Abstract
Prion diseases are associated with the conversion of the α-helix rich prion protein (PrPC) into a β-structure-rich insoluble conformer (PrPSc) that is thought to be infectious. The mechanism for the PrPC → PrPSc conversion and its relationship with the pathological effects of prion diseases are poorly understood, partly because of our limited knowledge of the structure of PrPSc. In particular, the way in which mutations in the PRNP gene yield variants that confer different susceptibilities to disease needs to be clarified. We report here the 2.5-Å-resolution crystal structures of three scrapie-susceptibility ovine PrP variants complexed with an antibody that binds to PrPC and to PrPSc; they identify two important features of the PrPC → PrPSc conversion. First, the epitope of the antibody mainly consists of the last two turns of ovine PrP second α-helix. We show that this is a structural invariant in the PrPC → PrPSc conversion; taken together with biochemical data, this leads to a model of the conformational change in which the two PrPC C-terminal α-helices are conserved in PrPSc, whereas secondary structure changes are located in the N-terminal α-helix. Second, comparison of the structures of scrapie-sensitivity variants defines local changes in distant parts of the protein that account for the observed differences of PrPC stability, resistant variants being destabilized compared with sensitive ones. Additive contributions of these sensitivity-modulating mutations to resistance suggest a possible causal relationship between scrapie resistance and lowered stability of the PrP protein.
Prion diseases are deadly neurodegenerative pathologies affecting numerous mammal species (1). They occur sporadically as well as after hereditary or infectious transmission; the high resistance of the infectious agent to classic inactivation techniques and the apparent absence of nucleic acid in its composition (1) are very intriguing. Of all the hypotheses on the nature of the infectious agent, the prion hypothesis, which states that the infection relies solely on a protein, is the most widely accepted. According to this proposition, the key event in the pathogenesis is the conversion of the α-helix-rich host prion protein (PrPC) into a pathogenic isoform (PrPSc) characterized by its insolubility, its high content in β-sheet, and its protease resistance (1). Consistent with that hypothesis, PrPC clearly plays a central role in transmissible spongiform encephalopathies (TSE) (2), and PrPSc formation is one of the main pathological features except in one reported case (3). However, despite the abundance of data now available on TSE, the prion hypothesis still is not fully validated: the infectious and pathological mechanisms of prion diseases are unclear, and the exact roles of PrPC and PrPSc in the brain dysfunctions caused by TSE have yet to be established. One of the main difficulties encountered in the confirmation of the prion hypothesis results from the heterogeneity of the best purified PrPSc samples, which makes the biochemical and structural characterizations of PrPSc problematic. A first step in such a characterization would be to identify and structurally define epitopes of antibodies that cross-react with PrPC and PrPSc. This would provide structural information directly derived from the infectious agent and help understand the mechanisms of PrPSc formation and spreading in infected organisms.
One of the important features of prion diseases is that mutations in the PRNP gene influence susceptibility. In sheep, a set of polymorphisms at positions 136, 154, and 171 of the PrP sequence (sheep numbering) are linked to scrapie susceptibility (4, 5). The homozygous genotype A136-R154-R171 (ARR) induces resistance, whereas V136-R154-Q171 (VRQ) confers high scrapie susceptibility. Between these two extremes the additional A136-R154-Q171 (ARQ) and A136-H154-Q171 (AHQ) variants are associated with medium and low susceptibility, respectively. Other PrP mutations and polymorphisms influence the susceptibility of humans (6) and mice (7) to transmissible spongiform encephalopathies or the incubation period duration, but no genotype is known to protect against the infectious agent in those species, as observed in sheep. The mechanisms by which such mutations influence the pathological process remain to be described. Because the PrPC → PrPSc conversion correlates well with the pathological evolution, it is expected that PrP mutations linked to prion disease have a direct effect on its thermodynamic stability or folding kinetics. Indeed, it was shown recently that ovine (Ov)PrP mutations V136A and Q171R, associated with a resistant phenotype in sheep, destabilize the recombinant prion protein (8). These results are in apparent contradiction with current models of the amyloid formation process (9) and need an explanation. It also remains to be established whether this correlation is coincidental or reflects a causal relationship between resistance and destabilization of PrPC.
Our objective was to gain deeper insight into the molecular mechanisms of PrPSc formation and to explain the influence of pathological mutations on this process. We have determined the x-ray structure of the C-terminal domain of scrapie-susceptibility OvPrP variants. We have cocrystallized this domain with a Fab fragment that cross-reacts with PrPC and PrPSc and report here the crystal structures of the C-terminal domain of sheep native recombinant prion protein variants ARQ, ARR, and VRQ complexed with this Fab. The structures provide side-chain positions, some of which have not been defined previously, and allow us to examine the structural correlates of scrapie-related sheep polymorphisms in the OvPrP structure. Most importantly, the PrP–Fab structure defines the epitope of the antibody. We provide evidence that this epitope is conserved in PrPC and PrPSc from brains of infected animals, which constitutes structural information on the pathological prion conformer directly derived from an infectious sample. On the basis of these results, we propose a model of the structure of the C-terminal domain of OvPrPSc.
