Abstract
In prokaryotes, two DNA glycosylases recognize and excise oxidized pyrimidines: endonuclease III (Nth) and endonuclease VIII (Nei). The oxidized purine 8-oxoguanine, on the other hand, is recognized by Fpg (also known as MutM), a glycosylase that belongs to the same family as Nei. The recent availability of the human genome sequence allowed the identification of three human homologs of Escherichia coli Nei. We report here the crystal structure of a human Nei-like (NEIL) enzyme, NEIL1. The structure of NEIL1 exhibits the same overall fold as E. coli Nei, albeit with an unexpected twist. Sequence alignments had predicted that NEIL1 would lack a zinc finger, and it was therefore expected to use a different DNA-binding motif instead. Our structure revealed that, to the contrary, NEIL1 contains a structural motif composed of two antiparallel β-strands that mimics the antiparallel β-hairpin zinc finger found in other Fpg/Nei family members but lacks the loops that harbor the zinc-binding residues and, therefore, does not coordinate zinc. This “zincless finger” appears to be required for NEIL1 activity, because mutating a very highly conserved arginine within this motif greatly reduces the glycosylase activity of the enzyme.
Oxidized DNA damages are recognized and removed by base excision repair processing (1–3), the first step of which is catalyzed by a DNA glycosylase. In general, the oxidative DNA glycosylases are either purine- or pyrimidine-specific. Two DNA glycosylases recognize and remove oxidized pyrimidines: endonuclease III (Nth), a member of the Nth superfamily, and endonuclease VIII (Nei), a member of the Fpg/Nei family (for reviews see refs. 4–7). Although Nth is widely distributed over all three kingdoms, Nei is only sparsely represented and found in some γ-proteobacteria, actinomycetes, and vertebrates (6). Escherichia coli lacking both Nth and Nei exhibit a high spontaneous mutation frequency and are hypersensitive to hydrogen peroxide and ionizing radiation (8–11).
The two members of the Fpg/Nei family share a common mechanism of action (for reviews see refs. 6 and 12). Catalysis by both E. coli Nei (EcoNei) and Fpg (EcoFpg) is by means of the N-terminal proline, which forms a Schiff base with the oxidized lesion (13–15). Both EcoNei and EcoFpg are trifunctional enzymes containing glycosylase, β,δ lyase, and 5′ phosphodiesterase activities (16–19). Fpg and Nei also share common structural motifs, including helix-two-turns-helix (H2TH) and antiparallel β-hairpin zinc finger motifs (6, 12). A comparison of EcoNei covalently complexed to DNA (20) with the Fpg structures (21–24) revealed that their overall folds are very similar; however, their substrate preferences are markedly different: EcoFpg prefers 8-oxoguanine and oxidized purines (25, 26), whereas EcoNei recognizes oxidized pyrimidines (19, 27–29).
Interestingly, several of the actinomycetes, including Mycobacterium tuberculosis and Streptomyces coelicolor, contain three Nei paralogs (6), and a search of the human sequence database revealed that Homo sapiens similarly contains three homologs of Nei, Nei-like (NEIL)1, NEIL2, and NEIL3 (30–32). All three genes have been cloned, and NEIL1 and NEIL2 have been expressed and partially characterized (6, 30–36). In nullizygous Nth–/– mice, the Nei-like activities apparently serve as a backup for mNth1 (37–40). Furthermore, when NEIL1 is knocked down by RNA interference, embryonic stem cells become hypersensitive to ionizing radiation (41).
Although they differ in their substrate preferences (6, 30–32, 34–36), NEIL1 and NEIL2 recognize oxidized pyrimidines, and both form a Schiff base with substrates containing oxidized pyrimidines (6, 32). Furthermore, NEIL1 with a site-directed mutation in the catalytic N-terminal proline or glutamic acid residues (P2T and E3Q) is inactive (32). NEIL1 and NEIL2 share the common catalytic proline at the N-terminal position, whereas NEIL3 has a valine at this position (6, 32). All three proteins share a H2TH DNA-binding motif (6), but only NEIL2 and NEIL3 contain a zinc finger motif (6, 32). Because NEIL1 is active on DNA-containing oxidized pyrimidines (30, 32), it has been assumed that NEIL1 must possess an alternative DNA-binding motif. Interestingly, two other members of the Fpg/Nei family, Arabidopsis thaliana Fpg and Candida albicans Fpg, also lack a zinc-finger motif (6, 12).
Here we report the structure of an enzymatically active deletion construct of NEIL1, lacking 56 C-terminal residues. Although NEIL1 shares the same fold as members of the Fpg/Nei family, the structure revealed some unexpected features. Although sequence alignments had predicted that NEIL1 would lack a zinc finger, a structural motif in that enzyme strongly resembles the zinc finger found in bacterial Fpg and Nei. Mutating a highly conserved arginine within the NEIL1 “zincless finger” motif strongly diminished that enzyme's glycosylase activity, underscoring the importance of this motif for enzyme activity.
