Abstract
The 70-kDa heat shock cognate (Hsc70) chaperone plays a crucial role in protein (re-)folding and triage in the mammalian cytosol. Here we study, by NMR, the 44-kDa nucleotide-binding domain (NBD) of this molecule, which allosterically regulates, by binding either ADP or ATP in a cleft between the two main lobes, the chaperoning affinity of the attached substrate-binding domain. The NBD is also a center of interaction with cochaperones that couple into the allostery. By measuring residual dipolar couplings by NMR, we show that the orientation of two lobes of the Hsc70 NBD in solution deviates up to 10° from their positions in 14 superimposing x-ray structures. Additional orientational differences of subdomains within the lobes unveil the Hsc70 NBD in solution as a flexible molecular machine that can adjust the relative positions of all of its four subdomains. Because the residues interacting with the nucleotide emanate from all four subdomains, adjustments in subdomain orientation should affect the nucleotide chemistry and vice versa. Our data suggest a hypothesis that cochaperone or substrate domain binding perturbs the relative subdomain orientations, thereby functionally and allosterically coupling to the nucleotide state of the NBD.
The 70-kDa heat shock protein (Hsp70) chaperones are conserved from bacteria to mammals and are the main facilitators of protein folding, refolding, and trafficking (1). In this process, they are assisted by several cochaperones. Mammalian Hsp70's have additional roles in activation of transcription factors, protein kinases, antigen presentation, tumor immunogenicity, and ribosome assembly (2). In humans, the constitutively expressed cytosolic form is Hsc70 (heat shock cognate). Hsc70 is functionally coupled to the proteolytic and apoptotic cascades placing it at the center of protein triage (3).
Hsc70 is a 70-kDa monomer, for which no complete structure has been obtained. It consists of three domains, which were solved individually. There is an N-terminal 44-kDa nucleotide-binding domain (NBD), which is solved by crystallography, and is the focus of this report (4). A 15-kDa substrate-binding domain (SBD), connected to the NBD by a 6–10 hydrophobic residue, harbors the substrate-binding cleft that binds exposed hydrophobic sequences in misfolded substrate proteins, and was solved in different forms and species by crystallography and NMR (5, 6). A subsequent 10-kDa domain of α-helical lid structure tunes the kinetics of substrate binding and release (7). ATP binding at the NBD promotes release of the protein substrate from the SBD; protein substrate binding at the SBD promotes the hydrolysis of ATP in the NBD. Hence, Hsc70 is a heterotropic allosteric system. The allosteric process is enhanced by the cochaperones HdJ (enhances protein substrate binding and ATP hydrolysis) (8) and the nucleotide factor BAG-1 (enhances ADP to ATP exchange; ref. 9). Many other factors bind to Hsc70 as well.
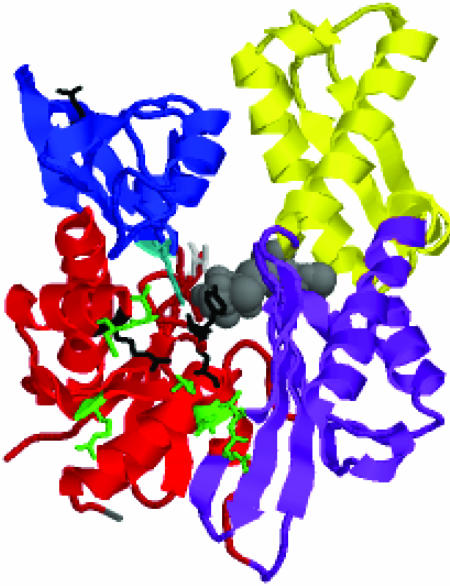
In the NBD, one recognizes two main lobes (see the legend to Fig. 1 for residue counts). Within the lobes, one further distinguishes the subdomains IA, IB, and IIA and IIB. Subdomains IA and IB, deriving from two different lobes, are in close contact and from a contiguous base. Domain IA harbors both the N terminus as well as the C terminus; the latter forms the link to the SBD, which is therefore expected to reside in the proximity of subdomain IA. Subdomains IB and IIB do not touch each other and line a wide cleft. A single nucleotide, either ADP or ATP, in conjunction with Mg2+ and two potassium ions, binds at the bottom of this cleft and makes contacts with residues from all four subdomains.
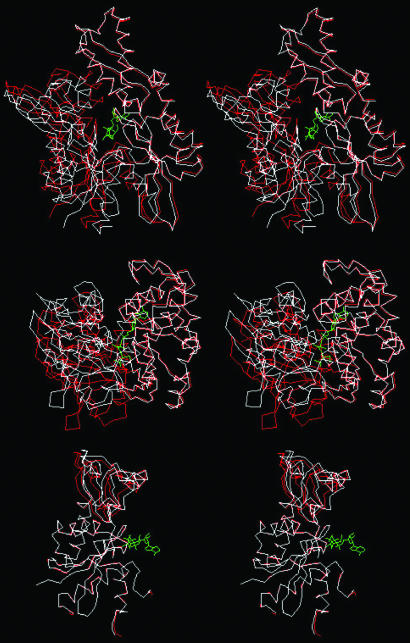
Fig. 1.
