Abstract
Delayed implantation (embryonic diapause) occurs when the embryo at the blastocyst stage achieves a state of suspended animation. During this period, blastocyst growth is very slow, with minimal or no cell division. Nearly 100 mammals in seven different orders undergo delayed implantation, but the underlying molecular mechanisms that direct this process remain largely unknown. In mice, ovariectomy before preimplantation ovarian estrogen secretion on day 4 of pregnancy initiates blastocyst dormancy, which normally lasts for 1–2 weeks by continued progesterone treatment, although blastocyst survival decreases with time. An estrogen injection rapidly activates blastocysts and initiates their implantation in the progesterone-primed uterus. Using this model, here we show that among ≈20,000 genes examined, only 229 are differentially expressed between dormant and activated blastocysts. The major functional categories of altered genes include the cell cycle, cell signaling, and energy metabolic pathways, particularly highlighting the importance of heparin-binding epidermal growth factor-like signaling in blastocyst–uterine crosstalk in implantation. The results provide evidence that the two different physiological states of the blastocyst, dormancy and activation, are molecularly distinguishable in a global perspective and underscore the importance of specific molecular pathways in these processes. This study has identified candidate genes that provide a scope for in-depth analysis of their functions and an opportunity for examining their relevance to blastocyst dormancy and activation in numerous other species for which microarray analysis is not available or possible due to very limited availability of blastocysts.
Successful implantation results from reciprocal interactions between an implantation-competent blastocyst and a receptive uterus. Highly coordinated cellular and molecular events, directed by ovarian estrogen and progesterone (P4), produce a favorable uterine environment, the receptive state, to support implantation. The blastocyst also functions as an active unit with its own molecular program of cell growth and differentiation (1, 2). It is difficult to distinguish embryonic and uterine events during normal pregnancy with respect to blastocyst activation and uterine receptivity because of the changing levels of ovarian hormones. Because estrogen is essential for on-time uterine receptivity and blastocyst activation in mice (3), ovariectomy before preimplantation estrogen secretion on the morning of day 4 results in implantation failure, initiating a state of blastocyst dormancy. This condition, referred as delayed implantation, can be maintained by continued P4 treatment. However, implantation with blastocyst activation rapidly occurs by an estrogen injection in P4-primed mice (3, 4). Delayed implantation also occurs naturally (facultative) during lactation after postpartum ovulation and successful mating in mice; implantation occurs after termination of the suckling stimulus (5). Lactational delay in mice occurs due to insufficient ovarian estrogen secretion. Mustelids, marsupials, and many other wild species also exhibit obligatory seasonal delayed implantation. Species-specific regulation of delayed implantation widely varies, ranging from hormonal changes, lactational to seasonal, or photoperiod to nutritional (6).
During delayed implantation, embryos develop into blastocysts and undergo zona hatching, albeit at a slower pace, with reduced or no cell division. These blastocysts are metabolically dormant and incompetent to initiate attachment in the uterus. Using this model, we have previously shown that the blastocyst's state of activity is also a determining factor in initiating implantation in the receptive uterus (3). However, the mechanisms by which blastocysts undergo dormancy and survive for an extended period and then resume activation to implantation competency when exposed to estrogen in utero remain largely unknown. The implantation process is a two-way interaction between blastocyst and uterus, and it is speculated that signals arising from the blastocyst target uterine cells to initiate this process. However, the identity and nature of the signals remain unknown.
Delayed implantation in mice provides a powerful model to address these issues at the molecular level by comparing global gene expression in blastocysts during dormancy and activation. Such an analysis has been hindered by the lack of a suitable microarray platform with genes unique to early embryos and the difficulties of obtaining a large number of embryos. Recent establishment of a 60-mer oligo microarray platform, enriched for genes expressed in stem cells and early embryos including preimplantation embryos (7), fulfills this objective. With an optimized labeling reaction for a small amount of RNA with two rounds of cRNA linear amplification (7, 8), we compared gene expression differences between dormant and activated blastocysts. The expression of several key genes at the protein expression level was validated by immunofluorescence. The results indicate that dormant and activated blastocysts are distinguishable at the gene expression level, suggesting specific molecular mechanisms are involved in blastocyst dormancy and activation for implantation.
Materials and Methods
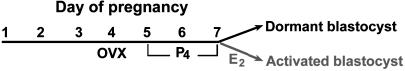
Animals and Embryo Collection. Adult CD-1 (Charles River Breeding Laboratories) female mice were mated with fertile males of the same strain to induce pregnancy (day 1 = vaginal plug). To induce delayed implantation, pregnant mice were ovariectomized on the morning (0800–0900 h) of day 4 and injected daily with P4 (2 mg per mouse, s.c.) from days 5–7. To activate dormant blastocysts, P4-primed delayed implanting pregnant mice were injected with estradiol-17β (25 ng per mouse) on day 7. Blastocysts were recovered by flushing uteri with Whitten's medium between 12 and 14 h after the last steroid injections (Fig. 1). They were washed six times in the same medium, flash-frozen in batches of 100 in each tube, and stored at –80°C until use for RNA extraction.
Fig. 1.
Experimental design to generate dormant and activated blastocysts. Pregnant mice were ovariectomized on the morning (0900 h) of day 4. Delayed implantation was maintained by daily injection of P4 from days 5–7. To activate dormant blastocysts, P4-primed implanting mice were injected with estradiol-17β(E2) on day 7. Blastocysts were recovered between 12 and 14 h after the last steroid injections. OVX, ovariectomy.