Materials and Methods
Crystal Structure Determination. Cloning, expression, purification, and crystallization have been reported elsewhere (42). A C-terminal deletion fragment of human NEIL1 missing the last 56 residues (NEIL1CΔ56) crystallizes in space group R3, with cell parameters (a = b = 132.2 Å and c = 51.1 Å) in the hexagonal setting. Activity assays of the wild-type full-length NEIL1 and C-terminal deletion construct (NEIL1CΔ56) showed that the deletion construct appears to be ≈4-fold more active than full-length NEIL1 on thymine glycol-containing double stranded oligonucleotides (data not shown). NEIL1CΔ56 crystals diffract to a resolution of 2.1 Å by using a rotating anode x-ray generator and MAR image plate detector (MAR Research, Hamburg, Germany). Data collection statistics are summarized in Table 1. Initial attempts at molecular replacement with several search models from other members of the Fpg/Nei family failed to produce a clear solution. The structure of NEIL1CΔ56 was therefore solved by multiple isomorphous replacement by using a native dataset and three derivatives [selenomethionyl (SeMet)-NEIL1CΔ56, iodide-soaked native crystals, and the double derivative iodinated SeMet-NEIL1CΔ56 (SeMet-I)] (43). All data were collected at 100 K on a rotating copper anode source. Four selenium sites were located by using cns (44), including that coming from the substituted N-terminal formylmethionine (fMet). Because the Fpg/Nei family members require processing of the N-terminal fMet to use the second proline residue for enzyme catalysis (13–15, 45), we expected to find only three selenium sites. A glycosylase assay revealed that SeMet-NEIL1CΔ56 exhibited a reduced activity on a 5,6-dihydrouracil-containing substrate (data not shown), suggesting partial or incomplete processing of N-terminal fMet in the SeMet enzyme.
Table 1. Data collection, phasing, and refinement statistics for human NEIL1.
| Native | SeMet | SeMet-I | I | |
|---|---|---|---|---|
| Data collection statistics | ||||
| Resolution, Å | 30-2.10 | 20-2.30 | 20-2.40 | 20-2.30 |
| Unique reflections | 19,225 | 14,770 | 12,854 | 14,357 |
| Redundancy | 12 | 5 | 6 | 3 |
| Rmerge* | 0.085 (0.383) | 0.095 (0.437) | 0.089 (0.292) | 0.078 (0.241) |
| Completeness,* % | 95.1 (100) | 99.9 (100) | 98.4 (91) | 97.0 (85.5) |
| Overall I/σ〈I〉* | 39.6 (5.6) | 13.7 (2.8) | 19.5 (2.8) | 15.8 (2.5) |
| MIR phasing statistics | ||||
| No. of sites | 4 Se | 4 Se, 5 I | 8 I | |
| Phasing power | 0.39 | 0.74 | 1.0 | |
| Figure of merit (solve) | 0.543 | |||
| Figure of merit (resolve) | 0.717 | |||
| Refinement statistics | ||||
| Rwork, % | 19.4 | |||
| Rfree, % | 23.1 | |||
| rms deviations | ||||
| Bond length, Å | 0.0053 | |||
| Bond angles, ° | 1.29 | |||
| B-factor, Å2 | ||||
| Protein | 28 | |||
| Water | 37 |
Rmerge = Σ |I - 〈I〉|/ Σ I, where 〈I〉 is the average intensity from multiple observations of symmetry-related reflections. Phasing power = ΣhklFH/Σ|FPH - FPH,calc|. Rwork and Rfree = Σ ||Fo| - |Fc||/Σ |Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated with ≈10% of the reflections not used in refinement. MIR, multiple isomorphous replacement; I, iodide; Se, selenium.
Values for the highest resolution shell are shown in parentheses.
Selenium phases were used to calculate isomorphous difference Fourier maps with the iodide datasets. solve (46) was used to refine the heavy atom positions. resolve substantially improved the electron density map, and generated an initial protein model (47). Model building was completed by using o (48). A side-by-side comparison of the solvent-flattened experimental map and 2Fo – Fc map illustrates the quality of the experimental phases (Fig. 6, which is published as supporting information on the PNAS web site). The final model was produced through iterative cycles of manual model building and refinement by using cns. The free R factor was calculated with 10% of the reflections set aside. Protein residues 2–202 and 208–290 as well as 307 water molecules were built into the electron density map. A tris(hydroxymethyl)aminomethane molecule from the crystallization buffer is positioned next to the N-terminal proline. All nonglycine and nonproline residues in the final model lie in the most favored and additionally allowed regions of the Ramachandran plot. There is a cis-proline (Pro-68) in the loop connecting β-strands 3 and 4. The final R factor is 19.4% (Rfree = 23.1%) for 19,225 reflections in the 30- to 2.10-Å resolution range.