Materials and Methods
Production and Purification of Recombinant Variants of OvPrP. Briefly, the cDNA encoding the VRQ, ARQ, or ARR variants of the prion C-terminal domain (residues 103–234) was cloned in pET-28a (Novagen) plasmid and expressed in the BL21 DE3 Escherichia coli strain after isopropyl β-d-thiogalactoside induction. The expressed truncated His-tagged prion proteins accumulated in inclusion bodies. After lysis, sonication, and solubilization of the inclusion bodies by 6 M urea, purification and renaturation of the prion protein were performed on a nickel Sepharose column by heterogeneous phase renaturation (10). The His tag was cleaved by using biotinylated thrombin (Novagen). After the removal of thrombin from the reaction mix by binding the enzyme to streptavidin agarose beads, the prion C-terminal domain was recovered in 10 mM Mops, pH 7.2/0.01% NaN3 from a HiPrep 26/10 desalting column (Pharmacia) by using an Akta fast protein liquid chromatograph (Pharmacia). Final protein concentration was measured by determining optical density at 280 nm using an extinction coefficient of 18,005 M–1·cm–1 deduced from the composition of the protein.
Fab Fragment Production. VRQ14 hybridoma resulted from immunization of Prnp0/0 mice with recombinant ovine variant VRQ and was selected for recognition of the immunization antigen on a BIAcore instrument (the full-length recombinant protein being linked to a carboxymethylated dextran chip through its N-terminal part). Ascitic fluids were produced in nude mice. After recovery of the ascitic fluid, the antibody was purified by protein A-Sepharose affinity chromatography. After dialysis in 0.1 M phosphate (pH 7.4) and concentration to 2 mg/ml, the purified antibody was subjected to papain-limited proteolysis by using a papain-to-antibody ratio of 1:100 (wt/wt). The reaction was stopped by using iodoacetamide at a final concentration of 10 mM. The Fc fragment was separated from the reaction mixture by protein A-Sepharose affinity chromatography, and the Fab fragment was further purified on a Sephacryl S100 HR (Pharmacia) gel-filtration column and stored in 150 mM sodium chloride/0.01% sodium azide at a concentration of 3 mg/ml.
Determination of the Primary Structure of the Fab. Total RNA was extracted from 5.107 hybridoma cells according to standard procedures. Both RT-PCR and 5′ rapid amplification of cDNA ends (RACE) amplification of the light- and heavy-chain variable domain sequences were performed with the Smart RACE cDNA amplification kit (Clontech). Briefly, RT-PCR was primed with the poly(T) and SMART-IIA (AAGCAGTGGTATCAACGCAGAGTCAC) oligonucleotides provided in the kit; in the following PCR step, selective amplification of the antibody cDNA sequences used the SMART-IIA primer in combination with the MKC primer (GTTCAGGACGCCATTTTGTCGTTCA) for the κ light chain and with the Cγ2 primer (GTGGATAGACCGATGGGGCTGTTGT) for the G2a heavy chain. PCRs were performed by incubation of cDNA with Taq polymerase in a Geneamp 2400 thermocycler (Perkin–Elmer) programmed for 25 cycles of the following temperature schedule: 94°C for 30 sec, 68°C for 30 sec, and 72°C for 3 min. After characterization on agarose gel and purification with the NucleoTrap gel-extraction kit (Clontech), PCR products were cloned into the pGEM-T vector (Promega) for transformation of DH10 E. coli and sequenced.
Purification and Crystallization of the PrP–Fab Complex. After a 15-min incubation of the Fab at a 70-μM concentration with a 2-fold excess of the OvPrP C-terminal domain, the OvPrP–Fab complex was eluted on a size-exclusion chromatography column (Sephacryl S100 HR). The purified complex was concentrated to 10 mg/ml before crystallization trials. Conditions to obtain crystals were first determined by using Crystal Screen I and II solution kits (Hampton Research, Riverside, CA). Monocrystals were finally obtained in 18–24% polyethylene glycol 8000/10000/0.1 M citrate/0.2 M ammonium acetate, pH 6.3. It was found by matrix-assisted laser desorption ionization/time-of-flight mass spectrometry and N-terminal sequencing that the crystals contain the 114–234 OvPrP fragment, most likely because of slow proteolysis. All three OvPrP–Fab complexes crystallize in the P21212 space group, with one complex per asymmetric unit.
Data Collection and Structure Determination. Diffraction data of the ARQ variant–Fab complex were collected on the BM30a beam line (European Synchrotron Radiation Facility, Grenoble, France), and those of the ARR and VRQ variant complexes were collected on the X06SA beam line (Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland). Integration and merging of the reflections was achieved with the denzo and scalepack programs (11). The structures of the ARQ–Fab and ARR–Fab complexes were determined by molecular replacement with amore software (12) by using as models Fab D2.3 (13) (PDB ID code 1YEH) constant and variable parts and dimeric human prion (huPrP) (residues 125–185) (PDB ID code 1I4M). The molecular replacement solution was then refined by torsion-angle molecular dynamics performed in simulated annealing cycles by using the cns-solve program (14). The stereochemistry was assessed with procheck (15). Buried residues are defined as those with side chains that have a solvent-accessible area <20 Å2 (16) (calculated by using a 1.4-Å probe). Data collection and refinement statistics are reported in Table 1. Figures were drawn by using molscript (17), raster3d (18), and bobscript (19).