RNA Extraction, Labeling, and Hybridization on the National Institute on Aging (NIA) 22,000 60-Mer Oligo Microarray. Three batches of 100 blastocysts were collected for each group of dormant and activated blastocysts, and mRNA was extracted from each batch by using a Quickprep micro polyA RNA Extraction Kit (Amersham Pharmacia Biosciences). Aliquots of mRNA equivalent to 24 blastocysts from each mRNA subset were labeled with Cy3 dyes by two rounds of linear amplification labeling reactions for cRNA targets using Fluorescent Linear Amplification Kits (Agilent Technologies, Palo Alto, CA). The quality and size distribution of targets were determined by the RNA 6000 Nano Lab-on-chip Assay (Agilent) and quantitated by using a microscale spectrophotometer (NanoDrop, Agilent). Hybridization of cRNA targets from dormant or activated blastocysts was performed on the NIA 22K 60-mer oligo microarray platform (Agilent). Extensive validation by quantitative real-time RT-PCR with rigorous standards confirmed the array results for essentially all genes tested (7).
Microarray Data Analysis. The data for 21,939 noncontrol probes per microarray were processed by feature extraction 5.1.1 software (Agilent) and further analyzed by ANOVA–false discovery rate (FDR) statistics (http://lgsun.grc.nia.nih.gov/ANOVA) (7). Statistical significance was determined by using the FDR (10%) method. A scatter plot was composed by using the NIA microarray analysis tool (http://lgsun.grc.nia.nih.gov/ANOVA). genmapp/mapp finder (9) was used for a functional annotation analysis with gene ontology terms (10).
Preparation of Beads Carrying Heparin-Binding EGF-Like Growth Factor (HB-EGF). The loading of Affi-Gel Blue beads (Bio-Rad, Hercules, CA, 100–200 mesh) of about the size of a blastocyst with HB-EGF or BSA (100 ng/μl) was achieved as described (11). Loaded beads (seven beads per horn) were transferred into uteri of day 4 pseudopregnant mice produced by mating with vasectomized males. Mice were killed between 0900 and 1000 h on day 5 after blue dye injection (3).
In Situ Hybridization. Frozen sections were hybridized with 35S-labeled cRNA probes to mouse Hegf1 as described (12).
Immunofluorescence Detection of Proteins in Blastocysts. To localize Brca1, HB-EGF, p21, and Igf2r, immunofluorescence was captured in a Zeiss LSM 510 confocal scanning laser microscope as described (13). Rabbit polyclonal antibodies specific to these antigens were used. Images shown depict FITC-labeled antigens as green, propidium iodide-labeled nuclei as red, and merge as yellow.
Results and Discussion
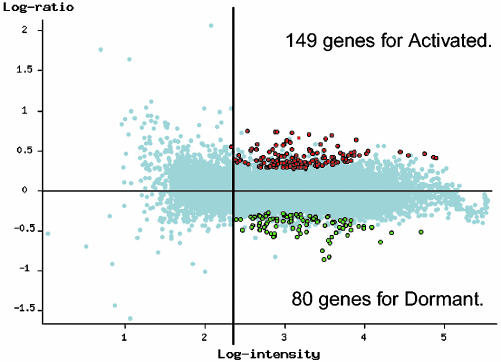
Genes Are Differentially Expressed in Dormant and Activated Blastocysts. A complete gene expression dataset, representing 21,939 gene features, is available at the NIA mouse cDNA project (http://lgsun.grc.nia.nih.gov/cDNA/cDNA.html), Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo), and ArrayExpress (www.ebi.ac.uk/arrayexpress) web sites. Microarray analysis revealed that only 229 genes (≈1.0%) were differentially expressed between dormant and activated blastocysts with statistical significance (P < 0.01 and FDR <0.1) (Fig. 2 and Table 3, which is published as supporting information on the PNAS web site). Thus, a relatively small number of genes were associated with two physiologically altered states of the blastocysts (dormancy vs. activation). Of the 229 genes, 80 were more highly expressed in dormant blastocysts, whereas 149 were more highly expressed in activated blastocysts (Table 1). Because 72 of the 229 (31.4%) genes were uncharacterized (unknown), we focused our attention on the known genes. The MAPPFinder-based (www.genmapp.org/MAPPFinder.html) gene ontology annotation was used to examine global characteristics of these genes (9, 10). Because such an analysis requires a sufficient number of genes, the genes that were most highly expressed in activated blastocysts were analyzed. Analysis of 89 such genes provides the following functional categories as characteristics of activated blastocysts: (i) cyclin-dependent kinase regulators and DNA replication initiators, (ii) glycolysis and energy pathway, and (iii) proteinnucleus import (Table 2 and Tables 4 and 5, which are published as supporting information on the PNAS web site). The analysis by another independent annotation method further showed that among 116 known genes expressed highly in activated blastocysts, the largest number (24) of genes were in the cell signaling category, including signaling by phosphatidylinositol-3-phosphate, Ca2+ and HB-EGF. The second (13), third (10) and fourth largest (9) numbers of genes were in metabolism, transcriptional regulation, and cell cycle categories, respectively. We also observed increased expression of a number of genes in the posttranscriptional modification, transcriptional regulation, and chromatin remodeling categories during blastocyst activation (Table 1).
Fig. 2.