Sequence Alignments and Phylogenetic Trees. Homologous sequences were identified by using psi-blast (49) and aligned by using clustal w (50). Alignments were trimmed manually to remove poorly aligned regions. Phylogenetic trees were constructed by using phylip (51).
Activity Assay and Figures. Double-stranded oligonucleotides containing either an abasic site, 5,6-dihydrouracil, or thymine glycol were used to assay the activities of NEIL1CΔ56, the SeMet variant, and site-directed mutants, as described in ref. 32. Figs. 1, 3, and 4 were generated with setor (52).
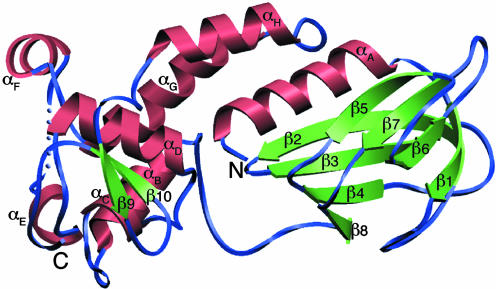
Fig. 1.
A ribbon diagram of human NEIL1. The model comprises residues 2–290; residues 203–207 are disordered and depicted as blue spheres. The secondary structure elements were defined by DSSP (62) and are as follows: αA (4–18), β1 (23–29), β2 (41–52), β3 (55–62), β4 (73–78), β5 (84–89), β6 (97–102), β7 (110–115), β8 (122–125), αB (141–150), αC (161–164), αD (176–186), αE (194–198), αF (212–218), αG (225–240), αH (249–259), β9 (269–272), and β10 (278–281).
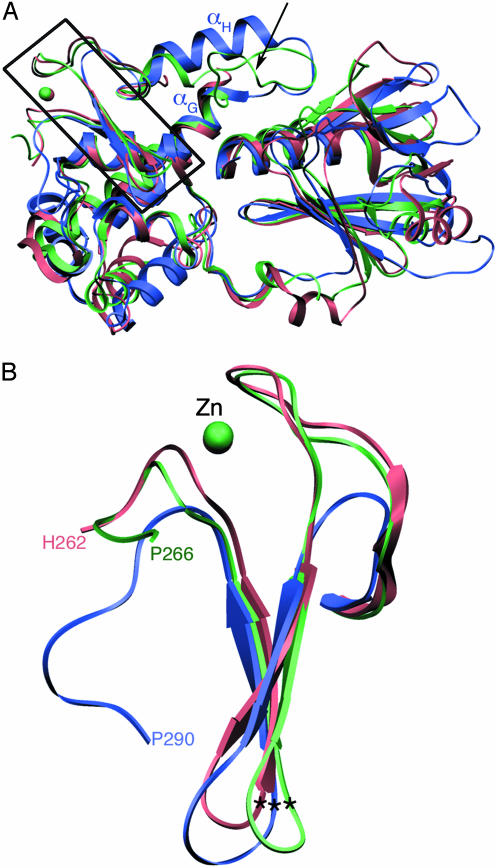
Fig. 3.
Comparison of human NEIL1 with other Fpg/Nei DNA glycosylases. (A) Superposition of human NEIL1 (blue) with EcoNei (pink; PDB ID code 1K3W) (20) and TthFpg (green; PDB ID code 1EE8) (21). The region encompassing the zinc-finger motif is boxed. An arrow points to the location of the αF-β10 loop in Fpg. (B) Close-up of the zinc-finger motif. Shown are residues 230–262 for EcoNei, 231–266 for TthFpg, and 263–290 for human NEIL1. The asterisks indicate the position of the Cα of the conserved arginine.
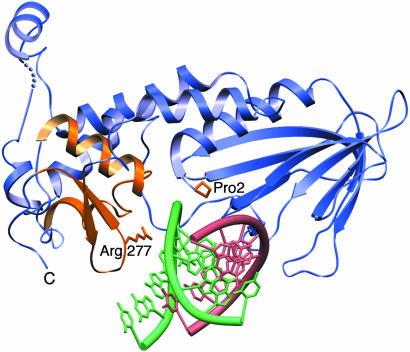
Fig. 4.
NEIL1–DNA model. DNA from EcoNei complex (lesion-containing strand in green and complementary strand in pink) was superimposed onto human NEIL1 (blue). The zincless finger, H2TH, catalytic proline, and conserved arginine are highlighted in gold.