Table 1. Data collection and refinement statistics.
| Complex | ARQ | ARR | VRQ |
|---|---|---|---|
| Data collection statistics | |||
| Cell dimensions, Å* | |||
| a | 91.77 | 90.96 | 92.05 |
| b | 145.59 | 144.23 | 145.68 |
| c | 43.02 | 42.93 | 43.20 |
| Resolution range, Å | 35-2.5 | 20-2.8 | 20-2.5 |
| I/σ(I) (outer shell)† | 55.1 (12) | 17.1 (4.2) | 32.1 (8.1) |
| Unique reflections | 18,975 | 15,056 | 19,765 |
| Redundancy | 6 | 4 | 7 |
| Completeness, (%) | 98.3 (94.6) | 99.5 (99.8) | 99.3 (91.8) |
| Overall (outer shell) | |||
| Rmerge (outer shell)‡ | 0.03 (0.10) | 0.087 (0.375) | 0.054 (0.164) |
| Refinement statistics | |||
| Rfactor (Rfree) | 0.22 (0.28) | 0.23 (0.29) | 0.22 (0.28) |
| Resolution range, Å | 15.0-2.5 | 15.0-2.8 | 15.0-2.5 |
| rmsd bond length, Å | 0.007 | 0.008 | 0.007 |
| rmsd bond angle, ° | 1.35 | 1.40 | 1.37 |
There is one Fab—OvPrP complex per asymmetric unit.
Outer shell: 2.61-2.55 Å (ARQ and VRQ) and 2.87-2.8 Å (ARR).
Rmerge = Σhkl Σi | Ii - 〈I〉 | /ΣhklΣi Ii.
ELISA. Prnp0/0 mouse brain homogenates and scrapie-infected and uninfected sheep brain homogenates were titrated by using the peroxidase-conjugated VRQ14 antibody as a detecting antibody in a sandwich ELISA test. We used 10% brain homogenates as such or subjected them to proteinase K digestion. The protease was used at a concentration of 50 μg/ml during 30 min at 37°C, and the reaction was stopped by boiling the samples at 100°C for 5 min. Serial 2-fold dilutions of samples then were mixed with an equal volume of the peroxidase-conjugated VRQ14 antibody (0.7 nM final concentration) and deposited in wells for 1 h at room temperature before washing and the addition of the Luminol (Pierce) substrate for 10 min, after which the plates were read. Competition ELISA of the Fab and the antibody was performed according to a similar protocol by using the same samples. Proteinase K-digested or undigested 1% brain homogenates were incubated with serial 2-fold dilutions of the Fab (0.2 μM starting concentration) for 15 min at room temperature, after which the mixtures were deposited in wells with an equal volume of peroxidase-conjugated VRQ14 antibody (0.7 nM final concentration) for 1 h at room temperature. Revelation and reading were as described above.
Mouse, hamster, and macaque brain homogenates, normal or infected, were a kind gift from Corinne Lasmézas (Commissariat 'l a'Energie Atomique, Fontenay Aux Roses, France). Mice were infected with C506M3 scrapie strain or 6PB1 bovine spongiform encephalopathy isolate, hamster with scrapie 263K, and macaque with bovine spongiform encephalopathy agent. ELISA was performed as described above.
Results
Structure of the OvPrP C-Terminal Domain. The overall fold of the crystallized part of the C-terminal domain (residues 114–234) of the three OvPrP variants analyzed consists of a short two-stranded β-sheet (residues 129–134 and 163–167) and three α-helices (residues 146–158, 174–196, and 203–228), linked by loops with no regular secondary structure (Fig. 1A); residues 114–126 and 229–234 are disordered and not seen in the electron density. This fold is identical to those revealed by previously determined NMR structures (20–23), as illustrated by the superpositions of bovine and huPrP Cαs to those of OvPrP, which yield rms deviation (rmsd) values of 1.1 and 1.2 Å, respectively. These values are within the rmsd between average structure and energetically equivalent conformers consistent with NMR data and are significantly larger than the uncertainty on Cα positions in this structure [0.4 Å, as determined from a Luzzati plot (24)], which demonstrates in particular that the antibody does not induce major changes in PrP other than restricting the flexibility of its epitope (see below).
Fig. 1.
Structure of the OvPrP C-terminal domain. (A) The OvPrP fold and the scrapie-sensitivity-associated mutations. Side chains of scrapie-sensitivity-related residues are represented as green ball-and-stick models in the OvPrP structure. (B Left) Structure of the huPrP crystallographic dimer (27) (one monomer is shown in salmon red, and the other is shown in blue). (B Right) Superimposition of the x-ray structures of OvPrP (this work, red) and of a huPrP domain (blue) constituted by the H3 helix of one monomer and of the rest of the sequence of the other monomer.