A scatter plot of 21,939 gene features on NIA 22,000 60-mer oligo microarray, comparing gene expression in dormant and activated blastocysts. Microarray data were analyzed by ANOVA-FDR statistics. The combined results of six hybridizations identified 229 differentially expressed genes (averaged log intensity ≥2.3, P ≤ 0.01, and FDR ≤0.1), including 80 genes highly expressed in dormant blastocysts (green circles) and 149 genes highly expressed in activated blastocysts (red circles).
Table 1. Functional categories of differentially expressed genes.
| Category | Total | Act | Dor |
|---|---|---|---|
| Adhesion molecule | 6 | 5 (3.4%) | 1 (1.3%) |
| Apoptosis/antiapoptosis | 4 | 2 (1.3%) | 2 (2.5%) |
| Cell cycle | 13 | 9 (6.0%) | 4 (5.0%) |
| Cell signaling | 38 | 24 (16.1%) | 14 (17.5%) |
| Chaperoning pathway | 4 | 4 (2.7%) | 0 (0.0%) |
| Chromatin remodeling | 7 | 6 (4.0%) | 1 (1.3%) |
| Cytoarchitecture | 8 | 7 (4.7%) | 1 (1.3%) |
| Genome stability | 3 | 3 (2.0%) | 0 (0.0%) |
| Ion transport/ion channel | 8 | 6 (4.0%) | 2 (2.5%) |
| Metabolism | 18 | 13 (8.7%) | 5 (6.3%) |
| Mitochondrial function | 3 | 2 (1.3%) | 1 (1.3%) |
| Posttranscriptional modification | 5 | 5 (3.4%) | 0 (0.0%) |
| Protein modification | 6 | 4 (2.7%) | 2 (2.5%) |
| Protein/amino acid transport | 5 | 4 (2.7%) | 1 (1.3%) |
| Proteolysis | 6 | 4 (2.7%) | 2 (2.5%) |
| Transcriptional regulation | 13 | 10 (6.7%) | 3 (3.8%) |
| Translation machinery | 7 | 5 (3.4%) | 2 (2.5%) |
| Others | 3 | 3 (2.0%) | 0 (0.0%) |
| Unknown | 72 | 33 (22.1%) | 39 (48.8%) |
| Total | 229 | 149 | 80 |
The number of genes assigned to a functional category and their prevalence (%) with respect to the total number of differentially expressed genes in activated (Act) or dormant (Dor) blastocysts are shown.
Table 2. Functional categories of selected genes differentially expressed in activated (Act) and dormant (Dor) blastocysts.
| (Act)/(Dor)
|
||||
|---|---|---|---|---|
| Functional category | Gene symbol | Fold change | P | FDR |
| Cell cycle | Ccne2 | 3.83 | 1E-05 | 0.004 |
| Cdc6 | 3.69 | 0 | 0 | |
| Ccnd1 | 3.07 | 2E-05 | 0.005 | |
| Ccne1 | 3.00 | 1E-05 | 0.004 | |
| Mcmd5 | 2.21 | 8E-04 | 0.067 | |
| Cdc45l | 2.20 | 4E-05 | 0.009 | |
| Orc11 | 1.96 | 7E-04 | 0.068 | |
| Brca1 | 1.89 | 9E-04 | 0.080 | |
| Cdkn1a/p21Cip1/Waf1 | 0.52 | 0.001 | 0.097 | |
| A530090O15Rik/Trb1 | 0.45 | 1E-04 | 0.018 | |
| Btg1 | 0.15 | 0 | 0 | |
| Nupr1 | 0.34 | 2E-04 | 0.026 | |
| Apbb1 | 0.49 | 5E-04 | 0.055 | |
| 0610043B10Rik | 0.47 | 3E-04 | 0.039 | |
| Pctk3 | 3.31 | 0 | 0 | |
| Vav3 | 3.77 | 0 | 0 | |
| Ranbp1 | 2.45 | 5E-04 | 0.052 | |
| Carbohydrate metabolism and energy pathway | Ldh1 | 4.03 | 0 | 0 |
| Pfkl | 3.40 | 4E-05 | 0.010 | |
| Eno1 | 2.59 | 1E-04 | 0.018 | |
| Pgk1 | 3.06 | 2E-04 | 0.028 | |
| Ctps | 2.90 | 2E-04 | 0.026 | |
| Pkm2 | 2.87 | 2E-04 | 0.028 | |
| Oxct | 0.35 | 5E-04 | 0.053 | |
| Igf2r | 0.30 | 2E-04 | 0.027 | |
| Irs1 | 0.44 | 3E-05 | 0.006 | |
| Calcium signaling | Pik3c2a | 2.76 | 1E-04 | 0.017 |
| Pitpn | 2.13 | 1E-04 | 0.023 | |
| Vav3 | 3.77 | 0 | 0 | |
| Pr1 | 4.42 | 0 | 0 | |
| Etohd4/Fgd6 | 5.46 | 0 | 0 | |
| Mtmr4 | 0.48 | 0.001 | 0.099 | |
| 2410005C22Rik/Pepp1 | 0.37 | 0 | 0.001 | |
| S100a13 | 0.36 | 3E-04 | 0.039 | |
| Tpd52l1 | 0.40 | 1E-05 | 0.002 | |
| Fer1l3 | 0.41 | 5E-05 | 0.010 | |
| Cytoplasmic-nuclear cargo | Kpna2 | 2.20 | 8E-05 | 0.016 |
| Kpna4 | 2.09 | 6E-04 | 0.060 | |
| Chromatine remodeling | Baz1a | 5.44 | 0 | 0 |
| Satb1 | 4.36 | 0 | 0 | |
| Sap30 | 2.17 | 0.001 | 0.090 | |
| Baf53a | 2.16 | 2E-04 | 0.027 | |
| Hells | 2.06 | 5E-04 | 0.049 | |
| Hdac5 | 0.51 | 5E-04 | 0.054 | |
| Adhesion molecule | Zyx | 2.99 | 2E-04 | 0.027 |
| Ocln | 2.88 | 0 | 0 | |
| Lgals4 | 2.70 | 0 | 0 | |
| Bysl | 1.96 | 0.001 | 0.086 | |
| Amotl1 | 0.43 | 7E-04 | 0.065 | |
| HB-EGF | Dtr/Hegfl/HB-EGF | 2.34 | 9E-04 | 0.080 |
A list of differentially expressed genes discussed in the text with fold changes and statistical significances is shown.