Results and Discussion
Structure Description. NEIL1 is composed of two domains connected by a linker (Fig. 1). The N-terminal domain comprises an α-helix followed by a two-layered β-sandwich, with each layer composed of four antiparallel β-strands. The C-terminal domain is mostly helical: It comprises seven α-helices, two of which are involved in the H2TH motif (helices C and D). Two antiparallel β-strands, immediately after the helical structure, form a structural motif mimicking an antiparallel β-hairpin zinc finger, despite the dearth of sequence similarity to Nei and Fpg homologs known to harbor a zinc finger and the absence of zinc (see discussion below). The approximate dimensions of the molecule are 60 × 35 × 25 Å3. Although the protein construct used for this study comprises residues 2–342 (including the C-terminal hexa-His tag), there is no identifiable density beyond residue 290. A search with the dali server (53) confirmed the expected structural similarity with other DNA glycosylases of the Fpg/Nei family, Thermus thermophilus Fpg (TthFpg; PDB ID code 1EE8; Cα rms deviation = 2.4 Å, 29% sequence identity, Z score = 21.0) (21) and EcoNei (PDB ID code 1K3W; Cα rms deviation = 3.1 Å, 25% sequence identity, Z score = 17.7) (20). The numbers reported by dali indicate that NEIL1 is structurally closer to Fpg than to Nei. For example, the sequence corresponding to the void-filling residues in EcoFpg is identical in NEIL1 (Met-81, Arg-118, and Phe-120) (Fig. 7, which is published as supporting information on the PNAS web site). Similarly, sequence–sequence distances calculated with either phylip (51) or puzzle (54) suggest that NEIL1 is more similar in sequence to TthFpg than to EcoNei, even though phylogenetic tree construction (6, 32) suggests that human NEIL1 shares a more recent common ancestor with EcoNei than with TthFpg. These two observations are consistent because the rate of evolution within the Nei clade is rapid compared with that among Fpg family members, as is apparent in the phylogenies.
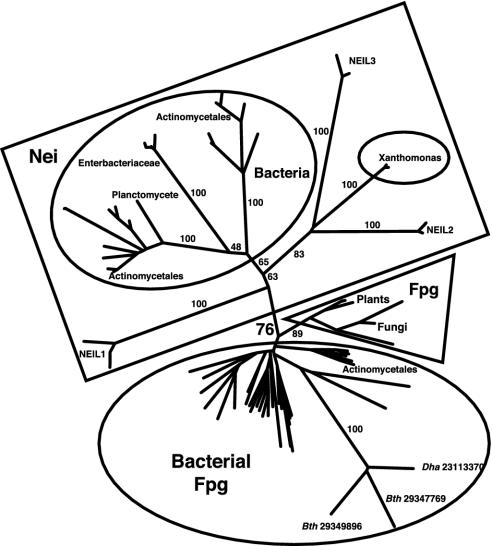
The phylogeny of the Fpg/Nei family (6, 32) exhibits bacterial (Fig. 2, largest ellipse) and plant/fungal (Fig. 2, triangle) “Fpg” clades, so named because at least one member has been studied biochemically and designated as Fpg. These clades are expected to contain mostly, but not exclusively, proteins with substrate specificity similar to EcoFpg. The clade designated as “Bacterial Fpg” comprises multiple representatives from Desulfitobacterium hafniense and Bacteroides thetaiotaomicron VPI-5482. Three members of the clade are unusually divergent in sequence (Fig. 2, long branch lengths). Interestingly, these proteins lack a proline at position 2 and an arginine homologous to EcoNei Arg-253 but possess clear zinc finger and H2TH sequence signatures.
Fig. 2.
Unrooted phylogeny of the Fpg/Nei family. Numbers associated with edges indicate bootstrap support percentages, whereas numbers associated with terminal nodes are GenBank identifiers. Ellipses enclose bacterial clades, the triangle encloses the plant/fungi clade, and the rectangle encloses sequences here designated Nei or NEIL. Dha and Bth represent Desulfitobacterium hafniense and Bacteroides thetaiotaomicron, respectively. GenBank accession nos. are as follows: Desulfitobacterium hafniense (23111783, 23113033, 23113370, and 23117013) and Bacteroides thetaiotaomicron VPI-5482 (29347769 and 29349896).
There is modest bootstrap support (76%) for a clade that contains the NEIL proteins as well as EcoNei, designated here as the Nei clade (Fig. 2, rectangle). Such a Nei clade is inconsistent with monophyly of proteins that do not coordinate zinc, suggesting that zinc coordination was lost independently in lineages leading to NEIL1 and to plants/fungi. The loss of metal binding in evolution is rare, although not unprecedented. Methionine sulfoxide reductases are divided into two classes that differ by the presence of zinc. Evolutionary analyses suggest that the metal was lost in Form 2 enzymes later in evolution (55). In addition, de novo protein design of a canonical zinc-finger motif (56) led to the engineering of peptides that faithfully reproduced the ββα architecture of the classical zinc finger, in the absence of metal ion (57, 58).