Structural Comparison of ARR, ARQ, and VRQ OvPrP Variants. The crystal structure of the OvPrP ARQ variant allows us to localize the side chains of the amino acids associated with these polymorphisms; together with the structures of the VRQ and ARR variants, it provides an opportunity to analyze the consequences of the A136V and Q171R sensitivity-modulating mutations in atomic detail. The ovine residues involved in scrapie-sensitivity polymorphism are on the protein surface, and their side chains are water-exposed. A comparison of the structures of the ARQ and VRQ variants reveals that the A → V substitution at position 136 causes the N162 side chain to rotate, perhaps as a result of steric hindrance between a γ methyl of the valine residue and the amide group of N162. In the VRQ variant, the N162 side-chain carbonyl oxygen establishes a hydrogen bond with residue R139 (Fig. 2 Upper), which stabilizes the VRQ variant as compared with ARQ (8). The Q → R substitution at position 171 was analyzed by comparing the structures of variants ARR and ARQ in 2Fo – Fc maps after refinement of the two structures, as Fourier difference analysis was precluded because of non-isomorphism between the ARR and ARQ PrP crystals. There is a hydrogen bond between residues R167 and Q171 in the ARQ variant (Fig. 2 Lower). Substitution of Q by R at position 171 displaces the R167 side chain because of the electrostatic repulsion between the two side-chain guanidinium groups; these two groups are distant by 5.6 Å, disordered, and hardly visible in the ARR variant electron-density maps. As a consequence, the ARR variant is destabilized compared with ARQ (8).
Fig. 2.
Structural consequences of scrapie-sensitivity-related mutations. The blue grid corresponds to the 1.0-σ level contour of the 2Fo – Fc omit map. Maps are calculated by using data in the 15.0- to 2.5-Å-resolution range for the ARQ and VRQ variants and the 15.0- to 2.8-Å-resolution range for the ARR variant. Hydrogen bonds are displayed as dashed lines, and distances of the atoms involved are reported. (Upper) A136V. The FoVRQ – FoARQ difference map, represented as red and green grids (contour levels: –5.0 σ and 5.0 σ, respectively) is superimposed on the ARQ 2Fo – Fc omit map. (Lower) Q171R.
The Epitope of the Antibody Is Conserved in PrPSc and PrPC. The structure of the complex shows that the epitope of the antibody consists mostly of PrP residues 188–199 (C-terminal part of helix H2 and N-terminal part of the H2–H3 loop; Fig. 3A). Only one face of helix H2 is buried in the Fab-combining site. Consistent with that, pepscan analysis of OvPrP antigenicity with overlapping 9-aa peptides do not identify the binding site of the antibody (data not shown). Most importantly, the antibody recognizes recombinant OvPrP and also the cellular and the pathological forms of PrP but not samples from Prnp0/0 mice brains, as shown by ELISA (Fig. 3B). This analysis was performed with both the antibody and the Fab to rule out avidity effects with aggregated PrPSc and was further supported by immunoprecipitation of both PrPC and PrPSc by the antibody (data not shown). An immunoblot confirms the presence of PrPC and PrPSc in the samples used for ELISA (Fig. 5, which is published as supporting information on the PNAS web site). The combined structural and immunochemical data are strong evidence that the C-terminal end of helix H2 and the N-terminal part of the H2–H3 loop are conserved in PrPSc. ELISA titration with normal or infected brain homogenates from mouse, hamster, and macaque indicate that the VRQ14 epitope is also found in these species and is conserved in both normal and pathological isoforms of their prion proteins (Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 3.
The epitope of the antibody is conserved in OvPrPC and OvPrPSc. (A) OvPrP–Fab structure (ARQ variant). Fab heavy and light chains are shown in green and blue, respectively. (Left) Overview of the complex. The part of the prion protein that, according to our model, is structurally conserved in the PrPC → PrPSc conversion is shown in red; the part that undergoes a secondary structure change is shown in orange. (Right) Close-up view of the boxed region of the complex. For clarity, only atoms that establish an intermolecular hydrogen bond are represented in ball-and-stick format. Hydrogen bonds are represented as dashed lines, and distances of the atoms involved (Å) are reported. (B) ELISA characterization of the binding properties of the antibody. PrPC and PrPC + PrPSc samples are recovered from brains of uninfected and infected sheep, respectively. PrPSc is produced by proteinase K (PK) treatment of PrPC + PrPSc. The proteinase K treatment of PrPC samples is used as a control for total digestion of PrPC. Prnp0/0 designates an extract from the brain of a Prnp0/0 mouse. (Upper) ELISA titration of OvPrP samples. (Lower) Competition ELISA of the Fab and the antibody. The lower signal for PrPSc in both the titration and the competition tests reflects the lower abundance of PrPSc compared with PrPC in infected sheep brains (41–43). A.U., arbitrary units.