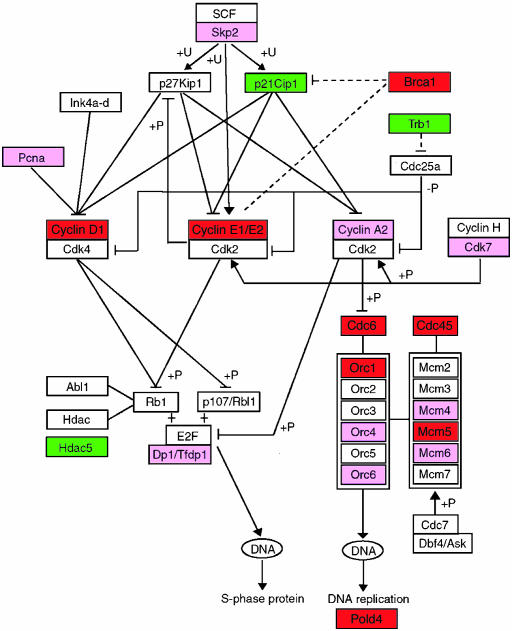
Cell Cycle-Related Genes Are Differentially Expressed in Dormant and Activated Blastocysts. Most significant differences in gene expression levels between dormant and activated blastocysts were observed for the cell cycle-related genes. Many genes involved in the G1/S checkpoint showed differential expressions. For example, p21cip1/WAF1 and Tribble/Trb1 were up-regulated in dormant blastocysts, whereas Ccnd1, Ccne1, and Ccne2 were down-regulated. Cdc6, Cdc45, Orc1, and Mcm5 involved in DNA replication and other cell-cycle-related genes, such as Brca1 and Pctk3, were also down-regulated in dormant blastocysts (Table 2, Fig. 3). These results indicate that the cell cycle is decelerated with blastocyst dormancy but is resumed with blastocyst activation. This is consistent with previous reports of increased cell number and uridine incorporation in dormant blastocysts on reactivation in utero by estrogen (14, 15).
Fig. 3.
A chart of cell cycle genes. Green and red boxes highlight for higher expression (FDR < 0.1) in dormant and activated blastocysts, respectively. The genes, which showed higher expression with P ≤ 0.05 and FDR ≥ 0.1 in activated blastocysts, are highlighted as pink boxes. The lines with arrowheads or T heads represent stimulation or suppression, respectively. The pathway figure is based on the Kyoto Encyclopedia of Genes and Genomes pathway database (49), with slight modification. Based on published data (16), the dashed lines from Brca1 represent increased expression of p21cip1/WAF1 and decreased expression of cyclin E in Brca1–/– embryos. +P, –P, and +U indicate phosphorylation, dephosphorylation, and ubiquitination, respectively.
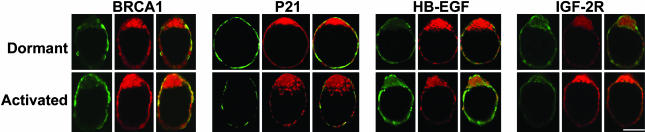
Specific regulatory pathways, especially those responding to estrogen, appear to be involved in regulating the cell cycle in blastocysts. For example, Brca1, down-regulated in dormant blastocysts (Figs. 3 and 4), is a known estrogen-responsive target. In Brca1–/– embryos, there is increased expression of p21 (16), which is also up-regulated in dormant blastocysts (Figs. 3 and 4). Collectively, our microarray results showing down-regulation of Brca1 with up-regulation of p21cip1/WAF1 in blastocyst dormancy are consistent with their known functions.
Fig. 4.
Localization of BRCA1, HB-EGF, p21, and IGF2R in dormant and activated blastocysts. Antigens (green) and nuclei (red) were visualized by using FITC-conjugated secondary antibodies and propidium iodide, respectively. Note higher signal intensity for BRCA1 and HB-EGF in activated blastocysts and of p21 and IGF2R in dormant blastocysts. (Bar = 50 μm.)
