Abstract
Acrp30/adiponectin is reduced in the serum of obese and diabetic individuals, and the genetic locus of adiponectin is linked to the metabolic syndrome. Recombinant adiponectin, administered to diet-induced obese mice, induced weight loss and improved insulin sensitivity. In muscle and liver, adiponectin stimulates AMP-activated protein kinase activation and fatty acid oxidation. To expression-clone molecules capable of binding adiponectin, we transduced a C2C12 myoblast cDNA retroviral expression library into Ba/F3 cells and panned infected cells on recombinant adiponectin linked to magnetic beads. We identified T-cadherin as a receptor for the hexameric and high-molecular-weight species of adiponectin but not for the trimeric or globular species. Only eukaryotically expressed adiponectin bound to T-cadherin, implying that posttranslational modifications of adiponectin are critical for binding. An adiponectin mutant lacking a conserved N-terminal cysteine residue required for formation of hexamer and high-molecular-weight species did not bind T-cadherin in coimmunoprecipitation studies. Although lacking known cellular functions, T-cadherin is expressed in endothelial and smooth muscle cells, where it is positioned to interact with adiponectin. Because T-cadherin is a glycosylphosphatidylinositol-anchored extracellular protein, it may act as a coreceptor for an as-yet-unidentified signaling receptor through which adiponectin transmits metabolic signals.
Adipose tissue is not only a storage depot for lipid but also a regulator of metabolism, through hormones known as adipokines. One adipokine is adiponectin (also denoted Acrp30, for adipocyte complement-related protein of 30 kDa), a molecule secreted exclusively by differentiated adipocytes (1). Adiponectin, which has homology to C1q, is found in the serum as three distinct oligomers, namely trimer, hexamer, and a high-molecular-weight (HMW) species (2). Adiponectin levels are decreased in the serum of obese and diabetic people (3) and animal models of obesity and diabetes. Replenishment by any of several methods induces weight loss and correction of insulin resistance (4–6). The mechanisms by which adiponectin influences metabolism are not fully understood but involve increasing fatty-acid oxidation in muscle through AMP-activated protein kinase (AMPK) activation, as well as synergizing with insulin in the liver to increase glycogen stores and to inhibit gluconeogenesis (6, 7). In tissue culture and isolated muscle, the trimeric isoform and a trimeric globular C-terminal subdomain activate AMPK (7, 8), whereas the hexamer and HMW isoforms activate NF-κB (9). Adiponectin also has been implicated in the inflammatory process of the metabolic syndrome, and reduced adiponectin levels have been correlated with impaired forearm blood flow, possibly linking endothelial dysfunction with adiponectin levels (10).
Analysis of the transmembrane pathways linking adiponectin to downstream signaling events has yielded conflicting results. Two recently described receptors that bind adiponectin (AdipoR1 and AdipoR2) (11) are distantly related to the family of seven-transmembrane spanning G protein-coupled receptors but have an inverted topology with the N terminus intracellular, which is distinct from other seven-transmembrane spanning receptors. In addition, the extracellular portion of these molecules is small, distinct from members of this class of receptors that bind peptide hormones.
To study how the biological activities of adiponectin are transmitted, we have performed a series of expression-cloning studies to identify cell-surface molecules capable of binding adiponectin, using a magnetic-bead panning method that may present higher-valency forms of the adiponectin ligand.
Methods
Cell Culture. C2C12 mouse myoblast and human embryonic kidney (HEK) cells were grown in DMEM with 10% FBS. Chinese hamster ovary (CHO) cells engineered to express the ecotropic retrovirus receptor (CHO-ER) were a gift of M. Krieger (Massachusetts Institute of Technology) and were grown in F12 medium with 10% FBS. Ba/F3 cells were grown in RPMI medium 1640 with 10% FBS and 5% Walter and Eliza Hall Institute-conditioned cell medium (12). Medium was from GIBCO.
Generation of Recombinant Proteins and Immunoprecipitation. The mouse adiponectin cDNA in the vector pcDNA (9) was modified by PCR mutagenesis to insert the Flag epitope (DYKDDDDK) between the signal sequence cleavage site and the N-terminal variable region; this construct was denoted 5′-Flag-Acrp30. The Flag epitope was inserted in a similar manner immediately before the stop codon of the adiponectin cDNA to generate a C-terminally tagged protein, 3′-Flag-Acrp30. The construct 5′-Flag-C22A-Acrp30, in which cysteine-22 is replaced by alanine, was generated by subcloning the 3′-Nhe-1 fragment of C22A-Acrp30 (2) into similarly digested 5′-Flag-Acrp30. To generate recombinant protein, plasmids were transiently transfected into HEK cells, and conditioned medium was generated as described in ref. 9. These supernatants were used either for fluorescence-activated cell-sorter (FACS) binding assays or further purified by ammonium sulfate precipitation followed by Hi-Q anion-exchange chromatography (Bio-Rad). After elution of 5′-Flag-Acrp30 by salt gradient, native gel electrophoresis and immunoblotting for the Flag epitope indicated resolution of the expressed protein into two species: the first contained predominantly trimeric protein, whereas the second contained hexameric and higher-molecular-weight 5′-Flag-Acrp30 (9). The purity of these preparations was judged to be ≈50% by SDS/PAGE and Coomassie staining (data not shown). Bacterially expressed globular or full-length adiponectin, tagged at the C terminus with the Flag epitope, was generated as described in ref. 9. Endotoxin was removed with an endotoxin-removal column (Pierce).