Discussion
OvPrP Crystallization and Structural Analysis. Crystallization trials with native recombinant PrP have been unsuccessful to date (1). This is probably because of the well documented high flexibility of the PrP protein N-terminal domain (25). Therefore, we have undertaken to determine the structure of the C-terminal part of OvPrP. We took advantage of the availability of several monoclonal antibodies that bind to this domain to attempt its crystallization as well as that of its complexes with the corresponding Fab fragments. Fab complexation offers possibilities for protein–protein contacts in a crystal that differ from those arising from the antigen alone, in addition to the well documented potential of Fab fragments to bury antigenic surfaces that render a protein aggregation-prone (26). Although we were not able to crystallize the PrP C-terminal domain alone (residues 114–234), we did obtain crystals of its complex with one Fab. The Fab epitope contains the N-terminal half of the loop joining OvPrP helix H2 to helix H3, which has been shown by NMR measurements of 15N relaxation times to be flexible (21) in uncomplexed PrP proteins. This loop does not establish any crystal contact and is stabilized in the complex (average B factor in the loop, 30.0 Å2; average B factor in OvPrP, 34.0 Å2). Exposure of this loop might have inhibited the growth of properly diffracting crystals of OvPrP and of other prion protein constructs corresponding to that used.
The determination of the OvPrP crystal structure provides an opportunity to analyze sequence variation among mammalian prion proteins in structural terms. Because the majority (19 of 25) of OvPrP buried residues are conserved in mammals, we conclude that the PrP C-terminal domain adopts the same fold in these species. This conclusion is consistent with the observation that all but one of the recombinant PrP folds determined thus far are nearly identical. Indeed, when comparing the OvPrP fold with those of previously studied PrP proteins, the only significant difference is with the huPrP crystal structure (27). In the dimer, the huPrP monomer structure differs from other PrP structures by the location of helix H3, which exchanges between the individual monomers in the dimer, in a process that involves a rearrangement of the single PrP intramolecular disulfide bridge into an intermolecular one (Fig. 1B). The only secondary structure difference between the huPrP monomer in the dimer and OvPrP is located in the hinge region of the swapped domain, constituted of the end of helix H2 and of the H2–H3 loop, which is most of the epitope recognized by the antibody. This hinge forms a small antiparallel β-sheet in the covalent dimer in which four main-chain hydrogen bonds compensate for the three main-chain H2 helix hydrogen bonds disrupted in the dimer caused by the H3 exchange. The energy cost of the conversion of native PrP into dimeric PrP comprises an entropy loss resulting from the dimerization of PrP, but besides that, it ought to be low because there is a balance between stabilizing interactions lost and gained in the dimer as compared with the monomer. By contrast, the kinetic barrier to dimerization is likely to be extremely large as a consequence of both the low probability of disulfide bond exchange and the high energy cost of the native core transient disruption required for H3 swapping. This explains why the proportion of PrP covalent dimer is very low in usual conditions and why it may be increased in conditions under which disulfide exchange is favored, such as at higher pH, where huPrP was crystallized (27).
Structural Correlates of Sheep Polymorphisms Associated to Scrapie Resistance. Focusing on the unique link between sheep polymorphisms at positions 136, 154, and 171 (sheep sequence numbering) and scrapie susceptibility (4, 5), it was shown recently that the resistant variants ARR and AHQ are destabilized compared with the susceptible variants VRQ and ARQ. The absolute difference of the unfolding free enthalpies of the extreme resistant (ARR) and susceptible (VRQ) variants is ≈10 kJ/mol, which is approximately one third of the unfolding free enthalpy for full-length prion protein (8). Structural comparisons of OvPrP variants ARR, ARQ, and VRQ show that mutations V136A and Q171R lead to disruption of hydrogen bonds located at the surface of the ovine recombinant prion protein. The measured unfolding free-enthalpy variations caused by the A136V and Q171R mutations (8), –5.9 kJ/mol and 4.9 kJ/mol, respectively, are within the range of those observed when a hydrogen bond is gained or lost in a protein, provided there is no unpaired buried ion formed (28).
What is the rationale for the variations in scrapie sensitivity induced by mutations at positions 136, 154, and 171? A correlation of scrapie susceptibility with mutation-induced changes of a single PrP function is unlikely, because the structural consequences of these mutations are local and the corresponding positions (Fig. 1 A) are distant from each other in the PrP structure by >17 Å. By contrast, the following observations are coherent with the hypothesis that variations in susceptibility are caused by changes of PrP stability: each of the scrapie-sensitivity mutations affects the protein stability, and the effect of a combination of these mutations is additive. How would stability changes alter PrP behavior in vivo? One mechanism involves the increased protease resistance that accompanies stabilization of scrapie-susceptibility variants of OvPrP (10). With a longer lifetime in the cell, the prion protein would become more susceptible to aggregation or to other pathological conformational changes, leading to an increased sensitivity to prion disease. In addition, the kinetic constant of amyloidogenesis of ARQ PrP is three times larger than that of ARR PrP at pH 7.0 and eight times larger at pH 4.0 (8). In the particular case of the ARR variant, a combination of increased protease sensitivity and slower amyloidogenesis would account for a switch from scrapie susceptibility to resistance associated with the single Q171R substitution. Nevertheless, other mechanisms for mutation-induced scrapie resistance cannot be ruled out (see below). Similar to observations in sheep, resistance has been observed in transgenic mice in which the Q → R mutation had been introduced in mouse PrP at the position corresponding to 171 in sheep (29) and in rabbit cells transfected with OvPrP scrapie-resistant variants (30), suggesting that the mechanism for resistance involved, whatever its exact nature, may occur in different mammalian contexts.