The crystal structure of the Hsc70 NBD (3–384) of bovine Hsc70 (4) (PDB ID code 3HSC), with bound ADP.PO4 (in gray). Indicated are the subdomains IA in red (residues 3–39,116–188, and 361–384); IB in blue (residues 40–115); IIA in purple (residues 119–228 and 307–360); and IIB in yellow (residues 229–306). IA and IB together form lobe I; IIA and IIB together form lobe II. Indicated in gray at the lower left corner is the C terminus (residue 384). The side chains of residues implied as being involved in DnaJ binding in E. coli DnaK are green at the homologous positions. They are: R171/167, N170/174, and T173/177 (Hsc70/DnaK count; ref. 31); and Y149/145, N151/147, D152/148, E218/217, and V219/218 (Hsc70/DnaK count; ref. 32). The side chain of T13, important for the ATP/ADP conformational change transduction, emanating from subdomain IA is in light gray. The side chain of K71, essential for ATP hydrolysis, emanating from domain IB is turquoise. Residues R155, Y149, and E175, identified by mutagenesis, as important for the allosteric coupling between the NBD and SBD (33), are black. Note that Y149 was also identified as a DnaJ site. Residue Q104 (the fluorescent probe W102 in DnaK) is marked in black at the top left corner of IB.
Although there is a large body of biochemical and biophysical data (10) on the allosteric coupling between substrate binding, and nucleotide binding, and of the stimulation of ATP hydrolysis by the cochaperone HdJ, the structural basis of these coupling processes remains mysterious. All 11 crystal structures of bovine Hsc70 NBD superimpose within an overall pairwise rms deviation (rmsd) of <0.5 Å, no matter which ligands it binds, including ADP and ATP. These studies thus depict the NBD as a rigid entity, not at all what is expected for an allosteric molecule. The first chink in this armor has come from a more recent x-ray structure of the NBD of the closely related Hsp70 (11), showing flexibility of the IIA–IIB junction. The other three subdomains, however, are identical with the Hsc70 NBD. Shifts of subdomain IIB have been associated with the nucleotide-release mechanism: an x-ray structure of Hsc70 NBD in the presence of the nucleotide-release factor BAG-1M (12), as well as a structure of DnaK NBD with GrpE (13), shows, exclusively, a movement of this domain with respect to the other three domains, which, again, superimpose. Consequently, domains IA, IB, and IIA superimpose within experimental error in fourteen different crystal structures.
Here, we present, to our knowledge, the first detailed solution NMR study of the conformation of the 44-kDa Hsc70 NBD (residues 1–386) of Bos taurus in the ADP.Pi state. By using transverse relaxation-optimized spectroscopy (TROSY)-triple resonance NMR spectroscopy at 800 MHz, the backbone resonances were assigned, allowing us to measure residual dipolar couplings in an alignment medium. The residual dipolar couplings assess the relative orientations of the subunits in Hsc70 with respect to the alignment medium, and hence with respect to each other (14). Basing our data analysis on the local coordinates of the subunits from the x-ray coordinates, we report a 6.7 ± 1.2° orientational difference in the Szz directions of subdomains IA and IIA in solution as compared with the Hsc70 crystal form. Overall single-axis rotation, accounting for all three components Szz, Syy, and Sxx, is 10° (A. M. Al-Hashimi, personal communication). The relative movement is rather a shearing than an opening or closing event. Equally interesting is an 8.8 ± 1.2° reorientation of Szz of subdomain IB with respect to subdomain IA, also not seen before in any crystal structure (single-axis rotation gives the same value). Surprisingly, we do not see much reorientation between domains IIA and IIB (3 ± 1.2°). Together with the available crystal structures, our data thus reveals that all four subdomains can perform substantial reorientational conformational changes, depending on experimental conditions. Hence, instead of an NBD that leaves no clue as how it could possibly drive allosteric transitions and communicate with cochaperones, the current studies characterize this molecule as an ensemble of states of different subunit orientations with ample possibilities to modulate its interactions with other domains and cofactors.
Materials and Methods
Protein Expression and Purification. The plasmid for wild-type Hsc70 NBD 1–386 was donated by D. B. McKay (Stanford University School of Medicine, Stanford, CA) and was expressed in Escherichia coli strain BL21(DE3) (15). The expression was induced by isopropylthio-β-d-galactoside to 1 mM at an OD600 of ≈1.0 in M9 medium containing 98% D2O, 2 g/liter protonated [13C]glucose, and 1 g/liter 15NH4Cl. Harvested cells were resuspended in 50 mM Tris·HCl (pH 8.0)/2 mM EDTA and disrupted by sonication. Cell debris was removed by centrifugation. The supernatant was loaded onto a DEAE52 column and eluted with 150 mM KCl. The Hsc70 fractions were dialyzed against 20 mM Hepes (pH 7.0)/25 mM KCl/10 mM EDTA. Then, EDTA was precipitated by adding 25 mM MgCl2 to the dialyzed Hsc70 pool, yielding 5 mM free Mg2+. The precipitate of (Mg)EDTA was removed by centrifugation. The supernatant was loaded onto an ATP-agarose affinity column (Sigma) and eluted with 3 mM ATP. The recombinant Hsc70 was purified to >95% homogeneity as judged from SDS/PAGE and was concentrated by ultrafiltration (Amicon Ultrafree15). The typical yield was 40 mg of triple-labeled Hsc70 NBD from a 1-liter culture.