The second example involves Btg1 (B cell translocation gene 1, antiproliferative), which negatively regulates cell proliferation. Its expression is maximal at the G0/G1 phases of the cycle but is down-regulated as cells progress through the G1 phase (17). Btg1 also regulates estrogen receptor-α (Esr1) functions via its interaction with Cnot7/Caf1/Pop2 (18). Thus up-regulation of Btg1 in dormant blastocysts provides further support for estrogen involvement in embryonic cell cycle regulation. In addition, Apbb1 [amyloid β (A4) precursor protein-binding], p8 Nupr1 (nuclear protein 1), and 0610043B10Rik (small ubiquitinated apoptotic protein), which presumably act as cell growth suppressors, were predominantly expressed in dormant blastocysts, whereas Pctk3 (PCTAIRE-motif protein kinase 3), a member of cdc2/CDC28 (Cks2)-related protein kinase gene family, was more highly expressed in activated blastocysts. Furthermore, Vav3, a guanine nucleotide exchange factor implicated in the regulation of Rho GTPases, and Ranbp1, a molecular partner of Rho GTPase, were up-regulated in activated blastocysts. There is evidence that Vav3 and Ranbp1 are involved in DNA replication (19). Heightened expression of Vav3 and Ranbp1 in activated blastocysts thus may be involved in increased cell cycle activity on resumption of activation from dormancy. Transcription of Trp-53 (p53), Mdm2, and Atm did not show significant differences between dormant and activated blastocysts, suggesting that blastocyst dormancy does not involve p53. Thus, selective cell cycle regulatory pathways are involved in inducing blastocyst dormancy.
Carbohydrate Metabolic and Energy Pathways Are Differentially Expressed in Dormant and Activated Blastocysts. Blastocysts remain metabolically dormant during delayed implantation. However, increased mitosis and cell number that occur between 8 and 12 h after reactivation of dormant blastocysts in vivo by estrogen are preceded by significant increases in uridine incorporation within 1 h and pyruvate uptake within 4 h (14, 15). In contrast, glucose uptake remains basal until 16 h, but increases thereafter after activation of dormant blastocysts (14), suggesting that pyruvate is a major source of energy during the early stages of blastocyst activation. In contrast, evidence showing increased activity of phosphofructokinase and pyruvate kinase with decreased lactate dehydrogenase activity in dormant blastocysts (20) suggests that energy requirement during blastocyst activation is glucose dependent. Our results of significant increases in Pfkl (phosphofructokinase, liver, B-type), Eno1 (enolase 1α, nonneuron), Pgk1 (phosphoglycerate kinase 1), Pkm2 (pyruvate kinase, muscle), and Ldh1 (lactate dehydrogenase 1, A chain) in activated blastocysts suggest that increased energy requirements during activation are satisfied by both glucose and pyruvate energy pathways. In contrast, Scot/Oxct (3-oxoacid CoA transferase), a key enzyme in the extrahepatic utilization of ketone bodies and a necessary step in ketolytic energy production, was down-regulated during activation.
Dauer larva in Caenorhabditis elegans resembles, in some respects, the blastocyst dormancy state. The best characterized gene that influences dauer life and aging in C. elegans is daf-2 that encodes an insulin/insulin-like growth factor I receptor-like protein (21). Insr, Irs1, Igf1r, and Igf2r are expressed in mouse and human preimplantation embryos, and Igf2 has an autocrine effect on preimplantation development (22–24). Igf2r and Irs1 showed higher expression in dormant blastocysts. It is interesting to note that Igf2r is more highly expressed in trophoblast stem (TS) cells than embryonic stem (ES) cells, whereas the situation is the reverse for Irs1 expression (25).
Igf2r, imprinted and maternally expressed in mice, encodes a transmembrane receptor that transports mannose-6-phosphate-tagged proteins and Igf2 to lysosomes. The implication of higher Igfr2 expression in dormant blastocysts is not clear. Inactivation of the Igf2r gene leads to increased systemic Igf2 levels, contributing to embryonic overgrowth and perinatal lethality (26, 27). Thus, it is possible that higher levels of Igf2r in dormant blastocysts (Table 2 and Fig. 4) contribute to halting growth and maintaining dormancy state during delayed implantation. Irs1 is a phosphorylated substrate for insulin, Igf1, and class I insulin receptors. Mice with null mutation for Irs1 are born alive but show retarded embryonic and postnatal growth. They are also resistant to the glucose-lowering effects of insulin, Igf1, and Igf2 (28, 29). It is possible that Igfr2 and Irs1 help in maintaining viability of blastocysts during dormancy.
Calcium Signaling Pathways Are Differentially Regulated in Dormant and Activated Blastocysts. A number of genes involved in inositol phosphate and Ca2+ signaling pathways showed significant alterations between dormant and activated blastocysts, primarily being up-regulated during activation (Tables 2 and 4). For example, Pik3c2a, Pitpn, Vav3, Pr1, and Etohd4/Fgd6 were up-regulated in activated blastocysts, whereas Mtmr4, Pepp1, S100a13, Tpd52l1, and Fer1l3/myoferlin were up-regulated in dormant blastocysts. Inositol 1,4,5-trisphosphate (phosphatidylinositol-3-phosphate) regulates cell growth and differentiation by modulating intracellular Ca2+ release and is involved in oocyte activation. Recent evidence suggests that Ca2+ signaling also regulates preimplantation embryo development and blastocyst adhesion during implantation (30, 31), and inhibition of Ca2+ channel activity attenuates blastocyst competency for implantation (13). Up- or down-regulation of many of the genes that participate in phosphatidyl inositol and Ca2+ signaling pathways suggests that they are important for blastocyst dormancy and activation.
Cytoplasmic–Nuclear Cargo Is Differentially Regulated in Dormant and Activated Blastocysts. Karyopherins, a family of transport factors, are also called importins or exportins, depending on the direction in which they carry their cargo to partition cellular components between the nucleus and cytoplasm. Transcripts of karyopherins (importin-α2 and -α4) were increased in activated blastocysts (Tables 2 and 4). Importin-α molecules also play critical roles in development. The interstitial deletion D14 affecting the importin-α2 gene in Drosophila causes recessive female sterility characterized by a block of nurse cell-oocyte transport during oogenesis. Furthermore, it appears to be involved in the regulation of growth and cell cycles, because its subcellular distribution depends on the phases of the cell cycle (32). Thus, increased transcript levels of karyopherins after transition from dormancy to implantation competency suggest their roles in blastocyst activation.