Implications for the Mechanism of PrPSc Formation. Taken together with biochemical and biophysical data, the observation that the epitope of the antibody is conserved in PrPC and PrPSc allows us to propose an identification of the part of OvPrP with an unchanged secondary structure in the PrPC → PrPSc conversion. Two lines of evidence suggest that, in addition to the two C-terminal turns of helix H2 and to the N-terminal part of loop H2–H3, a major part of helix H3 and the N-terminal part of helix H2 are also conserved in PrPC and PrPSc. First, the disulfide bond that connects the N terminus of H2 and the C-terminal half of helix H3 is intact in PrPSc and does not seem to be disrupted during the transition (31). Second, the isolated H2–H3 bundle does not have any intrinsic β-sheet-forming propensity, because truncated versions of mouse PrP corresponding to this region display high helical content, as shown by circular dichroism measurements (32). Binding of the antibody to OvPrPSc also implies that its epitope is accessible in PrPSc, a property shared with the epitope of a recombinant antibody that binds to the C-terminal part of helix H3 (33). This lends support to the hypothesis that the α-helices of PrPSc are exposed (34).
If the H2–H3 helical bundle is mostly conserved in PrPC and PrPSc, to account for the loss of α-helix content of PrPSc compared with PrPC (35), helix H1 will transform into a β-structure. This would be consistent with in vitro demonstration of the lability of the S1–H1–S2 region of the OvPrP protein (36). Lability of the N-terminal half of helix H1 is also suggested by the observation that residues in this part of the protein have the highest temperature factors in the OvPrP structure and a fast hydrogen-exchange rate compared with the other secondary structure elements of recombinant PrP (36). The fact that a synthetic peptide spanning helix H1 and β-strand S2 adopts a stable β-hairpin structure in solution (37) provides additional support to the suggestion that this region of the protein is labile. Finally, observations that the S1–H1–S2 region is targeted by antibodies that selectively recognize PrPSc (38) or affect prion propagation (39, 40) also support the notion that this region adopts different structures in PrPC and PrPSc.
On the basis of these data, including the antibody-binding studies summarized in Fig. 4, we propose a model of the PrPC → PrPSc transformation in which, in PrPSc, a β-sheet originating from the S1–S2 strands of PrPC replaces the S1–H1–S2 part of the PrPC structured domain, with H2 and H3 remaining largely unchanged. Because the PrPC structure is conserved in mammals, our results are likely to extend to other mammalian species, a conclusion supported by cross-reactivity of the VRQ14 antibody with PrPC and PrPSc in all of the species we have been able to test (Fig. 6).
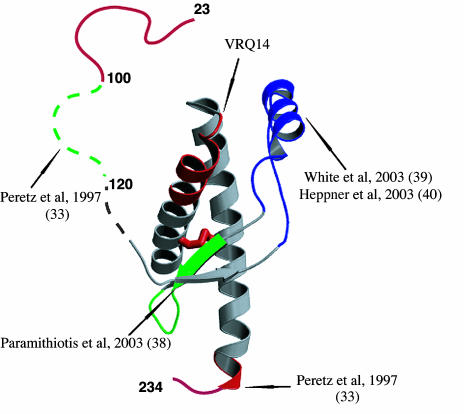
Fig. 4.
Antigenic mapping of the structural changes in the PrPC → PrPSc conversion. The epitopes are mapped on the OvPrP structure to which three residues have been added at the C terminus; their conformation is that determined by NMR. The epitopes of PrPSc-specific antibodies are shown in green, and those present in both PrPC and PrPSc are shown in red. The conserved disulfide is shown in red also. Antibody binding to the region in blue blocks PrPSc formation in vivo. Each displayed epitope is linked to its bibliographic input.
It is remarkable that the only group of mutations known to confer total resistance to scrapie solely affects the part of the protein that we propose to undergo secondary structure changes during PrPC → PrPSc conversion. In the two scrapie-sensitive variants (ARQ and VRQ) we studied, there are additional hydrogen bonds involving residues in the S1–H1–S2 region in OvPrPC as compared with the scrapie-resistant variant ARR; because our model for PrPSc formation proposes that S1–S2 serves as a template for β-sheet elongation, it is possible that these mutations confer scrapie susceptibility, because they support the extension of the S1–S2 β-sheet. This would be consistent with observations that the rate of amyloid formation is higher in susceptible than in resistant variants (8). Confirmation of this hypothesis must await the determination of the structure of the S1–H1–S2 region in OvPrPSc.
In conclusion, the combined immunochemical and structural characterizations of the interaction of OvPrPC and infectious OvPrPSc with an antibody have provided structural information on the PrPC → PrPSc conversion; the availability of additional antibodies that bind to PrPC and PrPSc will allow further structural characterization of this transformation.