Superposition of NEIL1 onto EcoNei and TthFpg illustrates the structural similarity among the three enzymes (Fig. 3). There are nonetheless significant differences: A segment comprising αH and the loop connecting αG and αH in NEIL1 corresponds to a region that has been shown to be disordered in the borohydride trapped covalent Nei–DNA (20) and Fpg–DNA (22, 24) complexes or complexes with DNA containing an abasic site (23, 24), whereas it is ordered in uncomplexed TthFpg (21). The difference, however, is not just due to the presence or absence of DNA, but seems to also hinge on the presence of a nucleobase lesion. The recent crystal structure of Bacillus stearothermophilus Fpg with lesion-containing DNA showed that this flexible loop, which is positioned to recognize 8-oxoguanine, is ordered (see discussion below) (59). The structure overlay also revealed a 19-residue insertion in human NEIL1 (residues 204–222), which comprises helix αF (Figs. 1 and 7). This insertion is unique to NEIL1, which has only been found in vertebrates, and might play a part in the interaction with protein partners, such as DNA ligase III, Pol β, or XRCC1 (7).
Previous sequence alignments had predicted that NEIL1 would lack a zinc-finger motif because of the absence of the canonical zinc-binding residues in the region after the H2TH motif. The highly positively charged C-terminal region in NEIL1 had been posited to play a part in binding DNA. Our structure revealed that NEIL1 does in fact contain a structural motif that mimics an antiparallel β-hairpin zinc finger. The two loops that ligate the zinc atom in bacterial Fpg and Nei are absent, but the two β-strands (β9 and β10) superimpose quite well onto those of EcoNei and TthFpg (Fig. 3B). Two other Fpg/Nei glycosylases predicted to lack the characteristic zinc-finger motif, A. thaliana Fpg and C. albicans Fpg, share some sequence similarity with NEIL1; in particular, they exhibit sequence features consistent with a zincless finger, including the conserved arginine.
The crystal structure of unliganded EcoNei was reported to be in an open conformation, differing from that of the DNA-bound complex of the same enzyme by an angle of about 50° (12). Interestingly, the crystal structure of uncomplexed NEIL1 reported here is in the “closed” conformation and is superimposable onto either DNA-bound EcoNei (20) or unliganded TthFpg (21) structures (Fig. 3A), in accordance with a previous study that described that no conformational change accompanies substrate binding in Fpg (24). We note that our structural superpositions could be complicated by the fact that they compare homologous enzymes from different species. A detailed comparison of domain movements in NEIL1 upon substrate binding will have to await the structure of the enzyme bound to its DNA substrate.
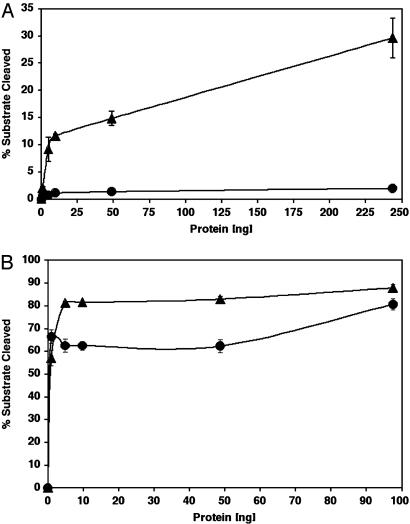
A Model for DNA Binding. In the absence of a NEIL1–DNA complex, the DNA from the EcoNei covalent complex can be superimposed onto the NEIL1 structure (20). The overlay showed that minor adjustments of protein side chains would be required to accommodate the DNA (Fig. 4). The superposition places the DNA in proximity of Pro-2, the catalytic N-terminal proline; Met-81, a residue that would fill the void created by the eversion of the damaged base; and two DNA binding motifs: the H2TH and zincless finger motifs. The H2TH motif (helices C and D in NEIL1) is characteristic of the Fpg/Nei family. In fact, a search for similar folds revealed that it is only found in members of the Fpg/Nei family (12). In EcoNei, residues located in the loop of the H2TH motif contact the phosphate of the lesion and the two phosphates on either side (20). The other DNA binding motif in the C-terminal region of enzymes of the Fpg/Nei family is the zinc finger, a motif absolutely required for DNA binding: When the cysteine residues that coordinate the zinc atom in the zinc finger of EcoFpg are mutated, DNA binding is ablated (60, 61). In EcoNei, the DNA binding region of the zinc finger motif is concentrated in the loop connecting the two β-strands (20). Our structural alignment (Fig. 7) combined with sequence alignments indicates that Arg-277, a residue located in the loop connecting the two β-strands of the zincless finger, is very highly conserved among the Fpg/Nei family members, including NEIL1. The guanidinium group of the corresponding arginine in EcoNei, Arg-253, contacts the phosphates on both sides of the DNA lesion (20). R253A was shown to be defective in cleaving 5,6-dihydrouracil-containing DNA. Its activity on abasic sites, on the other hand, was close to wild type (20). Arg-277 in NEIL1CΔ56 was similarly mutated to alanine: The protein variant showed a marked decrease in glycosylase activity (Fig. 5A), whereas the lyase activity was similar to that of wild type (Fig. 5B). Site-directed mutagenesis thereby confirms the importance of Arg-277 and that of the zincless finger in the glycosylase activity of NEIL1.