After the protein was purified, it was unfolded in 4.5 M guanidine hydrochloride to back-exchange the amide protons; thereafter, the protein was refolded by fast dilution in buffer. Extensive buffer exchange with 5 mM MgCl2/25 mM KCl/20 mM Tris·HCl (pH 7.2) prepared the protein in the apo (nucleotide-free) form. The refolded protein had identical ATPase functionality as measured by 31P NMR experiments as protein that was never refolded; moreover, the 1H-15N TROSY chemical shifts of the refolded protein were identical to those of a sample 15N-labeled Hsc70 NBD that was never refolded (data not shown). Together, this procedure demonstrates that the refolding did not affect the protein structure or function.
NMR Assignment Measurements. Samples for NMR spectroscopy were prepared at a protein concentration of 0.4–0.5 mM containing 5 mM MgCl2, 25 mM KCl, 20 mM Tris·HCl, 10 mM ADP, 0.005% sodium azide, and 10% (vol/vol) D2O (pH 7.2). NMR experiments for backbone assignments were obtained from an inhouse written suite of 2H-decoupled, TROSY-based 3D HNCO and HN(CA)CO, HNCA and HN(CO)CA, and HNCACB and HN(CO)CACB, based on ref. 16, as well as 3D NOESY-TROSY. One native sample and one refolded sample were used for spectra recording; the spectra were identical except for intensity differences. All NMR experiments were at 30°C on a Varian INOVA 800-MHz spectrometer equipped with a triple-resonance gradient probe. Total instrument time used for the assignments was ≈555 h; experimental parameters are given in Table 2, which is published as supporting information on the PNAS web site. The sample showed no signs of degradation after the experiments were completed. All spectra were processed with nmrpipe (17) and analyzed with the assistance of autoassign (18) as well as manual connectivity tracking by using sparky (19).
Measurement of Residual Dipolar Couplings. The nonionic liquid-crystalline medium (20) of C12E6 polyethylene glycol (PEG)/hexanol (Sigma) at a molar ratio of 0.64 was compatible with Hsc70 NBD in the ADP.Pi (20 mM Pi) state. PEG (C12E6) was dissolved in the desired buffer at 10% wt/vol and titrated with hexanol to form a lamellar phase. A concentrated protein solution (≈1 mM) was titrated into the mixture. Three samples were made in this way at PEG concentrations of 4%, 5%, and 6.5%. The D2O quadrupolar spectrum showed pure doublets, indicating homogeneously aligned samples.
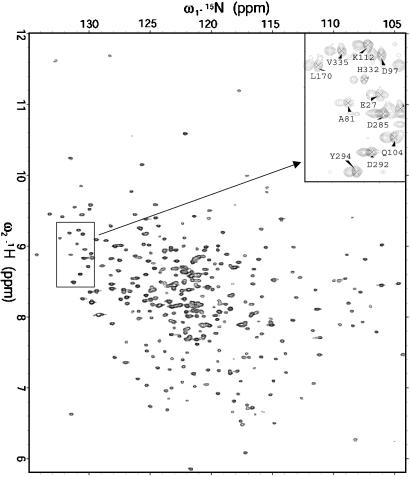
Because the Hsc70 NBD TROSY spectrum is exceptionally well resolved (see Fig. 2), we could use a 2D experiment to obtain the residual dipolar couplings (RDCs). We incorporated a κt1/2–180(N,H)-κt1/2 sequence at the beginning of the 15N evolution period of the 2D TROSY, following ideas by Yang et al. (21). With κ = 0, one essentially obtains the undisturbed TROSY spectrum; with κ = 2, the 15N resonance is shifted by J + D Hz, but also acquires an unfavorable anti-TROSY relaxation behavior. We found a good compromise by using κ values 0 and 0.5, and 1 as a control to resolve cases of overlap (Fig. 2 Inset). All spectra were processed by nmrpipe. RDCs were measured from the κ-shifted spectra from a single aligned sample by using the 2D peak-fitting routine provided by the program sparky. On the basis of repeat experiments, we estimate an accuracy of approximately plus or minus 4–5 Hz for the RDCs.
Fig. 2.
15N-1H TROSY of 0.4 mM 44-kDa Hsc70 NBD at 800 MHz. (Inset) An area of the κ = 0 (labeled resonances) and κ = 0.5 (unlabeled resonances) shifted TROSY spectra (see Materials and Methods) in 4% PEG-hexanol.
Measurement of NH Relaxation. 15N R1 and R2 relaxation experiments with TROSY detection were recorded at 800 MHz. Average T2 and T1 at 30°C were 25 ms and 2.1 s, respectively, indicating a rotational correlation time of 19 ns, in the expected range for a 44-kDa spherical monomer. A lack of sensitivity precluded further analysis in terms of order parameters.
