hum Mutat 1995; 5: 101–102. [DOI] [PubMed] [Google Scholar]
- Wagner JA , Vassilakis A , Yee K , Li M , Hurlock G , Krouse ME et al. Two novel mutations in a cystic fibrosis patient of Chinese origin. Hum Genet 1999; 104: 511–515. [DOI] [PubMed] [Google Scholar]
- Wu C , Shu S , Zielenski J , Chiang C , Tsui L . Novel cystic fibrosis mutation (2215insG) in two adolescent Taiwanese siblings. J Formos Med Assoc 2000; 99: 564–567. [PubMed] [Google Scholar]
- Alper O , Shu S-G , Lee M-H , Wang B-T , Lo S-Y , Lin K-L et al. Detection of novel CFTR mutations in Taiwanese cystic fibrosis patients. J Formos Med Assoc 2003; 102: 287–291. [PubMed] [Google Scholar]
- Chen H-J , Lin S-P , Lee H-C , Chen C-P , Chiu N-C , Hung H-Y et al. Cystic fibrosis with homozygous R553X mutation in a Taiwanese child. J Hum Genet 2005; 50: 674–678. [DOI] [PubMed] [Google Scholar]
- Li N , Pei P , Bu D-F , He B , Wang G-F . A novel CFTR mutation found in a Chinese patient with cystic fibrosis. Chin Med J (Engl) 2006; 119: 103–109. [DOI] [PubMed] [Google Scholar]
- Liu J , Peng Y , Zhao Y , Wang W , Guo Y , He J et al. [Clinical manifestations and gene analysis of 2 Chinese children with cystic fibrosis]. Zhonghua Er Ke Za Zhi 2012; 50: 829–833. [PubMed] [Google Scholar]
- Gaildrat P , Killian A , Martins A , Tournier I , Frebourg T , Tosi M . Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol Biol 2010; 653: 249–257. [DOI] [PubMed] [Google Scholar]
- Li H , Ji CY , Zong XN , Zhang YQ . [Height and weight standardized growth charts for Chinese children and adolescents aged 0 to 18 years]. Zhonghua Er Ke Za Zhi 2009; 47: 487–492. [PubMed] [Google Scholar]
- HGMD® Professional 2015, 1; Available at http://www.hgmd.cf.ac.uk/.
- Macek M Jr , Mackova A , Hamosh A , Hilman BC , Selden RF , Lucotte G et al. Identification of common cystic fibrosis mutations in African-Americans with cystic fibrosis increases the detection rate to 75%. Am J Hum Genet 1997; 60: 1122. [PMC free article] [PubMed] [Google Scholar]
- Morral N , Bertranpetit J , Estivill X , Nunes V , Casals T , Gimenez J et al. The origin of the major cystic fibrosis mutation (ΔF508) in European populations. Nat Genet 1994; 7: 169–175. [DOI] [PubMed] [Google Scholar]
- Lu S , Yang X , Cui Y , Li X , Zhang H , Liu J et al. Different cystic fibrosis transmembrane conductance regulator mutations in Chinese men with congenital bilateral absence of vas deferens and other acquired obstructive azoospermia. Urology 2013; 82: 824–828. [DOI] [PubMed] [Google Scholar]
- Wine JJ , Kuo E , Hurlock G , Moss RB . Comprehensive mutation screening in a cystic fibrosis center. Pediatrics 2001; 107: 280–286. [DOI] [PubMed] [Google Scholar]
- Castellani C , Cuppens H , Macek M Jr , Cassiman JJ , Kerem E , Durie P et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J Cyst Fibros 2008; 7: 179–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CFTR 2 Database. Available at http://www.cftr2.org/browse.php.
- Zielenski J . Genotype and phenotype in cystic fibrosis. Respiration 2000; 67: 117–133. [DOI] [PubMed] [Google Scholar]
- Schram CA . Atypical cystic fibrosis Identification in the primary care setting. Can Fam Physician 2012; 58: 1341–1345. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.