Coimmunoprecipitation of 5′-Flag-Acrp30 or 5′-Flag-C22A-Acrp30 and hemagglutinin (HA)-T-cadherin was performed in HEK cells transfected with single or double combinations of plasmids (six-well plates with 2 μg of plasmid per well). Medium was changed 1 day after transfection. One day later, the cells were washed once in ice-cold PBS++ (PBS with 0.9 mM CaCl2 and 0.5 mM MgCl2) and lysed on ice in 150 mM NaCl/50 mM Tris (pH 7.5)/0.9 mM CaCl2/0.5 mM MgCl2/0.1 mM PMSF/1 mM benzamidine/1% Triton X-100, transferred to microfuge tubes, and centrifuged at 15,000× g for 10 min. Supernatants were immunoprecipitated on anti-Flag resin, resolved on duplicate 4–12% gradient SDS/PAGE under reducing conditions by using the Mes buffer system (Invitrogen), and analyzed by immunoblotting with horseradish peroxidase-conjugated antibodies specific for the HA or Flag epitope in a similar manner to that described in ref. 12.
FACS Binding Assay. Adherent cells (C2C12 and CHO) or suspension cells (Ba/F3) were transferred to serum-free medium for 2 h and then incubated at 4°C for 30 min to block endocytosis. The cells were incubated in 1% bovine serum albumin (BSA) with PBS++ at 4°C for 30 min to block nonspecific binding sites. All subsequent steps were at 4°C. The cells were incubated for 2 h in conditioned serum-free medium from HEK cells transiently transfected with different expression constructs diluted with an equal volume of blocking solution, washed twice in PBS++, and incubated for 1 h with allophycocyanin (APC)-conjugated anti-Flag Ab diluted 1:100 in blocking solution (Prozyme, San Leandro, CA). After washing twice in PBS++, adherent cells were removed by scraping, resuspended in PBS containing 10% FBS and 0.5 μg/ml propidium iodide (Sigma), and analyzed by FACS (Becton Dickinson). The median fluorescence of live, propidium iodide-negative cells was determined.
Preparation of Magnetic Beads. Activated magnetic beads (M280, Dynal, Great Neck, NY) were incubated with anti-Flag Ab (M2, Sigma). After coupling, the beads were blocked in a solution of PBS++ containing 1% BSA for 1 h, added to 40 ml of conditioned medium from HEK cells transfected with 5′-Flag-Acrp30, and incubated overnight at 4°C. Control beads were incubated with the supernatants of HEK cells transfected with the empty vector pcDNA. After binding to the supernatants, the beads were washed twice in PBS++ containing 0.1% BSA and stored in the same buffer.
C2C12 cDNA Library Construction. An undifferentiated C2C12 cDNA expression library was made in the bicistronic retroviral vector pBI-GFP by using standard techniques (13). Characterization of the unamplified plasmid library revealed ≈0.5 × 107 independent transformants, and 95% contained inserts with an average size of 1.5 kb. Infectious cDNA viral particles were produced by transfection of the plasmid library into a packaging cell line (14). The resulting virus-containing supernatant was used to transduce ≈5% of a population of 2 × 108 naive Ba/F3 cells, as determined by FACS analysis of GFP expression.
Magnetic Bead Panning. Transduced cells were expanded for 2 days before being subjected to binding on the magnetic beads. The cells were prepared as described for the FACS analysis with the addition of 0.1 mg/ml mouse IgG (Sigma) in the blocking step; all subsequent steps were carried out at 4°C or on ice. The cells were precleared by incubating 30 ml of cells (2.4 × 107 per ml) with 30 μl of control magnetic beads for 1 h. Bound cells were separated by use of a magnet, and nonadherent cells were subjected to two additional rounds of binding to control beads. Nonadherent cells were incubated with 75 μl of 5′-Flag-Acrp30 beads for 1 h, after which adherent cells were separated with a magnet and washed three times for 5 min each in 10 ml of PBS++ with 0.1% BSA. The bound cells were expanded in culture. In subsequent rounds of binding, preclearing was performed twice with 15 μl of control beads before adding 30 μl of 5′-Flag-Acrp30 beads. Aliquots of cells were analyzed by FACS for GFP expression after each round of binding and expansion in culture.
Amplification of Enriched cDNA Insert. Genomic DNA was prepared from the cell pools after the third sort and subjected to PCR amplification by using retroviral-specific primers flanking the cDNA cloning site of the retroviral vector pBI-GFP. PCR products were separated by agarose gel electrophoresis and visualized with EtdBr staining. Specific bands were excised, eluted (QIAquick, Qiagen, Valencia, CA), subcloned in the vector pTOPO-II (Invitrogen), and sequenced.