Results and Discussion
Structure and Dynamics. The TROSY NMR spectrum of the Hsc70 ATPase domain is very well resolved (See Fig. 2). In both ATP and ADP states, multiple signals occur for at least 50 residues. A very stable conformation was obtained by adding excessive KPi (≈20 mM) and ADP to form the ADP-Pi form, which gives a single set of signals for most of the protein. Nearly complete backbone 1H, 13C, and 15N NMR assignments were obtained by using TROSY versions of 3D triple-resonance experiments. By using the chemical shift index of Wishart and Sykes (22), we find that the locations of the secondary structure elements coincide with the crystal structure; moreover, analysis of a 3D NOESY-TROSY spectrum confirms the connectivity patterns in helices and sheets (Fig. 6, which is published as supporting information on the PNAS web site). Thus, the solution and crystal structures are closely related, bestowing confidence that the x-ray coordinates can be used for the fitting of the residual dipolar couplings. Due to the multiple conformations, a grand total of 431 NH resonances was assigned; at the same time, no assignments were obtained for 30 residues. Five noticeable fragments that are not completely assigned are 36–39 (linkage of IA to IB), 124–129 (linkage of IB to IA), 181–182 (linkage of IB to IIA), 200–205 (ligand-binding site), and 365–366 (linkage of IIA to IA). There are additional residues in these linkage regions showing very weak or multiple peaks, suggesting that these residues are undergoing conformational exchange. The linkage regions from IIA to IIB (residues 222–227) and from IIB to IIA (residues 303–309) are also undergoing conformational exchange. Taken together, one can clearly conclude that these linkage regions are flexible in solution. Additional evidence to probe the flexible regions came from T1, T2 relaxation measurements. The limited results show there is very flexible linkage around residues 189–193 (connection lobe 1 and lobe 2), because the T1, T2 values for them are similar to those of the completely flexible N and C termini (data not shown).
Use of RDCs to Determine the Subdomain Orientation of Hsc70 ATPase. In recent years, RDCs between the 15N and 1H amide nuclei have been successfully used to validate structures determined by NMR, x-ray crystallography, or homology modeling, to refine structures determined by conventional NMR approaches, to characterize complexes, and to determine relative domain orientations in multidomain proteins and nucleic acids (23, 24). In some cases, deviations between x-ray and solution structures have been detected by these methods (25, 26). The NH RDCs are obtained in a slightly anisotropic environment, created by magnetically oriented liquid crystalline suspensions, which induce a small (steric) alignment of the otherwise freely tumbling protein with respect to the magnetic field. When the structure is known, the RDCs determine the orientation of the molecule with respect to the magnetic field. In a multidomain protein or complex, the orientations of all domains can be obtained with respect to the field, and hence with respect to each other.
The RDC between nuclei m and n is given by ref. 14:
 |
[1] |
where 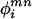 represent the angles between the mn-internuclear vector and the x′, y′, or z′ axes of the molecular frame (often taken as the PDB frame). The quantity in the brackets is called the Saupe order matrix, which depends on the time-averaged angles θi of the molecular axes x′, y′, or z′ with respect to the magnetic field. δi is the Kronecker delta. When the protein structure is known (i.e., all or most angles
represent the angles between the mn-internuclear vector and the x′, y′, or z′ axes of the molecular frame (often taken as the PDB frame). The quantity in the brackets is called the Saupe order matrix, which depends on the time-averaged angles θi of the molecular axes x′, y′, or z′ with respect to the magnetic field. δi is the Kronecker delta. When the protein structure is known (i.e., all or most angles  are known) five or more noncoaxial RDCs suffice to obtain the time-average orientation of the molecular axes x′, y′, or z′ with respect to the magnetic field. We use the program pales (27), which utilizes singular value decomposition to solve the order matrix on the basis of a known structure and the obtained RDCs (14). After order matrix diagonalization to a new x,y,z frame, one obtains the three Euler angles (α,β,γ), which describe the orientations of the principal axes x,y,z of the alignment frame with respect to the PDB frame and Szz, Syy, and Sxx, which represent the amount of alignment along the principal axes of the alignment frame (of the order 10–3 – 10–4). By convention, |Szz| > |Syy| > |Sxx|.
are known) five or more noncoaxial RDCs suffice to obtain the time-average orientation of the molecular axes x′, y′, or z′ with respect to the magnetic field. We use the program pales (27), which utilizes singular value decomposition to solve the order matrix on the basis of a known structure and the obtained RDCs (14). After order matrix diagonalization to a new x,y,z frame, one obtains the three Euler angles (α,β,γ), which describe the orientations of the principal axes x,y,z of the alignment frame with respect to the PDB frame and Szz, Syy, and Sxx, which represent the amount of alignment along the principal axes of the alignment frame (of the order 10–3 – 10–4). By convention, |Szz| > |Syy| > |Sxx|.