Note. While this work was being reviewed, an x-ray structure of a similar construct of OvPrP appeared (44).
Supplementary Material
Acknowledgments
We dedicate this paper to the memory of Dr. D. Dormont; without his support we would not have been able to carry out the work described. We thank Dr. J.-L. Ferrer for helping us with the use of the BM30 beamline; Dr. P. Boudinot for assistance with Fab sequencing; Dr. Corinne Lasmézas for supplying prion-infected brains; Dr. M. Moudjou for help with immunoprecipitations; and Mr. P. Pourquier for letting us use his equipment. Diffraction data were collected at the European Synchrotron Radiation Facility (Grenoble, France) and the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland). We are grateful to the machine and beam line groups whose outstanding efforts have made these experiments possible. We thank Dr. Siobhán Staunton for help with the manuscript. Support from the Groupement d'Intérêt Scientifiques “Infections à prions,” the Centre National de la Recherche Scientifique, and the Institut National de la Recherche Agronomique is gratefully acknowledged. F.E. is a recipient of a fellowship from the French Ministry for Research.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ARR, A136-R154-R171; VRQ, V136-R154-Q171; ARQ, A136-R154-Q171; AHQ, A136-H154-Q171; Ov, ovine; huPrP, human prion; rmsd, rms deviation.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1TPX, 1TQB, and 1TQC).
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bueler, H., Aguzzi, A., Sailer, A., Greiner, R. A., Autenried, P., Aguet, M. & Weissmann, C. (1993) Cell 73, 1339–1347. [DOI] [PubMed] [Google Scholar]
- 3.Lasmezas, C. I., Deslys, J. P., Robain, O., Jaegly, A., Beringue, V., Peyrin, J. M., Fournier, J. G., Hauw, J. J., Rossier, J. & Dormont, D. (1997) Science 275, 402–405. [DOI] [PubMed] [Google Scholar]
- 4.Goldmann, W., Hunter, N., Foster, J. D., Salbaum, J. M., Beyreuther, K. & Hope, J. (1990) Proc. Natl. Acad. Sci. USA 87, 2476–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldmann, W., Martin, T., Foster, J., Hughes, S., Smith, G., Hughes, K., Dawson, M. & Hunter, N. (1996) J. Gen. Virol. 77, 2885–2891. [DOI] [PubMed] [Google Scholar]
- 6.Liemann, S. & Glockshuber, R. (1999) Biochemistry 38, 3258–3267. [DOI] [PubMed] [Google Scholar]
- 7.Moore, R. C., Hope, J., McBride, P. A., McConnell, I., Selfridge, J., Melton, D. W. & Manson, J. C. (1998) Nat. Genet. 18, 118–125. [DOI] [PubMed] [Google Scholar]
- 8.Rezaei, H., Choiset, Y., Eghiaian, F., Treguer, E., Mentre, P., Debey, P., Grosclaude, J. & Haertle, T. (2002) J. Mol. Biol. 322, 799–814. [DOI] [PubMed] [Google Scholar]
- 9.Kelly, J. W. (1998) Curr. Opin. Struct. Biol. 8, 101–106. [DOI] [PubMed] [Google Scholar]
- 10.Rezaei, H., Marc, D., Choiset, Y., Takahashi, M., Hui Bon Hoa, G., Haertle, T., Grosclaude, J. & Debey, P. (2000) Eur. J. Biochem. 267, 2833–2839. [DOI] [PubMed] [Google Scholar]
- 11.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–325. [DOI] [PubMed] [Google Scholar]
- 12.Navaza, J. (1994) Acta Crystallogr. A 50, 157–163. [Google Scholar]
- 13.Gigant, B., Charbonnier, J. B., Eshhar, Z., Green, B. S. & Knossow, M. (1997) Proc. Natl. Acad. Sci. USA 94, 7857–7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 15.Laskowski, R. A., McArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 16.Shrake, A. & Rupley, J. A. (1973) J. Mol. Biol. 79, 351–371. [DOI] [PubMed] [Google Scholar]
- 17.Kraulis, P. (1991) J. Appl. Crystallogr. 24, 924–950. [Google Scholar]
- 18.Merritt, E. A. & Murphy, M. E. P. (1994) Acta Crystallogr. D 50, 869–873. [DOI] [PubMed] [Google Scholar]
- 19.Esnouf, R. M. (1997) J. Mol. Graphics 15, 133–138. [DOI] [PubMed] [Google Scholar]
- 20.Riek, R., Hornemann, S., Wider, G., Billeter, M., Glockshuber, R. & Wüthrich, K. (1996) Nature 382, 180–182. [DOI] [PubMed] [Google Scholar]