Fig. 5.
Activities of NEIL1CΔ56 wild type (▴) and the NEIL1CΔ56 R277A mutant (•). (A) DNA glycosylase/lyase activity on 5,6-dihydrouracil-containing, double-stranded oligonucleotides. (B) Lyase activity on a substrate containing an abasic site.
Lesion Recognition. NEIL1 has been shown to have a substrate specificity similar to that of Nei, because oxidized pyrimidines are better substrates than 8-oxoguanine (32). The ring-saturated pyrimidines thymine glycol (both 5R and 5S), dihydrothymine, and dihydrouracil; the oxidized pyrimidines 5-hydroxyuracil and 5-hydroxycytosine; formamidopyrimidines; the ring fragmentation product urea; and abasic sites are all good substrates for NEIL1 (30, 32, 35, 36). High-resolution structural information is not yet available for bacterial Nei complexed with lesion-containing DNA. Recent crystal structures of B. stearothermophilus Fpg in complex with DNA containing either 8-oxoguanine or dihydrouracil revealed that the lesion is recognized by residues located in the αF-β10 loop (for Fpg nomenclature, see ref. 59). It was further suggested that the mobility of the loop might play a part in lesion recognition and catalysis (59). Interestingly, as mentioned above, in our structure of unliganded NEIL1, the corresponding segment is ordered similarly to what was described for uncomplexed TthFpg, although this region in NEIL1 is composed of a helix and a loop (helix αH and loop preceding it), rather than a loop. Superposition of NEIL1 onto the B. stearothermophilus Fpg dihydrouracil-containing DNA complex shows that NEIL1 not only lacks the αF-β10 loop but that there are no residues in the vicinity of the modeled dihydrouracil lesion that might explain the enzyme's preference for this substrate. It is plausible that upon NEIL1's binding to a DNA substrate, a conformational change occurs that brings protein residues in contact with the lesion-containing strand. A model for damaged base recognition by NEIL1 will have to await the structure of a complex with DNA containing an oxidized pyrimidine.
Supplementary Material
Acknowledgments
We thank Wendy Cooper for technical assistance, Drs. Mark Rould and Scott Morrical for helpful discussions, and Jeffrey Blaisdell and Dr. Mark Rould for critical reading of the manuscript. This work was supported by National Institutes of Health Grant R37 CA33657 (to S.S.W.), an award to the University of Vermont under the Howard Hughes Medical Institute Biomedical Research Support Program for Medical Schools, an initiative in structural and computational biology funded by the U.S. Department of Energy Experimental Program to Stimulate Competitive Research, the Vermont Cancer Center, and the Vermont Genetics Network. S.D. is a Pew Scholar in the biomedical sciences.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: H2TH, helix-two-turns-helix; Nth, endonuclease III; Nei, endonuclease VIII; NEIL, Nei-like; EcoNei, Escherichia coli Nei; EcoFpg, Escherichia coli Fpg; SeMet, selenomethionyl; TthFpg, Thermus thermophilus Fpg.
Data deposition: The coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1TDH).