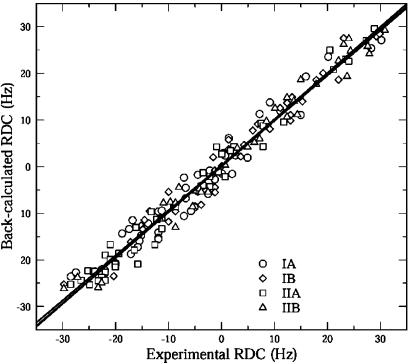
A total of 241 RDCs were measured by using the PEG/hexanol alignment medium (20). Eliminating RDCs that belonged to residues that displayed peak overlap or to residues located in flexible loops, we used 173 dipolar couplings of this set (see Fig. 6). Among all available crystal structures of Hsc70 ATPase (PDB ID code 3HSC) was found to give the best fit to the experimental couplings, as judged by the rmsd of measured and back-calculated dipolar couplings. The fitting of the couplings by pales is illustrated in Fig. 3 and Table 1. The first row in Table 1 shows the overall alignment tensor when treating the whole molecule as a single rigid entity. It yielded an rmsd of fit of 3.2 Hz. Guided by our findings that the intersubdomain loops are likely flexible, we investigated whether better results could be obtained by fitting the dipolar couplings of each subdomain (IA, IB, IIA, and IIB) independently. From the next four rows, one sees that the latter actually gives much better fitting results for all four subdomains; especially noteworthy is that for IIA the quality of the data fitting has improved to an rmsd of 2.2 Hz.
Fig. 3.
Correlation between the experimentally measured (Fig. 2) HN-N dipolar couplings versus the back-calculated values on the basis of crystal structure (PDB ID code 3HSC) for all four subdomains of Hsc70 NBD. The lines represent the least-squares fits for the individual sub domains: IA, Y = 0.158 + 0.964X; IB, Y = 0.365 + 0.966X; IIA, Y = 0.256 + 0.989X; and IIB, Y = 0.035 + 0.979X.
Table 1. Singular value decomposition analysis of HN-N (RDC) for Hsc70 NBD (residues 3-384).
| Results | No. of RDCs | Szz(e — 3) | α, ° | β, ° | γ, ° | rms, Hz | Rsvd | Q | General degree of order | Condition no. |
|---|---|---|---|---|---|---|---|---|---|---|
| All | 173 | 1.39 | 101.09 | 96.21 | 104.92 | 3.02 | 0.98 | 0.19 | ||
| IA | 50 | 1.37 | 99.20 | 93.99 | 109.64 | 2.90 | 0.98 | 0.20 | 1.287 | 1.90 |
| IB | 39 | 1.39 | 95.54 | 93.43 | 101.47 | 2.53 | 0.98 | 0.18 | 1.335 | 1.95 |
| IIA | 48 | 1.40 | 103.78 | 98.10 | 102.17 | 2.26 | 0.99 | 0.13 | 1.334 | 1.92 |
| IIB | 36 | 1.37 | 104.45 | 99.87 | 105.28 | 2.41 | 0.99 | 0.14 | 1.331 | 1.89 |
| Lobe 1 | 89 | 1.37 | 97.32 | 94.34 | 105.40 | 3.03 | 0.98 | 0.21 | 1.300 | 1.74 |
| Lobe 2 | 84 | 1.38 | 104.59 | 99.14 | 104.36 | 2.42 | 0.99 | 0.14 | 1.330 | 1.53 |
Results include individual domain (4) fitting and lobe (2) fitting. Four subdomains are defined as in Fig 1. Lobe 1 includes IA and IB, and lobe 2 includes IIA and IIB. Szz, the largest component of the alignment tensor; α, β, and γ, the Euler angles, which define the orientation of the alignment tensor in the frame of the PDB x-ray structure coordinates (PDB ID code 3HSC); rms, rms deviation of the |Dcal — Dexp|; Rsvd, the correlation coefficient of singular value decomposition analysis; Q, the quality factor (24).
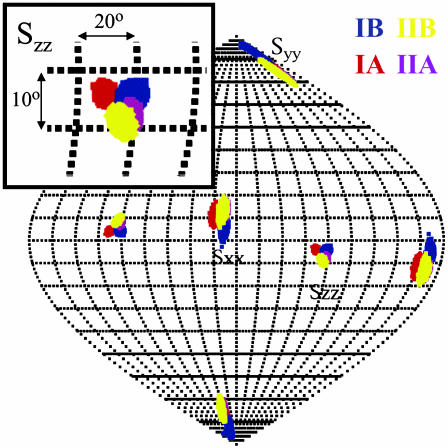
The results of the calculations of the orientation for the four subdomains are shown in Fig. 3. The map is the result of 10,000 pales calculations in which the experimental dipolar couplings were varied by as much as 3.5 Hz in a random fashion to simulate experimental error. The orientational distribution of the Szz principal axes is nearly Gaussian, with an rmsd of plus or minus 0.8°. The plot shows two well defined, nonoverlapping spots for the ensemble of the Szz axes of subdomains IA and IIA; they are separated by 6.7 ± 1.6°. Because the overlap between the distributions is nonexistent, the statistical significance of the difference is beyond question. The Szz orientations of Domains IIA and IIB overlap almost completely. The Szz axis distribution of subdomain IB partially overlaps with that of IA and IIA (Fig. 4). Nevertheless, the statistical significance of the difference of the orientation of IB with either IA or IIA, as analyzed by a 2D t test (28) on the two distributions, exceeds 99%. The differences and correspondences in tensor orientations for the different subdomains are also reproduced by the Sxx direction. We calculated the condition number (the ratio between the largest and the smallest singular value), which is also shown in Table 1. Our condition numbers (all <2) indicate that the dipolar data set is linearly independent, resulting in a very good determination of the relative subdomain orientations.