Genes Involved in Chromatin Remodeling Are Differentially Regulated in Dormant and Activated Blastocysts. Although the “dormancy vs. activation” analysis was performed by using whole blastocysts, it is desirable to distinguish whether the genes are expressed in the inner cell mass (ICM) or trophectoderm (TE), two distinctive cell types in the blastocyst. However, this is technically difficult, and thus we used the previous microarray data comparing ES and TS cells (25). The ES and TS cells are derived from the ICM and TE, respectively, and may well serve as surrogates for the ICM and TE.
We observed increased expression of a number of genes involved in posttranscriptional modification, transcriptional regulation, and chromatin remodeling during blastocyst activation (Tables 1 and 2). Most of these chromatin-remodeling genes were differentially expressed in ES and TS, representing the ICM and TE, respectively. With the exception of Hdac5, genes such as Baf53a, Sap30, and Lysh/Hells were up-regulated in ES, whereas Baz1a and Satb1 were up-regulated in TS. BAF53 requires BRG1 for making a complex with chromatin/matrix (33). Lysh/Hells is highly expressed in proliferating cells but down-regulated during apoptosis. Lysh/Hells–/– mice die perinatally (34), suggesting that this gene is important for cell proliferation and function during embryogenesis. In contrast, Baz1a and Satb1 up-regulated in TS are involved in chromatin remodeling, suggesting their role in TE differentiation and implantation.
Genes Encoding Adhesion Molecules Are Up-Regulated in Blastocysts with Activation. Blastocyst adhesiveness is essential for trophectoderm attachment to the uterine luminal epithelium for implantation. Primary adhesion molecules that are implicated in implantation are selectins, galectins, heparan sulfate proteoglycans, integrins, cadherins, and the trophinin–bystin complex (35). Activation of dormant blastocysts in utero by estrogen makes them adhesion competent for implantation. This is consistent with our observation of up-regulation of Ocln, Zyx, Lgals4, and Bysl in activated blastocysts (Tables 2 and 4). Ocln encoding occludin is important for the formation and maintenance of the blastocoel cavity (36), and Bysl that encodes bystin is involved in trophinin-mediated cell adhesion between trophoblast and uterine epithelial cells via interaction with trophinin, tastin, and cytokeratin (37). Although mice lacking zyxin function normally (38), and mice carrying galectin1/3 double mutations (39) are viable and compatible with implantation, the function of Lgals4 (galectin4) is not known. It is possible that galectin4 plays a role in blastocyst adhesiveness and compensates for the lack of galectins1/3.
In contrast, Amotl1, encoding a peripheral membrane protein, was up-regulated in dormant blastocysts. This tight junction protein is specifically expressed in exocrine epithelial cells and regulates paracellular permeability and cell polarity (40). It acts within subregions of visceral endoderm to regulate morphogenetic movements required for embryo viability and growth. Thus, up-regulation of Amotl1 during dormancy may be involved in maintaining blastocoel cavity for an extended period. It is interesting to note that, whereas Amotl1 is down-regulated, occludin is up-regulated during activation. This may suggest that these two molecules have different functions during implantation.
HB-EGF Signaling Participates in Embryo–Uterine Dialogue During Implantation. In mice, the earliest change in gene expression so far reported before implantation at the site of a blastocyst is an increased transcription of Hegf1, the gene encoding HB-EGF, in the luminal epithelium. Transcription is detectable around 1600 h on day 4, which is 6–7 h before the increased localized vascular permeability (12). Hegf1 induction is not triggered by a dormant blastocyst during delayed implantation (12, 41), suggesting that a factor secreted by an implantation-competent blastocyst induces this event, although the identity of this factor is not known.
The EGF-like growth factors interact with the receptor subtypes of the erbB gene family, ErbB1, -2, -3, and -4. Genetic and biochemical studies have highlighted the roles of embryonic ErbB1 and ErbB4 in interacting with uterine HB-EGF in implantation in mice (42, 43). HB-EGF is present as soluble and transmembrane forms in the luminal epithelium at the site of a blastocyst, suggesting paracrine and/or juxtacrine interactions with embryonic ErbBs, as well as autocrine, paracrine, and/or juxtacrine interactions with uterine ErbBs during implantation (12, 44). For example, the expression of both ErbB1 and ErbB4 is down-regulated in dormant blastocysts during delayed implantation but is readily up-regulated with blastocyst activation (43, 45). Furthermore, although soluble HB-EGF promotes blastocyst growth and differentiation (12), cells that express the transmembrane form of HB-EGF adhere to active, but not dormant, blastocysts in vitro (43), suggesting paracrine and juxtacrine functions of HB-EGF. In addition, by directing an HB-EGF–toxin conjugate toward wild-type and erbB1–/– blastocysts, it was shown that HB-EGF interacts with embryonic ErbB4 (42). Recent evidence also shows that trafficking of HB-EGF to the blastocyst surface is regulated by Ca2+ signaling (46). Collectively, these results suggest that an interaction between uterine HB-EGF and blastocyst ErbBs is important for the attachment reaction.