Overexpression of T-Cadherin in Cells. A full-length mouse T-cadherin cDNA clone (IMAGE ID 3987627) was subcloned in the vector pcDNA to generate pcDNA-Tcad. The insert from this clone was used for subsequent manipulations. By using PCR mutagenesis, the coding sequence for the HA epitope (YPYD-VPDYA) was inserted after the predicted signal sequence of T-cadherin after amino acid residue 24. The unmodified cDNA was cloned into the retroviral vector pBI-GFP to generate the construct pBI-GFP-Tcad, and infectious transducing viral particles were generated as described in ref. 12. Naive Ba/F3 and CHO-ER cells were transduced with this construct to generated Ba/F3-GFP-Tcad and CHO-GFP-Tcad. Cell pools expressing T-cadherin were generated by FACS sorting of cell populations expressing high GFP levels. Control cells (CHO-GFP) were generated by infection of naive cells with retroviral supernatants encoding pBI-GFP alone.
Plate Binding Assay to Detect Adiponectin and T-Cadherin Interactions. CHO-GFP-Tcad and CHO-GFP were plated (1.5 × 104 per well) in 96-well plates (Becton Dickinson). The next day, the cells were incubated in serum-free medium for 1 h, placed at 4°C for 30 min, and blocked in PBS++ containing 4% dried milk for 30 min. All subsequent steps were at 4°C. Bacterially expressed 3′-Flag-tagged globular adiponectin, bacterially expressed 3′-Flag-tagged full-length adiponectin, and eukaryotically expressed trimeric or hexameric and HMW 5′-Flag-Acrp30 produced in HEK cells were incubated with the cells for 1 h in blocking buffer at the indicated concentrations. The protein concentration of partially purified eukaryotic protein was determined by BSA assay (Pierce), and the concentration of purified bacterially expressed proteins was determined by the absorbance at 280 nm and the calculated extinction coefficient (DNASTAR, Madison, WI). After washing twice in PBS++, 4 μg/ml anti-Flag M2 Ab was added for 1 h in blocking solution, two washes were performed with PBS ++, and then an incubation with secondary Ab (horseradish peroxidase-donkey anti-mouse; Jackson ImmunoResearch) diluted 1:1,000 in blocking solution and developed with colorimetric 3,3′,5,5′-tetramethylbenzidine substrate (Pierce). The absorbance at 450 nm was measured and plotted versus the concentration of added ligand, expressed as the concentration of trimer species.
Results
Eukaryotic Production of Adiponectin. A plasmid containing the entire coding sequence for adiponectin was modified by PCR to insert the Flag epitope at either the N or C terminus of the mature polypeptide, 5′-Flag-Acrp30 and 3′-Flag-Acrp30, respectively. After transient transfection of these constructs into HEK cells, recombinant protein accumulated at high concentration in the tissue culture supernatant, as determined by immunoblotting of conditioned medium with antibodies specific either for the Flag epitope or the globular domain of adiponectin (data not shown). These supernatants were used either for FACS binding assays as described below or further partially purified by ammonium sulfate precipitation followed by anion-exchange chromatography. After chromatography, native gel electrophoresis showed resolution of the expressed protein detected by immunoblotting for the Flag epitope into two species: the first contained predominantly trimeric 5′-Flag-Acrp30, whereas the second contained hexameric and HMW 5′-Flag-Acrp30. The purity of these preparations was judged to be ≈50% by SDS/PAGE and Coomassie staining (data not shown).
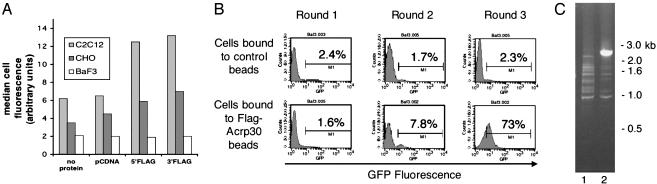
Adiponectin Specifically Binds to C2C12 Cells. As demonstrated in Fig. 1A, Ba/F3 cells did not bind substantially to either 5′-Flag-Acrp30 (lane 9) or 3′-Flag-Acrp30 (lane 12); the signal obtained was similar to that obtained with either control transfected supernatants (lane 6) or when only blocking solution (lane 3) was added. CHO cells yielded a slightly greater signal when incubated in conditioned medium containing either 5′-Flag-Acrp30 (lane 8) or 3′-Flag-Acrp30 (lane 11), compared with the control of only blocking protein (lane 3) or control transfected supernatant (lane 5). C2C12 cells, which respond to adiponectin, demonstrated a 2-fold increase in signal when cells were incubated in conditioned medium containing either 5′-Flag-Acrp30 (lane 7) or 3′-Flag-Acrp30 (lane 10), compared with blocking solution (lane 1) or control transfected supernatant (lane 4). These results indicate that C2C12 cells, but not Ba/F3 cells, specifically bind adiponectin.
Fig. 1.