Fig. 4.
Sauson–Flamsteed projection of the directions of ordering (Szz, Syy, and Sxx) for oriented Hsc70 NBD in PEG/hexanol (4%). IA is red, IB is blue, IIA is purple, and IIB is yellow. (Inset) The area of the Szz orientations and degree scale.
The degrees of orientation, given by general degrees of order (14) are almost equal for all subdomains involved (Table 1). This result is compatible with either a static structure or a dynamic molecule where the individual mobilities of the subdomains are about equal. The fact that many of the resonances in the connecting loops are missing, as well as that the connector between lobes I and II is very mobile, would at least allow the possibility of an ensemble of dynamically readjusting subdomains. Because the subdomains have approximately the same molecular weights, relative mobility, if existent, would be expected to yield equal individual degrees of order.
We have also used the pales program to predict the alignment tensor of the 3HSC crystal structure. We find that the three predicted Euler angles (α,β,γ) are very close to what we obtained from the experiment; especially the β angle (which defines the angle between Szz and molecular frame) is 95°, and the experimental range is 94–100°. The predicted rhombicity factor η [(Syy-Sxx)/Szz] is 0.88, and the experimental factors ranged from 0.63 to 0.79. We believe that this is firm proof that the alignment of Hsc70 NBD in the neutral PEG/hexanol medium is governed by steric exclusion only, and not by interaction with the medium. Therefore, we conclude that the differences between the crystallographic structure and the solution conformation are accurate, and are most likely due to the larger degree of conformation freedom enjoyed by the protein in its natural state.
Crossvalidation of RDCs. We wished to investigate whether the obtained differences in orientation for the subdomains could be due to local structural changes in the domains as compared to the crystal structure, or to the particular selection of dipolar couplings available for analysis. To this end, we selected, at random, 50% of the RDCs available per subdomain, and performed the complete pales-fitting procedure. This process was repeated 10 times with different random selections for each domain. The average Euler angles based on the selected RDCs are within 1° of the all-data fitting per subdomain. The value of Szz (degree of order) is also close to the all-data fitting result. We conclude that the measured differences in subdomain orientations are not biased by the choice of RDCs.
Solution Structure Model from RDCs and Crystal Structure. The molecular modeling program insightii was used to adapt the crystal structure coordinates of the Hsc70 NBD to the solution conditions based on the RDCs results. First, the PDB file was split in the four subdomains and each individual subdomain was rotated according to the experimental Euler angles. They were subsequently reassembled with translational motions only until the linker residues were close enough (<3 Å) to form peptide bonds. insightii was used to link the subunits by peptide bonds. This procedure was followed by energy minimization to optimize the local geometry for the linkage regions. We verified that the energy minimization did not change relative domain orientation. Superpositions of the x-ray structure and the RDC-refined structure are shown in Fig. 5. Coordinates for the final hybrid x-ray-NMR model are available as Data Set 1, which is published as supporting information on the PNAS web site.
Fig. 5.
Stereo superpositions of x-ray (PDB ID code 3HSC; in red) and dipolar-refined solution NMR structure (in white) of Hsc70 NBD (1–386) in the ADP.Pi state. (Top) Domains IIA and IIB (lobe II) were superimposed, showing that lobe I (domains IA and IB) is sheared by 10°.(Middle) Detail of the same overlay showing that the shearing is based at the IA and IIA interface and affects the cleft between the domains. (Bottom) Domains IA were overlaid, illustrating the 8.8° rotation of domain IB.
Functional Implications. The NBD of the Hsp70 proteins is at the center of many of the functional interactions of the Hsp70s. In the ATP state, it is closely associated with the SBD and drives it to a low-substrate affinity; in the ADP state, the association between the two domains is less intricate, and the SBD is in a high-affinity form (29). The NBD by itself hydrolyzes ATP to ADP. The NBD contains a major binding interface for the cochaperone Hsp40 (DnaJ), which enhances the ATP hydrolysis rate. NBD also binds to nucleotide exchange factors GrpE (Coli) and BAG, and with the proteins HIP, CD40, and likely, also HOP.
The structural basis of the allosteric coupling remains mysterious; in addition, most of the reversible binding interfaces with the cofactors and how they perform their functions have not been identified. SAX data (29) indicate that the SBD and NBD are most closely associated in the ATP and least closely in the ADP state. This finding would suggest that major conformational changes take place, likely at surface areas, within the domains that would modulate this change in interaction. Indeed, solution NMR studies of the SBD by itself have indeed demonstrated that major and widespread structural and dynamical differences prevail in this domain when peptide substrate is present or absent (30). DnaJ binding reportedly occurs at a site located at the bottom cleft area of the NBD between domains IA and IIA remotely from the ATP-binding site (31, 32) yet it enhances ATP hydrolysis (see Fig. 1). All these functional data thus strongly suggest an NBD that can dynamically alter its conformation upon allosteric and cofactor binding and transduce surface-binding events to the nucleotide binding site and vice versa.