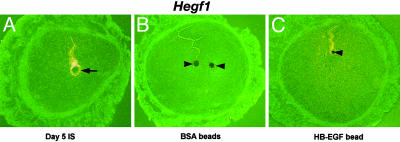
Our present microarray data show that Hegf1 is significantly up-regulated during blastocyst activation, suggesting its role in implantation. This was further validated by immunofluorescence analysis (Fig. 4). Because there is evidence that EGF family members can induce the expression of their own genes in an autoinduction manner (47), we speculated that increased HB-EGF levels in activated blastocysts would induce Hegf1 expression in the uterine epithelium at the sites of their apposition. This was tested by transferring blastocyst-size Affi-gel blue beads presoaked in HB-EGF into uterine lumens of pseudopregnant mice on day 4. We previously showed that HB-EGF-loaded beads induce discrete local implantation-like responses with the expression of COX-2 and BMP2 genes in the uterine stroma similar to those elicited by living blastocysts (2, 11), but the possibility that HB-EGF induces its own gene expression in the uterine epithelium was not explored. As shown here in Fig. 5, in situ hybridization clearly detected the expression of Hegf1 in the regions of the uterus surrounding the beads presoaked in HB-EGF, not BSA, 24 h after the bead transfer, similar to that seen during normal implantation. This suggests that HB-EGF produced by blastocysts and secreted by ectodomain shedding of proHB-EGF (48) induces its own expression in the uterus in a paracrine manner (47). Thus, the microarray data further elucidate an important role of HB-EGF on both sides of the blastocyst and uterus during implantation. These results provide evidence that one of the signaling molecules involved in establishing a hierarchy of events between the embryo and uterus for implantation is HB-EGF that originates first in implantation-competent blastocysts. This autoinduction loop of HB-EGF is an example of a molecular crosstalk between these two different entities that leads to the initiation of the implantation process.
Fig. 5.
In situ hybridization of Hegf1 at the implantation or bead sites. (A) A representative section of a day 5 implantation site from natural mating. (B and C) A representative uterine section containing beads preabsorbed in BSA (B) or in HB-EGF (C). Arrow indicates the location of a blastocyst, whereas arrowheads indicate the location of beads (×100).
Perspectives. This study is an attempt to elucidate the molecular mechanisms by which blastocysts undergo dormancy and resume activation in a global perspective. This is an important area of research, because delayed implantation occurs in a wide range of laboratory and wild animals (6). Our results of microarray analysis reported here are relative to changes in blastocysts in dormancy vs. blastocysts undergoing activation with respect to the uterine status. Whether changes in uterine gene expression under these conditions influence changes in gene expression in the blastocyst or vice versa is not currently known. It is also not known whether delayed implantation occurs in humans. However, better insights into the molecular basis of blastocyst dormancy and activation are likely to provide meaningful information regarding the mechanism by which blastocysts become implantation-competent and initiate a reciprocal interaction with the receptive uterus for implantation, having clinical implications for improving fertility in women. In conclusion, our present studies in blastocysts at two different physiological states provide information that pathways involving cell cycle, energy metabolism, and Ca2+ signaling participate in the resumption of growth and metabolic activation of blastocysts returning from their dormancy state, whereas chromatin remodeling and signaling involving adhesion molecules and HB-EGF participate in embryo–uterine interactions required for implantation. We believe our present investigation provides a landscape of gene interaction in the blastocyst when it journeys through different states of activity. This study also presents opportunities to examine candidate genes of specific molecular pathways relevant to blastocyst dormancy and activation in numerous other species for which microarray analysis is not available or possible due to very restricted availability to blastocysts.
Supplementary Material
Acknowledgments
We thank Jian Tan for help with in situ hybridization. Michael Klagsbrun (Harvard Medical School, Boston) kindly provided purified HB-EGF. This work was supported in part by grants from the National Institutes of Health (HD12304, HD33994, and DA 06668). T.H., H.W., and M.G.C. were supported by fellowships from the Serono Foundation, the Lalor Foundation, and the National Institute of General Medical Sciences Pharmacology Research Associate Program, respectively.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: P4, progesterone; EGF, epidermal growth factor; FDR, false discovery rate; NIA, National Institute on Aging; TE, trophectoderm; ES, embryonic stem; TS, trophoblast stem.