Cloning of T-cadherin as an adiponectin receptor. (A) FACS binding assay of tissue culture cells by using unpurified tissue culture supernatants containing Flag-tagged adiponectin. Adherent (C2C12, CHO) or suspension (Ba/F3) cells were incubated with control blocking solution or supernatants from mock vector-transfected HEK cells (left two columns) or unpurified cell supernatants containing 5′-or3′-FlagAcrp30 (right two columns; protein as depicted). Bound protein was detected by incubating cells with an APC-conjugated anti-Flag mAb and analyzed by FACS. Live cells were identified by exclusion of propidium iodide staining. The average of the median APC staining for each sample is shown; only C2C12 cells, and not CHO or Ba/F3 cells, demonstrate binding (n = 2). (B) FACS analysis of GFP expression of sequentially enriched pools of Ba/F3 cells transduced with a retroviral C2C12 cDNA expression library coexpressing GFP from an internal ribosome entry site. Naive Ba/F3 cells were infected with the cDNA library and enriched for binding to anti-Flag magnetic beads previously incubated with either 5′-Flag-Acrp30 or mock vector-transfected HEK cell supernatants. Three rounds of enrichment were performed as described in the text. After each round of binding and expansion of adherent cells, aliquots of cells were analyzed for GFP expression by FACS. After each round of sorting, the percentages of cells expressing GFP are indicated in the figure, compared with uninfected cells. (C) Genomic PCR amplification of integrated proviral cDNA insert of enriched Ba/F3 cell pools. Genomic DNA was prepared after the third round of enrichment from control bead-enriched Ba/F3 cells (lane 1; B Upper Right) or 5′-Flag-Acrp30 bead-enriched Ba/F3 cells (lane 2; B Lower Right). Primer pairs flanking the cDNA cloning site of the parental vector pBI-GFP were used to amplify the integrated proviral cDNA. A single 2.5-kb band was amplified from cells enriched for binding to 5′-Flag-Acrp30 beads, as shown by agarose gel electrophoresis and EtBr staining. Standards are indicated in kb.
Expression Cloning of T-Cadherin by Panning Transduced Ba/F3 Cells on Adiponectin-Linked Magnetic Beads. We used anti-Flag Ab covalently linked to magnetic beads to immunopurify 5′-Flag-Acrp30 from conditioned HEK medium; in parallel, we made control beads by incubating anti-Flag coupled magnetic beads with the supernatants of HEK cells transfected with the empty vector pcDNA. We constructed a cDNA expression library in the bicistronic retroviral vector pBI-GFP by using mRNA from undifferentiated C2C12 cells. This vector contains the coding sequence of GFP under the control of an internal ribosome entry site (15). The cDNA expression library was transduced into naive Ba/F3 cells. Transduced cells were expanded for 2 days before being subjected to binding on the magnetic beads. The bound cells were expanded in culture and repanned in a similar method three times. After each round of binding and expansion, aliquots of cells were analyzed by FACS for GFP expression to follow enrichment of cells containing an integrated retrovirus. Enrichment in GFP expression in a cell population after binding to 5′-Flag-Acrp30 beads would indicate enrichment of a cDNA that conferred binding (16). As shown in Fig. 1B, after one round of binding to the beads, 2.4% of cells purified on control beads and 1.6% of cells purified on 5′-Flag-Acrp30 beads were GFP-positive, relative to uninfected cells (Fig. 1B). After the second round of binding, 1.7% of cells purified on control beads but 7.8% of cells purified on 5′-Flag-Acrp30 beads were GFP-positive (Fig. 1B). By the third sort, cells binding to control beads had not enriched compared with the first sort, 2.3% being GFP-positive. However, 73% of cells binding to the 5′-Flag-Acrp30 beads were GFP-positive, indicating that the population had enriched for cells containing an integrated retroviral cDNA clone (Fig. 1B). Thus the enriched cell population likely contains a cDNA(s) encoding a receptor for adiponectin. Genomic DNA was prepared from the different cell pools after the third sort and subjected to PCR amplification by using retroviral-specific primers flanking the cDNA cloning site of the retroviral vector pBI-GFP. In cells incubated with 5′-Flag-Acrp30-containing beads, but not with control beads, a single specific band of 2.5 kb was seen (Fig. 1C). We subcloned and sequenced this band, which encoded a full-length cDNA identical to mouse T-cadherin (GenBank accession no. BC021628).
Binding of Adiponectin to T-Cadherin. To confirm that T-cadherin binds adiponectin, we made stable cell lines overexpressing T-cadherin. The T-cadherin cDNA was cloned into the retroviral vector pBI-GFP, and naive Ba/F3 and CHO-ER cells were transduced with infectious virus derived from this construct. Cell pools (Ba/F3-GFP-Tcad and CHO-GFP-Tcad) expressing T-cadherin were generated by FACS selection of highly GFP-expressing cell lines.
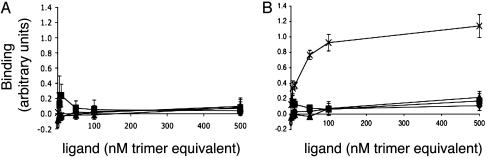
We used an ELISA to show direct binding of adiponectin to adherent cells expressing T-cadherin. Fig. 2A shows that CHO-ER cells infected with control vector expressing only GFP did not bind any of the tested proteins. As shown in Fig. 2B, CHO cells expressing T-cadherin bound only hexameric and HMW oligomers of 5′-Flag-Acrp30 but did not bind trimeric 5′-Flag-Acrp30. The estimated half-maximal binding concentration was 25 nM, expressed as adiponectin trimer equivalents. There was no binding of either globular or full-length bacterially produced protein to CHO cells expressing T-cadherin, implying that recognition of adiponectin by T-cadherin may require posttranslational modifications of adiponectin; additionally, the globular head of adiponectin is not sufficient for binding but may participate in conjunction with other domains. In separate experiments, 3′-Flag-Acrp30 produced in eukaryotic cells produced similar results (data not shown), indicating that the location of the epitope tag is not important for binding and confirming that only eukaryotic derived protein binds T-cadherin.
Fig. 2.