References
- 1.Wallace, S. S. (1998) Radiat. Res. 150, Suppl., S60–S79. [PubMed] [Google Scholar]
- 2.Wallace, S. S. (2002) Free Radical Biol. Med. 33, 1–14. [DOI] [PubMed] [Google Scholar]
- 3.Wilson, D. M., III, Sofinowski, T. M. & McNeill, D. R. (2003) Front. Biosci. 8, 963–981. [DOI] [PubMed] [Google Scholar]
- 4.Krokan, H. E., Standal, R. & Slupphaug, G. (1997) Biochem. J. 325, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCullough, A. K., Dodson, M. L. & Lloyd, R. S. (1999) Annu. Rev. Biochem. 68, 255–285. [DOI] [PubMed] [Google Scholar]
- 6.Wallace, S. S., Bandaru, V., Kathe, S. & Bond, J. P. (2003) DNA Repair 2, 441–453. [DOI] [PubMed] [Google Scholar]
- 7.Izumi, T., Wiederhold, L. R., Roy, G., Roy, R., Jaiswal, A., Bhakat, K. K., Mitra, S. & Hazra, T. K. (2003) Toxicology 193, 43–65. [DOI] [PubMed] [Google Scholar]
- 8.Jiang, D., Hatahet, Z., Blaisdell, J. O., Melamede, R. J. & Wallace, S. S. (1997) J. Bacteriol. 179, 3773–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito, Y., Uraki, F., Nakajima, S., Asaeda, A., Ono, K., Kubo, K. & Yamamoto, K. (1997) J. Bacteriol. 179, 3783–3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace, S. S., Harrison, L., Jiang, D., Blaisdell, J. O., Purmal, A. A. & Hatahet, Z. (1999) in Advances in DNA Damage and Repair: Oxygen Radical Effects, Cellular Protection, and Biological Consequences, eds. Dizaroglu, M. & Karakaya, A. E. (Plenum, New York), Vol. A302, pp. 419–430. [Google Scholar]
- 11.Blaisdell, J. O., Hatahet, Z. & Wallace, S. S. (1999) J. Bacteriol. 181, 6396–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zharkov, D. O., Shoham, G. & Grollman, A. P. (2003) DNA Repair 2, 839–862. [DOI] [PubMed] [Google Scholar]
- 13.Zharkov, D. O., Rieger, R. A., Iden, C. R. & Grollman, A. P. (1997) J. Biol. Chem. 272, 5335–5341. [DOI] [PubMed] [Google Scholar]
- 14.Sidorkina, O. M. & Laval, J. (2000) J. Biol. Chem. 275, 9924–9929. [DOI] [PubMed] [Google Scholar]
- 15.Rieger, R. A., McTigue, M. M., Kycia, J. H., Gerchman, S. E., Grollman, A. P. & Iden, C. R. (2000) J. Am. Soc. Mass Spectrom. 11, 505–515. [DOI] [PubMed] [Google Scholar]
- 16.Bailly, V., Verly, W. G., O'Connor, T. & Laval, J. (1989) Biochem. J. 262, 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connor, T. R. & Laval, J. (1989) Proc. Natl. Acad. Sci. USA 86, 5222–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graves, R. J., Felzenszwalb, I., Laval, J. & O'Connor, T. R. (1992) J. Biol. Chem. 267, 14429–14435. [PubMed] [Google Scholar]
- 19.Jiang, D., Hatahet, Z., Melamede, R. J., Kow, Y. W. & Wallace, S. S. (1997) J. Biol. Chem. 272, 32230–32239. [DOI] [PubMed] [Google Scholar]
- 20.Zharkov, D. O., Golan, G., Gilboa, R., Fernandes, A. S., Gerchman, S. E., Kycia, J. H., Rieger, R. A., Grollman, A. P. & Shoham, G. (2002) EMBO J. 21, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugahara, M., Mikawa, T., Kumasaka, T., Yamamoto, M., Kato, R., Fukuyama, K., Inoue, Y. & Kuramitsu, S. (2000) EMBO J. 19, 3857–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilboa, R., Zharkov, D. O., Golan, G., Fernandes, A. S., Gerchman, S. E., Matz, E., Kycia, J. H., Grollman, A. P. & Shoham, G. (2002) J. Biol. Chem. 277, 19811–19816. [DOI] [PubMed] [Google Scholar]
- 23.Serre, L., Pereira de Jesus, K., Boiteux, S., Zelwer, C. & Castaing, B. (2002) EMBO J. 21, 2854–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromme, J. C. & Verdine, G. L. (2002) Nat. Struct. Biol. 9, 544–552. [DOI] [PubMed] [Google Scholar]
- 25.Chung, M. H., Kasai, H., Jones, D. S., Innoue, H., Ishikawa, H., Ohtsuka, E. & Nishimura, S. (1991) Mutat. Res. 254, 1–12. [DOI] [PubMed] [Google Scholar]
- 26.Tchou, J., Kasai, H., Shibutani, S., Chung, M. H., Laval, J., Grollman, A. P. & Nishimura, S. (1991) Proc. Natl. Acad. Sci. USA 88, 4690–4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melamede, R. J., Hatahet, Z., Kow, Y. W., Ide, H. & Wallace, S. S. (1994) Biochemistry 33, 1255–1264. [DOI] [PubMed] [Google Scholar]
- 28.Purmal, A. A., Lampman, G. W., Bond, J. P., Hatahet, Z. & Wallace, S. S. (1998) J. Biol. Chem. 273, 10026–10035. [DOI] [PubMed] [Google Scholar]