- 21.López Garcia, F., Zahn, R., Riek, R. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8334–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, H., Farr-Jones, S., Ulyanov, N. B., Llinas, M., Marqusee, S., Groth, D., Cohen, F. E., Prusiner, S. B. & James, T. L. (1999) Biochemistry 38, 5362–5377. [DOI] [PubMed] [Google Scholar]
- 23.Calzolai, L., Lysek, D. A., Güntert, P., von Schroetter, C., Riek, R., Zahn, R. & Wüthrich, K. (2000) Proc. Natl. Acad. Sci. USA 97, 8340–8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luzzati, V. (1952) Acta Crystallogr. 5, 802–810. [Google Scholar]
- 25.Donne, D. G., Viles, J. H., Groth, D., Mehlhorn, I., James, T. L., Cohen, F. E., Prusiner, S. B., Wright, P. E. & Dyson, H. J. (1997) Proc. Natl. Acad. Sci. USA 94, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovari, L. C., Momany, C. & Rossmann, M. G. (1995) Structure (London) 3, 1291–1293. [DOI] [PubMed] [Google Scholar]
- 27.Knaus, K., Morillas, M., Swietnicki, W., Malone, M., Surewicz, W. K. & Yee, V. C. (2001) Nat. Struct. Biol. 8, 770–774. [DOI] [PubMed] [Google Scholar]
- 28.Fersht, A. R. (1987) Trends Biochem. Sci. 12, 301–304. [Google Scholar]
- 29.Perrier, V., Kaneko, K., Safar, J., Vergara, J., Tremblay, P., DeArmond, S. J., Cohen, F. E., Prusiner, S. B. & Wallace, A. C. (2002) Proc. Natl. Acad. Sci. USA 99, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sabuncu, E., Petit, S., Le Dur, A., Lan Lai, T., Vilotte, J. L., Laude, H. & Vilette, D. (2003) J. Virol. 77, 2696–2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welker, E., Raymond, L. D., Scheraga, H. A. & Caughey, B. (2002) J. Biol. Chem. 277, 33477–33481. [DOI] [PubMed] [Google Scholar]
- 32.Eberl, H. & Glockshuber, R. (2002) Biophys. Chem. 96, 293–303. [DOI] [PubMed] [Google Scholar]
- 33.Peretz, D., Williamson, R. A., Matsunaga, Y., Serban, H., Pinilla, C., Bastidas, R. B., Rozenshteyn, R., James, T. L., Houghten, R. A., Cohen, F. E., et al. (1997) J. Mol. Biol. 273, 614–622. [DOI] [PubMed] [Google Scholar]
- 34.Wille, H., Michelitsch, M. D., Guenebaut, V., Supattapone, S., Serban, A., Cohen, F. E., Agard, D. A. & Prusiner, S. B. (2002) Proc. Natl. Acad. Sci. USA 99, 3563–3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan, K. M., Baldwin, M., Nguyen, J., Gasset, M., Serban, A., Groth, D., Mehlhorn, I., Huang, Z., Fletterick, R. J., Cohen, F. E., et al. (1993) Proc. Natl. Acad. Sci. USA 90, 10962–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calzolai, L. & Zahn, R. (2003) J. Biol. Chem. 278, 35592–35596. [DOI] [PubMed] [Google Scholar]
- 37.Kozin, S. A., Bertho, G., Mazur, A. K., Rabesona, H., Girault, J. P., Haertle, T., Takahashi, M., Debey, P. & Hoa, G. H. (2001) J. Biol. Chem. 276, 46364–46370. [DOI] [PubMed] [Google Scholar]
- 38.Paramithiotis, E., Pinard, M., Lawton, T., LaBoissiere, S., Leathers, V. L., Zou, W. Q., Estey, L. A., Lamontagne, J., Lehto, M. T., Kondejewski, L. H., et al. (2003) Nat. Med. 9, 893–899. [DOI] [PubMed] [Google Scholar]
- 39.White, A. R., Enever, P., Tayebi, M., Mushens, R., Linehan, J., Brandner, S., Anstee, D., Collinge, J. & Hawke, S. (2003) Nature 422, 80–83. [DOI] [PubMed] [Google Scholar]
- 40.Heppner, F. L., Musahl, C., Arrighi, I., Klein, M. A., Rulicke, T., Oesch, B., Zinkernagel, R. M., Kalinke, U. & Aguzzi, A. (2001) Science 294, 178–182. [DOI] [PubMed] [Google Scholar]
- 41.Madec, J. Y., Groschup, M. H., Buschmann, A., Belli, P., Calavas, D. & Baron, T. (1998) J. Virol. Methods 75, 169–177. [DOI] [PubMed] [Google Scholar]
- 42.Beekes, M., Baldauf, E., Cassens, S., Diringer, H., Keyes, P., Scott, A. C., Wells, G. A., Brown, P., Gibbs, C. J., Jr., & Gajdusek, D. C. (1995) J. Gen. Virol. 76, 2567–2576. [DOI] [PubMed] [Google Scholar]
- 43.Moudjou, M., Frobert, Y., Grassi, J. & La Bonnardiere, C. (2001) J. Gen. Virol. 82, 2017–2024. [DOI] [PubMed] [Google Scholar]
- 44.Haire, L. F., Whyte, S. M., Vasisht, N., Gill, A. C., Verma, C., Dodson, E. J., Dodson, G. G. & Bayley, P. M. (2004) J. Mol. Biol. 336, 1175–1183. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.