ELISA-based binding assay. Flag-tagged recombinant proteins were incubated with CHO-GFP cells (A) or CHO-GFP-Tcad-expressing cells (B). Bound protein was detected by subsequent binding of anti-Flag Ab and labeled secondary Ab. ×, HEK-produced Hexamer/HMW 5′-Flag-Acrp30; ▴, HEK-produced trimeric 5′-Flag-Acrp30; ♦, bacterially produced globular adiponectin 3′-Flag-tagged; ▪, bacterially produced full-length adiponectin 3′-Flag-tagged. The concentration of added protein is plotted vs. the average binding signal (arbitrary units) ± SD (n = 4).
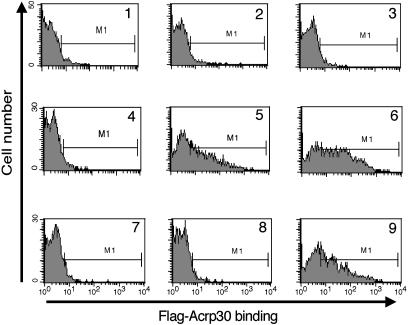
We used a FACS-based assay (Fig. 3) to show that 6 nM (graph 5) or 60 nM (graph 6) hexameric 5′-Flag-Acrp30 binds specifically to Ba/F3 cells expressing T-cadherin, although there is no detectable binding to control Ba/F3 cells (graphs 2 and 3). Background binding to either cell line in the absence of adiponectin is low (graphs 1 and 4). Graph 7 shows that the binding is completely competed by a 10-fold excess of untagged hexameric adiponectin. Addition of EDTA to the binding reaction completely blocks binding as well (graph 8); thus, we conclude that divalent cations are required for binding of T-cadherin and adiponectin. C1q, a molecule that shares homology with adiponectin, does not affect binding even at a 20-fold excess by weight (graph 9), implying that these molecules do not share receptors. In parallel experiments, there was significant binding neither of bacterially expressed Flag-tagged globular adiponectin nor of a preparation of trimeric mammalian-cell-produced 5′-Flag-Acrp30 to T-cadherin-expressing cell lines (data not shown), extending the results depicted in Fig. 2. These observations confirm that T-cadherin is not binding to the Flag epitope, thus ruling out a trivial explanation for the binding to 5′-Flag-Acrp30; additionally, the observations confirm that T-cadherin is a specific receptor for full-length hexameric and HMW forms of adiponectin but does not bind the globular or trimeric forms of adiponectin.
Fig. 3.
FACS-based binding assay. Cells (Ba/F3 or Ba/F3-GFP-Tcad) were incubated with or without Flag-tagged adiponectin; bound protein was detected by subsequent incubation with APC-labeled anti-Flag Ab and analyzed by FACS. Shown are histograms of APC fluorescence. Naive Ba/F3 cells (graphs 1–3) and Ba/F3-GFP-Tcad cells (graphs 4–9) were incubated without added protein (graphs 1 and 4), or 6 nM hexamer 5′-Flag-Acrp30 (graphs 2, 5, and 7–9) or 60 nM hexamer 5′-Flag-Acrp30 (graphs 3 and 6). The incubation reactions also included 60 nM hexameric untagged adiponectin (graph 7), 10 mM EDTA (graph 8), or a 20-fold excess of C1q by weight (graph 9).
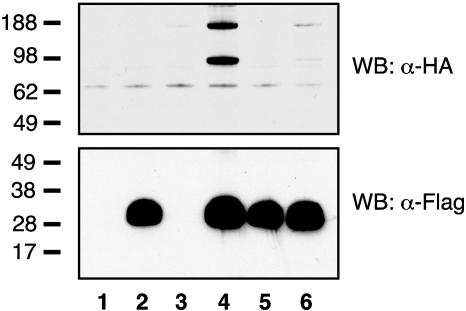
Lane 4 of Fig. 4 shows that after cotransfection of HEK cells with plasmids expressing HA-T-cadherin and 5′-Flag-Acrp30, HA-T-cadherin is immunoprecipitated from the Triton X-100 soluble lysate on anti-Flag resin (Upper) and migrates both as monomer and dimer under these conditions. Because HA-T-cadherin alone did not bind anti-Flag resin (lane 3, Upper), but 5′-Flag-Acrp30 expressed alone was efficiently immunoprecipitated (lane 2, Lower), we conclude that T-cadherin directly binds adiponectin. A mutant adiponectin lacking the N-terminal cysteine (5′-Flag-C22A-Acrp30) (2) and forming only the trimeric species is made in similar amounts as WT adiponectin and is efficiently immunoprecipitated on anti-Flag resin (lanes 2 and 5, Lower). In contrast with WT adiponectin, however, T-cadherin does not coimmunoprecipitate with this trimeric adiponectin mutant (lane 6, Upper); thus, we conclude that T-cadherin binds only to hexameric and HMW adiponectin.
Fig. 4.
Immunoprecipitation of cotransfected epitope-tagged T-cadherin and adiponectin. HEK cells were transfected with pcDNA alone (lane 1) or pcDNA expressing 5′-Flag-Acrp30 (lanes 2 and 4), 5′-Flag-C22A-Acrp30 (lanes 5 and 6), or HA-T-cadherin (lanes 3, 4, and 6). Forty-eight hours after transfection, aliquots of the Triton X-100 soluble cell lysates were immunoprecipitated on anti-Flag resin, electrophoresed on duplicate 4–12% polyacrylamide gels, and analyzed by immunobloting with anti-HA (Upper) or anti-Flag (Lower) mAbs conjugated to horseradish peroxidase. Standards are in kDa.