- 29.Dizdaroglu, M., Burgess, S. M., Jaruga, P., Hazra, T. K., Rodriguez, H. & Lloyd, R. S. (2001) Biochemistry 40, 12150–12156. [DOI] [PubMed] [Google Scholar]
- 30.Hazra, T. K., Izumi, T., Boldogh, I., Imhoff, B., Kow, Y. W., Jaruga, P., Dizdaroglu, M. & Mitra, S. (2002) Proc. Natl. Acad. Sci. USA 99, 3523–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hazra, T. K., Kow, Y. W., Hatahet, Z., Imhoff, B., Boldogh, I., Mokkapati, S. K., Mitra, S. & Izumi, T. (2002) J. Biol. Chem. 277, 30417–30420. [DOI] [PubMed] [Google Scholar]
- 32.Bandaru, V., Sunkara, S., Wallace, S. S. & Bond, J. P. (2002) DNA Repair 1, 517–529. [DOI] [PubMed] [Google Scholar]
- 33.Morland, I., Rolseth, V., Luna, L., Rognes, T., Bjørås, M. & Seeberg, E. (2002) Nucleic Acids Res. 30, 4926–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dou, H., Mitra, S. & Hazra, T. K. (2003) J. Biol. Chem. 278, 49679–49684. [DOI] [PubMed] [Google Scholar]
- 35.Miller, H., Fernandes, A. S., Zaika, E., McTigue, M. M., Torres, M. C., Wente, M., Iden, C. R. & Grollman, A. P. (2004) Nucleic Acids Res. 32, 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katafuchi, A., Nakano, T., Masaoka, A., Terato, H., Iwai, S., Hanaoka, F. & Ide, H. (2004) J. Biol. Chem. 279, 14464–14471. [DOI] [PubMed] [Google Scholar]
- 37.Takao, M., Kanno, S. I., Shiromoto, T., Hasegawa, R., Ide, H., Ikeda, S., Sarker, A. H., Seki, S., Xing, J. Z., Le, X. C., et al. (2002) EMBO J. 21, 3486–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elder, R. H. & Dianov, G. L. (2002) J. Biol. Chem. 277, 50487–50490. [DOI] [PubMed] [Google Scholar]
- 39.Ocampo, M. T., Chaung, W., Marenstein, D. R., Chan, M. K., Altamirano, A., Basu, A. K., Boorstein, R. J., Cunningham, R. P. & Teebor, G. W. (2002) Mol. Cell. Biol. 22, 6111–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takao, M., Kanno, S. I., Kobayashi, K., Zhang, Q. M., Yonei, S., van der Horst, G. T. J. & Yasui, A. (2002) J. Biol. Chem. 277, 42205–42213. [DOI] [PubMed] [Google Scholar]
- 41.Rosenquist, T. A., Zaika, E., Fernandes, A. S., Zharkov, D. O., Miller, H. & Grollman, A. P. (2003) DNA Repair 2, 581–591. [DOI] [PubMed] [Google Scholar]
- 42.Bandaru, V., Cooper, W., Wallace, S. S. & Doublié, S. (2004) Acta Crystallogr. D 60, 1142–1144. [DOI] [PubMed] [Google Scholar]
- 43.Dauter, Z. & Dauter, M. (2001) Structure (London) 9, R21–R26. [DOI] [PubMed] [Google Scholar]
- 44.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 45.Tchou, J. & Grollman, A. P. (1995) J. Biol. Chem. 270, 11671–11677. [DOI] [PubMed] [Google Scholar]
- 46.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Terwilliger, T. C. (2002) Acta Crystallogr. D 59, 34–44. [Google Scholar]
- 48.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 49.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Felsenstein, J. (1989) Cladistics 5, 164–166. [Google Scholar]
- 52.Evans, S. V. (1993) J. Mol. Graphics 11, 134–138. [DOI] [PubMed] [Google Scholar]
- 53.Holm, L. & Sander, C. (1998) Nucleic Acids Res. 26, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strimmer, K. & von Haeseler, A. (1996) Mol. Biol. Evol. 13, 964–969. [Google Scholar]
- 55.Kumar, R. A., Koc, A., Cerny, R. L. & Gladyshev, V. N. (2002) J. Biol. Chem. 277, 37527–37535. [DOI] [PubMed] [Google Scholar]
- 56.Miller, J., McLachlan, A. D. & Klug, A. (1985) EMBO J. 4, 1609–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Struthers, M. D., Cheng, R. P. & Imperiali, B. (1996) Science 271, 342–345. [DOI] [PubMed] [Google Scholar]
- 58.Dahiyat, B. I. & Mayo, S. L. (1997) Science 278, 82–87. [DOI] [PubMed] [Google Scholar]
- 59.Fromme, J. C. & Verdine, G. L. (2003) J. Biol. Chem. 278, 51543–51548. [DOI] [PubMed] [Google Scholar]
- 60.O'Connor, T. R., Graves, R. J., de Murcia, G., Castaing, B. & Laval, J. (1993) J. Biol. Chem. 268, 9063–9070. [PubMed] [Google Scholar]
- 61.Tchou, J., Michaels, M. L., Miller, J. H. & Grollman, A. P. (1993) J. Biol. Chem. 268, 26738–26744. [PubMed] [Google Scholar]
- 62.Kabsch, W. & Sander, C. (1983) Biopolymers 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.