In contrast to the x-ray studies cited in Introduction, the current solution NMR study of Hsc70 NBD is indeed unveiling a much more dynamic molecule. More importantly, we find that the Hsc70 NBD in solution shows significant subdomain orientation differences as compared with all available crystal structures referenced above. Most significantly, we detect a 10° relative shearing motion between subdomains IA and IIA as compared with the crystal structure (Fig. 5 Middle). Equally interesting is an 8.8° reorientation of subdomain IB with respect to subdomain IA, also not seen before in any crystal structure (Fig. 5 Bottom). Together with the available crystal structures, our solution NMR dipolar data thus reveal that all four subdomains and lobes can perform substantial reorientational conformational changes, depending on experimental conditions. Hence, the current studies sketch this molecule as an ensemble of states of different subunit orientations with ample possibilities to modulate its interactions with other domains and cofactors.
Of great functional significance is the shearing motion of domains IA and IIA. Mutagenesis studies imply that residues on both of these subdomains are involved in the binding of the cochaperone DnaJ (31, 32). These residues are indicated on the structure in Fig. 1 in green. Our results suggest that DnaJ binding to the residues in both of these subdomains domains is likely to perturb (the ensemble of) relative subdomain orientations. This effect would be directly transduced to the nucleotide, which sits at the interface of all four subdomains: the nucleotide base and deoxyribose are bound by residues from subdomains IIA (majority) and IIB (minority), whereas the polyphosphate moiety is complexed by residues from subdomains IA (majority) and IB (minority). A change in relative orientations of IA and IIA should thus affect the local environment of the nucleotide and affect hydrolysis. Conversely, a change in the nucleotide state could by the same mechanism affect the relative orientations of the subdomains IA and IIA, and thus bind or release the factors that lodge in the cleft area between them.
Although the interaction interface of the NBD with the SBD is still unknown, there have been reports that mutations of residues R151, Y145, and E171 in DnaK (R155, Y149, and E175 in Hsc70) affect the allosteric coupling between the two domains.† These residues all lie in the same area as the (putative) HdJ-binding site in subdomain IA (black in Fig. 1), but are so close to IIA, that a relative reorientation of these subdomains would likely affect their accessibility. As suggested by the position of the C terminus of the NBD, the SBD could position itself into this same lower cleft as HdJ. As such, the relative shearing of subdomains IA and IB may also be of direct relevance to the allosteric coupling of the NBD and SBD.
Our finding of changes in the relative orientation of domains IA and IB is likely also of functional consequence. In particular, the large offset of domain IB from the entire lobe 2 and its subdomain IIB implies that nucleotide binding and release is likely regulated by the relative movement of both domains IB and IIB. This result adds to previous studies (12, 13) suggesting that the process is modulated by reorientation of domain IIB only. Second, residue K71, which is essential to ATP hydrolysis (33), emanates from subdomain IB, whereas residue T13, from which the OH group is essential to the couple ATP hydrolysis to the overall allosteric change (34), emanates from subdomain IA. Hence, a reorientation of subdomains IA and IB would also couple to the ATP site and chemistry, and, vice versa, the nucleotide state into the relative positioning of these subdomains. Another interesting fact is that residue W102 in DnaK resides in domain IB (not conserved; Q104 in Hsc70) and displays a dramatic change in fluorescence upon ATP or ADP binding (7). Actually, the site W102 is remarkably remote from the ATP-binding site itself (see Fig. 1), and how its environment might change upon ATP/ADP binding is not at all clear. Our current data on the relative adaptability of domain IB should be kept in mind when searching for an explanation of these facts.
Supplementary Material
Acknowledgments
We thank Dr. David B. McKay for the plasmid of bovine Hsc70 NBD, Dr. Matt Revington for many discussions, Dr. Hashim M. Al Hashimi for in-depth discussions with respect to the dipolar coupling data analysis, and Ms. Valentyna Semenchenko and Dr. Alexander V. Kurochkin for assistance with protein purification. This work was supported by National Institutes of Health Grant RO1 GM063027.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Hsp70, 70-kDa heat shock protein; Hsc70, heat shock cognate; NBD, nucleotide-binding domain; SBD, substrate-binding domain; TROSY, transverse relaxation-optimized spectroscopy; RDC, residual dipolar coupling; rmsd, rms deviation; PEG, polyethylene glycol.
Footnotes
Mayer, M., Conference on Molecular Chaperones and the Heat Shock Response, May 1–5, 2002, Cold Spring Harbor, NY.