Discussion
By using expression cloning, we identified T-cadherin as a previously uncharacterized receptor for eukaryotically produced hexameric and HMW isoforms of adiponectin. This result provides insights into how adipose tissue may participate in the regulation of metabolism through binding of an adipokine to a receptor expressed on endothelial and muscle cells and establishes a framework to characterize other potential components of the adiponectin-signaling complex.
Relationship to the Metabolic Syndrome. Adiponectin, an adipokine initially identified in a screen for proteins secreted exclusively by differentiated adipocytes, is reduced in the serum of obese and diabetic individuals and has been proposed to participate in the development of the metabolic syndrome (17). The globular C-terminal domain and the trimeric forms of adiponectin activate AMPK and lead to increased fatty acid oxidation and reduction of serum glucose by several mechanisms (6–8), whereas the hexameric and HMW forms of adiponectin activate NF-κB pathways (9). Although the significance of the latter activity is unclear, the hexameric and HMW isoforms of adiponectin do inhibit apoptosis of endothelial cells (18). Here, NF-κB activity was not examined, but NF-κB has well described antiapoptotic activities (19).
There is sexual dimorphism in the distribution of adiponectin isoforms, with female mice having a greater fraction of HMW isoforms. Furthermore, HMW isoforms are preferentially increased after thiazolidinedione treatment (20) and after weight loss in obese individuals (18). Therefore, the finding that T-cadherin specifically binds the hexamer and HMW isoforms of adiponectin provides the first step in characterizing the activation of downstream signaling pathways.
The cellular mechanisms linking adiponectin to cellular signaling pathways are not understood. A report describing the expression cloning of the adiponectin receptors 1 and 2 involved bacterially produced protein preparations in which the oligomeric state of full-length adiponectin was uncharacterized (11). Subsequent to this report, similar sequences with homology to the proposed adiponectin receptors and to proteins functioning as progestin receptors (21) have been deposited and annotated in the GenBank database as the PAQR, or progestin/adiponectin/adipoQ receptor, family.
Bacterially expressed adiponectin did not bind T-cadherin, implying that eukaryotic posttranslational modifications of adiponectin are involved in the binding. This finding is consistent with results in which only eukaryotically expressed adiponectin was active in reducing hepatic glucose production (6); mutations in eukaryotically expressed adiponectin that removed conserved lysines critical for glycosylation had reduced activity (22). However, other modifications, such as proline hydroxylation of the collagen domain, may be involved in binding to T-cadherin. Because neither the trimeric species nor the globular C-terminal domain bound T-cadherin and both have activity in several assays measuring AMPK activity and fatty acid oxidation, there may be other proteins binding these isoforms of adiponectin. Alternatively, a serum reductase and a protease have been postulated to generate trimeric and globular adiponectin from hexameric and HMW isoforms (5, 23). T-cadherin may function as a receptor to present hexamer and HMW adiponectin sequentially to such processing and signaling molecules.
Cadherins Have Multiple Structures and Functions. The cadherins comprise a large family of cell-surface proteins involved in calcium mediated cell–cell interactions and signaling. With the exception of T-cadherin, all members of the cadherin superfamily contain a transmembrane domain, linking the extracellular portion of the molecule with intracellular signaling pathways. T-cadherin is anchored to the plasma membrane by a GPI anchor (24). Like classical or type-I cadherins, the extracellular portion of T-cadherin contains five ectodomains. Antibodies directed against ectodomain-1, which is most distal from the plasma membrane, block T-cadherin-mediated adhesion, implicating this portion of the molecule in cellular functions and possibly regulation of cell migration (25). The C-terminal GPI anchor binds lipoproteins, although the significance of this finding is uncharacterized (26). Whether cleavage of the GPI anchor is involved in regulating T-cadherin is unknown.
Divalent Cations Are Required for Adiponectin Binding to T-Cadherin. Binding of calcium at conserved residues contained within adjacent ectodomains of E-cadherin increases the rigidity of the molecule and promotes formation of dimers (27); calcium may have a similar role in T-cadherin structure. Notably, EDTA abrogated binding of adiponectin to T-cadherin, possibly by altering the structure of T-cadherin. Alternatively, the structure of adiponectin itself may depend on calcium because collagen X, a homologue of adiponectin, contains calcium cations chelated by residues that are conserved in the globular C-terminus of adiponectin, although the structure of adiponectin did not reveal bound calcium (28, 29).