References
- 1.Bukau, B. & Horwich, A. L. (1998) Cell 92, 351–366. [DOI] [PubMed] [Google Scholar]
- 2.Morimoto, R. I. (2002) Cell 110, 281–284. [DOI] [PubMed] [Google Scholar]
- 3.Wickner, S., Maurizi, M. & Gottesman, S. (1999) Protein Sci. 286, 1888–1893. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty, K. M., DeLuca-Flaherty, C. & McKay, D. B. (1990) Nature 346, 623–628. [DOI] [PubMed] [Google Scholar]
- 5.Morshauser, R. C., Hu, W., Wang, H., Pang, Y., Flynn, G. C. & Zuiderweg, E. R. (1999) J. Mol. Biol. 289, 1387–1403. [DOI] [PubMed] [Google Scholar]
- 6.Zhu, X., Zhao, X., Burkholder, W. F., Gragerov, A., Ogata, C. M., Gottesman, M. E. & Hendrickson, W. A. (1996) Science 272, 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slepenkov, S. V. & Witt, S. N. (2002) Biochemistry 41, 12224–12235. [DOI] [PubMed] [Google Scholar]
- 8.Qian, Y. Q., Patel, D., Hartl, F. U. & McColl, D. J. (1996) J. Mol. Biol. 260, 224–235. [DOI] [PubMed] [Google Scholar]
- 9.Bimston, D., Song, J., Winchester, D., Takayama, S., Reed, J. C. & Morimoto, R. I. (1998) EMBO J. 17, 6871–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayer, M. P., Brehmer, D., Gassler, C. S. & Bukau, B. (2001) Adv. Protein Chem. 59, 1–44. [DOI] [PubMed] [Google Scholar]
- 11.Osipiak, J., Walsh, M. A., Freeman, B. C., Morimoto, R. I. & Joachimiak, A. (1999) Acta Crystallogr. D. 55, 1105–1107. [DOI] [PubMed] [Google Scholar]
- 12.Sondermann, H., Scheufler, C., Schneider, C., Hohfeld, J., Hartl, F. U. & Moarefi, I. (2001) Science 291, 1553–1557. [DOI] [PubMed] [Google Scholar]
- 13.Harrison, C. J., Hayer-Hartl, M., Di Liberto, M., Hartl, F. & Kuriyan, J. (1997) Science 276, 431–435. [DOI] [PubMed] [Google Scholar]
- 14.Prestegard, J. H., Al-Hashimi, H. M. & Tolman, J. R. (2000) Q. Rev. Biophys. 33, 371–424. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien, M. C. & McKay, D. B. (1993) J. Biol. Chem. 268, 24323–24329. [PubMed] [Google Scholar]
- 16.Loria, J. P., Rance, M. & Palmer, A. G., III (1999) J. Magn. Reson. 141, 180–184. [DOI] [PubMed] [Google Scholar]
- 17.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J. & Bax, A. (1995) J. Biomol. NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- 18.Zimmerman, D. E., Kulikowski, C. A., Huang, Y., Feng, W., Tashiro, M., Shimotakahara, S., Chien, C., Powers, R. & Montelione, G. T. (1997) J. Mol. Biol. 269, 592–610. [DOI] [PubMed] [Google Scholar]
- 19.Goddard, T. D. & Kneller, D. G., sparky 3, A Graphical NMR Assignment Program (University of California, San Francisco).
- 20.Rückert, M. & Otting, G. (2000) J. Am. Chem. Soc. 122, 7793–7797. [Google Scholar]
- 21.Yang, D., Venters, R. A., Mueller, G. A., Choy, W. Y. & Kay, L. E. (1999) J. Biomol. NMR 14, 333–343. [Google Scholar]
- 22.Wishart, D. S. & Sykes, B. D. (1994) J. Biomol. NMR 4, 171–180. [DOI] [PubMed] [Google Scholar]
- 23.Prestegard, J. H. (1998) Nat. Struct. Biol. 5, Suppl., 517–522. [DOI] [PubMed] [Google Scholar]
- 24.Bax, A. (2003) Protein Sci. 12, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukin, J. A., Kontaxis, G., Simplaceanu, V., Yuan, Y., Bax, A. & Ho, C. (2003) Proc. Natl. Acad. Sci. USA 100, 517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Millet, O., Hudson, R. P. & Kay, L. E. (2003) Proc. Natl. Acad. Sci. USA 100, 12700–12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zweckstetter, M. & Bax, A. (2000) J. Am. Chem. Soc. 122, 3791–3792. [Google Scholar]
- 28.Press, W. H., Flannery, B. P., Teukolsky, S. A. & Vetterling, W. T. (1989) in Numerical Recipes: The Art of Scientific Computing (Cambridge Univ. Press, Cambridge, U.K.), pp. 472–475.
- 29.Wilbanks, S. M., Chen, L., Tsuruta, H., Hodgson, K. O. & McKay, D. B. (1995) Biochemistry 34, 12095–12106. [DOI] [PubMed] [Google Scholar]
- 30.Pellecchia, M., Montgomery, D. L., Stevens, S. Y., Vander Kooi, C. W., Feng, H. P., Gierasch, L. M. & Zuiderweg, E. R. (2000) Nat. Struct. Biol. 7, 298–303. [DOI] [PubMed] [Google Scholar]
- 31.Suh, W. C., Lu, C. Z. & Gross, C. A. (1999) J. Biol. Chem. 274, 30534–30539. [DOI] [PubMed] [Google Scholar]
- 32.Gassler, C. S., Buchberger, A., Laufen, T., Mayer, M. P., Schroder, H., Valencia, A. & Bukau, B. (1998) Proc. Natl. Acad. Sci. USA 95, 15229–15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien, M. C., Flaherty, K. M. & McKay, D. B. (1996) J. Biol. Chem. 271, 15874–15878. [DOI] [PubMed] [Google Scholar]
- 34.Sousa, M. C. & McKay, D. B. (1998) Biochemistry 37, 15392–15399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.