T-Cadherin and Adiponectin Are Found in the Same Tissue Compartments. T-cadherin was initially described in the nervous system, but its tissue distribution is more widespread, with highest expression in the cardiovascular system and lower levels in muscle. In the vasculature, T-cadherin is localized to the intima and media and is expressed on endothelial and smooth muscle cells; expression is up-regulated in the neointima after injury of the mouse carotid artery by a balloon catheter (30, 31). The histochemical localization of T-cadherin is intriguing because it is similar to the localization of adiponectin. Adiponectin binds to collagens exposed during vessel-wall injury and localizes to injured vessel walls (32). Additionally, two of the three published adiponectin mouse gene knockouts have abnormal cellular proliferation in blood vessel walls after vessel injury, leading to a thickening of the vessel neointima by growth of smooth muscle cells (33, 34). This cellular proliferation is blocked by virus-mediated adiponectin expression (34). Interestingly, T-cadherin can regulate growth of cells in the nervous system (35). T-cadherin suppressed the growth of astrocytes and was reduced in a glioblastoma cell line (36), and reexpression of T-cadherin in this cell line induced growth arrest, which depended on p21 (CIP1/WAF1) expression. T-cadherin may participate in signaling through its association with other membrane proteins and incorporation into specific lipid domains of the cell membrane (37, 38), and it may restrict proliferation of the intima through its interactions with adiponectin. T-cadherin is not highly expressed in the hepatocyte, which is one of the major sites of activity of adiponectin in suppressing gluconeogenesis (6, 23); however, this finding does not exclude the participation of other cell types in the liver, such as fibroblast, endothelial, or blood-derived cells.
Acknowledgments
We thank Drs. C.-Z. Chen, L. Huang, B. Luo, B. Ranscht, J. Su, W. Tong, and G. Wong for discussion and advice. This work was supported in part by National Institutes of Health Grant R37DK47618 (to H.F.L.). C.H. was supported by a training grant from the National Institutes of Health to the Division of Respiratory Diseases at Children's Hospital and is the recipient of a Charles Hood Award from The Medical Foundation. J.S.B. is supported in part by a Career Development Award from the American Diabetes Association.
Abbreviations: AMPK, AMP-activated protein kinase; APC, allophycocyanin; FACS, fluorescence-activated cell sorter; HA, hemagglutinin; HEK, human embryonic kidney; HMW, high-molecular-weight.
References
- 1.Scherer, P. E., Williams, S., Fogliano, M., Baldini, G. & Lodish, H. F. (1995) J. Biol. Chem. 270, 26746–26749. [DOI] [PubMed] [Google Scholar]
- 2.Tsao, T. S., Tomas, E., Murrey, H. E., Hug, C., Lee, D. H., Ruderman, N. B., Heuser, J. E. & Lodish, H. F. (2003) J. Biol. Chem. 278, 50810–50817. [DOI] [PubMed] [Google Scholar]
- 3.Hara, K., Boutin, P., Mori, Y., Tobe, K., Dina, C., Yasuda, K., Yamauchi, T., Otabe, S., Okada, T., Eto, K., et al. (2002) Diabetes 51, 536–540. [DOI] [PubMed] [Google Scholar]
- 4.Yamauchi, T., Kamon, J., Waki, H., Terauchi, Y., Kubota, N., Hara, K., Mori, Y., Ide, T., Murakami, K., Tsuboyama-Kasaoka, N., et al. (2001) Nat. Med. 7, 941–946. [DOI] [PubMed] [Google Scholar]
- 5.Fruebis, J., Tsao, T. S., Javorschi, S., Ebbets-Reed, D., Erickson, M. R., Yen, F. T., Bihain, B. E. & Lodish, H. F. (2001) Proc. Natl. Acad. Sci. USA 98, 2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berg, A. H., Combs, T. P., Du, X., Brownlee, M. & Scherer, P. E. (2001) Nat. Med. 7, 947–953. [DOI] [PubMed] [Google Scholar]
- 7.Yamauchi, T., Kamon, J., Minokoshi, Y., Ito, Y., Waki, H., Uchida, S., Yamashita, S., Noda, M., Kita, S., Ueki, K., et al. (2002) Nat. Med. 8, 1288–1295. [DOI] [PubMed] [Google Scholar]
- 8.Tomas, E., Tsao, T. S., Saha, A. K., Murrey, H. E., Zhang Cc., C., Itani, S. I., Lodish, H. F. & Ruderman, N. B. (2002) Proc. Natl. Acad. Sci. USA 99, 16309–16313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsao, T. S., Murrey, H. E., Hug, C., Lee, D. H. & Lodish, H. F. (2002) J. Biol. Chem. 277, 29359–29362. [DOI] [PubMed] [Google Scholar]
- 10.Ouchi, N., Ohishi, M., Kihara, S., Funahashi, T., Nakamura, T., Nagaretani, H., Kumada, M., Ohashi, K., Okamoto, Y., Nishizawa, H., et al. (2003) Hypertension 42, 231–234. [DOI] [PubMed] [Google Scholar]
- 11.Yamauchi, T., Kamon, J., Ito, Y., Tsuchida, A., Yokomizo, T., Kita, S., Sugiyama, T., Miyagishi, M., Hara, K., Tsunoda, M., et al. (2003) Nature 423, 762–769. [DOI] [PubMed] [Google Scholar]
- 12.Huang, L. J., Constantinescu, S. N. & Lodish, H. F. (2001) Mol. Cell 8, 1327–1338. [DOI] [PubMed] [Google Scholar]
- 13.Bogan, J. S., Hendon, N., McKee, A. E., Tsao, T. S. & Lodish, H. F. (2003) Nature 425, 727–733. [DOI] [PubMed] [Google Scholar]
- 14.Naviaux, R. K., Costanzi, E., Haas, M. & Verma, I. M. (1996) J. Virol. 70, 5701–5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogan, J. S., McKee, A. E. & Lodish, H. F. (2001) Mol. Cell. Biol. 21, 4785–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, X., Constantinescu, S. N., Sun, Y., Bogan, J. S., Hirsch, D., Weinberg, R. A. & Lodish, H. F. (2000) Anal. Biochem. 280, 20–28. [DOI] [PubMed] [Google Scholar]
- 17.Matsuzawa, Y., Funahashi, T., Kihara, S. & Shimomura, I. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 29–33. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, H., Ouchi, N., Kihara, S., Walsh, K., Kumada, M., Abe, Y., Funahashi, T. & Matsuzawa, Y. (2004) Circ. Res. 94, 27–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang, C. Y., Mayo, M. W., Korneluk, R. G., Goeddel, D. V. & Baldwin, A. S., Jr. (1998) Science 281, 1680–1683. [DOI] [PubMed] [Google Scholar]
- 20.Pajvani, U. B., Hawkins, M., Combs, T. P., Rajala, M. W., Doebber, T., Berger, J. P., Wagner, J. A., Wu, M., Knopps, A., Xiang, A. H., et al. (2004) J. Biol. Chem. 279, 12152–12162. [DOI] [PubMed] [Google Scholar]
- 21.Zhu, Y., Bond, J. & Thomas, P. (2003) Proc. Natl. Acad. Sci. USA 100, 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang, Y., Xu, A., Knight, C., Xu, L. Y. & Cooper, G. J. (2002) J. Biol. Chem. 277, 19521–19529. [DOI] [PubMed] [Google Scholar]
- 23.Pajvani, U. B., Du, X., Combs, T. P., Berg, A. H., Rajala, M. W., Schulthess, T., Engel, J., Brownlee, M. & Scherer, P. E. (2003) J. Biol. Chem. 278, 9073–9085. [DOI] [PubMed] [Google Scholar]
- 24.Ranscht, B. & Dours-Zimmermann, M. T. (1991) Neuron 7, 391–402. [DOI] [PubMed] [Google Scholar]
- 25.Ivanov, D., Philippova, M., Tkachuk, V., Erne, P. & Resink, T. (2004) Exp. Cell Res. 293, 207–218. [DOI] [PubMed] [Google Scholar]
- 26.Niermann, T., Kern, F., Erne, P. & Resink, T. (2000) Biochem. Biophys. Res. Commun. 276, 1240–1247. [DOI] [PubMed] [Google Scholar]
- 27.Nagar, B., Overduin, M., Ikura, M. & Rini, J. M. (1996) Nature 380, 360–364. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro, L. & Scherer, P. E. (1998) Curr. Biol. 8, 335–338. [DOI] [PubMed] [Google Scholar]
- 29.Bogin, O., Kvansakul, M., Rom, E., Singer, J., Yayon, A. & Hohenester, E. (2002) Structure (London) 10, 165–173. [DOI] [PubMed] [Google Scholar]
- 30.Ivanov, D., Philippova, M., Antropova, J., Gubaeva, F., Iljinskaya, O., Tararak, E., Bochkov, V., Erne, P., Resink, T. & Tkachuk, V. (2001) Histochem. Cell Biol. 115, 231–242. [DOI] [PubMed] [Google Scholar]
- 31.Kudrjashova, E., Bashtrikov, P., Bochkov, V., Parfyonova, Y., Tkachuk, V., Antropova, J., Iljinskaya, O., Tararak, E., Erne, P., Ivanov, D., et al. (2002) Histochem. Cell Biol. 118, 281–290. [DOI] [PubMed] [Google Scholar]
- 32.Okamoto, Y., Arita, Y., Nishida, M., Muraguchi, M., Ouchi, N., Takahashi, M., Igura, T., Inui, Y., Kihara, S., Nakamura, T., et al. (2000) Horm. Metab. Res. 32, 47–50. [DOI] [PubMed] [Google Scholar]
- 33.Kubota, N., Terauchi, Y., Yamauchi, T., Kubota, T., Moroi, M., Matsui, J., Eto, K., Yamashita, T., Kamon, J., Satoh, H., et al. (2002) J. Biol. Chem. 277, 25863–25866. [DOI] [PubMed] [Google Scholar]
- 34.Matsuda, M., Shimomura, I., Sata, M., Arita, Y., Nishida, M., Maeda, N., Kumada, M., Okamoto, Y., Nagaretani, H., Nishizawa, H., et al. (2002) J. Biol. Chem. 277, 37487–37491. [DOI] [PubMed] [Google Scholar]
- 35.Takeuchi, T., Misaki, A., Liang, S. B., Tachibana, A., Hayashi, N., Sonobe, H. & Ohtsuki, Y. (2000) J. Neurochem. 74, 1489–1497. [DOI] [PubMed] [Google Scholar]
- 36.Huang, Z. Y., Wu, Y., Hedrick, N. & Gutmann, D. H. (2003) Mol. Cell. Biol. 23, 566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyle, D. D., Goings, G. E., Upshaw-Earley, J., Page, E., Ranscht, B. & Palfrey, H. C. (1998) J. Biol. Chem. 273, 6937–6943. [DOI] [PubMed] [Google Scholar]
- 38.Philippova, M. P., Bochkov, V. N., Stambolsky, D. V., Tkachuk, V. A. & Resink, T. J. (1998) FEBS Lett. 429, 207–210. [DOI] [PubMed] [Google Scholar]