References
- 1.Dey, S. K., Lim, H., Das, S. K., Reese, J., Paria, B. C., Daikoku, T. & Wang, H. (2004) Endocr. Rev. 25, 341–373. [DOI] [PubMed] [Google Scholar]
- 2.Paria, B. C., Reese, J., Das, S. K. & Dey, S. K. (2002) Science 296, 2185–2188. [DOI] [PubMed] [Google Scholar]
- 3.Paria, B. C., Huet-Hudson, Y. M. & Dey, S. K. (1993) Proc. Natl. Acad. Sci. USA 90, 10159–10162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoshinaga, K. & Adams, C. E. (1966) J. Reprod. Fertil. 12, 593–595. [DOI] [PubMed] [Google Scholar]
- 5.McLaren, A. (1968) J. Endocrinol. 42, 453–463. [DOI] [PubMed] [Google Scholar]
- 6.Renfree, M. B. & Shaw, G. (2000) Annu. Rev. Physiol. 62, 353–375. [DOI] [PubMed] [Google Scholar]
- 7.Carter, M. G., Hamatani, T., Sharov, A. A., Carmack, C. E., Qian, Y., Aiba, K., Ko, N. T., Dudekula, D. B., Brzoska, P. M., Hwang, S. S. & Ko, M. S. (2003) Genome Res. 13, 1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamatani, T., Carter, M. G., Sharov, A. A. & Ko, M. S. (2004) Dev. Cell 6, 117–131. [DOI] [PubMed] [Google Scholar]
- 9.Dahlquist, K. D., Salomonis, N., Vranizan, K., Lawlor, S. C. & Conklin, B. R. (2002) Nat. Genet. 31, 19–20. [DOI] [PubMed] [Google Scholar]
- 10.Gene Ontology Consortium (2001) Genome Res. 11, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paria, B. C., Ma, W., Tan, J., Raja, S., Das, S. K., Dey, S. K. & Hogan, B. L. (2001) Proc. Natl. Acad. Sci. USA 98, 1047–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das, S. K., Wang, X. N., Paria, B. C., Damm, D., Abraham, J. A., Klagsbrun, M., Andrews, G. K. & Dey, S. K. (1994) Development (Cambridge, U.K.) 120, 1071–1083. [DOI] [PubMed] [Google Scholar]
- 13.Wang, H., Matsumoto, H., Guo, Y., Paria, B. C., Roberts, R. L. & Dey, S. K. (2003) Proc. Natl. Acad. Sci. USA 100, 14914–14919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spindler, R. E., Renfree, M. B. & Gardner, D. K. (1996) J. Exp. Zool. 276, 132–137. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, P. V. & Dickson, A. D. (1975) J. Anat. 119, 453–459. [PMC free article] [PubMed] [Google Scholar]
- 16.Hakem, R., de la Pompa, J. L., Sirard, C., Mo, R., Woo, M., Hakem, A., Wakeham, A., Potter, J., Reitmair, A., Billia, F., et al. (1996) Cell 85, 1009–1023. [DOI] [PubMed] [Google Scholar]
- 17.Rouault, J. P., Rimokh, R., Tessa, C., Paranhos, G., Ffrench, M., Duret, L., Garoccio, M., Germain, D., Samarut, J. & Magaud, J. P. (1992) EMBO J. 11, 1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prevot, D., Morel, A. P., Voeltzel, T., Rostan, M. C., Rimokh, R., Magaud, J. P. & Corbo, L. (2001) J. Biol. Chem. 276, 9640–9648. [DOI] [PubMed] [Google Scholar]
- 19.Battistoni, A., Guarguaglini, G., Degrassi, F., Pittoggi, C., Palena, A., Di Matteo, G., Pisano, C., Cundari, E. & Lavia, P. (1997) J. Cell Sci. 110, 2345–2357. [DOI] [PubMed] [Google Scholar]
- 20.Sakhuja, D., Sengupta, J. & Manchanda, S. K. (1982) J. Endocrinol. 95, 283–286. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, K. D., Tissenbaum, H. A., Liu, Y. & Ruvkun, G. (1997) Science 277, 942–946. [DOI] [PubMed] [Google Scholar]
- 22.Rappolee, D. A., Sturm, K. S., Behrendtsen, O., Schultz, G. A., Pedersen, R. A. & Werb, Z. (1992) Genes Dev. 6, 939–952. [DOI] [PubMed] [Google Scholar]
- 23.Watson, A. J., Hogan, A., Hahnel, A., Wiemer, K. E. & Schultz, G. A. (1992) Mol. Reprod. Dev. 31, 87–95. [DOI] [PubMed] [Google Scholar]
- 24.Lighten, A. D., Hardy, K., Winston, R. M. & Moore, G. E. (1997) Mol. Reprod. Dev. 47, 134–139. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka, T. S., Kunath, T., Kimber, W. L., Jaradat, S. A., Stagg, C. A., Usuda, M., Yokota, T., Niwa, H., Rossant, J. & Ko, M. S. (2002) Genome Res. 12, 1921–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang, Z. Q., Fung, M. R., Barlow, D. P. & Wagner, E. F. (1994) Nature 372, 464–467. [DOI] [PubMed] [Google Scholar]
- 27.Lau, M. M., Stewart, C. E., Liu, Z., Bhatt, H., Rotwein, P. & Stewart, C. L. (1994) Genes Dev. 8, 2953–2963. [DOI] [PubMed] [Google Scholar]
- 28.Araki, E., Lipes, M. A., Patti, M. E., Bruning, J. C., Haag, B., 3rd, Johnson, R. S. & Kahn, C. R. (1994) Nature 372, 186–190. [DOI] [PubMed] [Google Scholar]
- 29.Tamemoto, H., Kadowaki, T., Tobe, K., Yagi, T., Sakura, H., Hayakawa, T., Terauchi, Y., Ueki, K., Kaburagi, Y., Satoh, S., et al. (1994) Nature 372, 182–186. [DOI] [PubMed] [Google Scholar]
- 30.Stachecki, J. J. & Armant, D. R. (1996) Development (Cambridge, U.K.) 122, 2485–2496. [DOI] [PubMed] [Google Scholar]
- 31.Wang, J., Mayernik, L., & Armant, D. R. (2002) Dev. Biol. 245, 270–279. [DOI] [PubMed] [Google Scholar]
- 32.Kussel, P. & Frasch, M. (1995) J. Cell Biol. 129, 1491–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao, K., Wang, W., Rando, O. J., Xue, Y., Swiderek, K., Kuo, A. & Crabtree, G. R. (1998) Cell 95, 625–636. [DOI] [PubMed] [Google Scholar]
- 34.Geiman, T. M. & Muegge, K. (2000) Proc. Natl. Acad. Sci. USA 97, 4772–4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimber, S. J. & Spanswick, C. (2000) Semin. Cell Dev. Biol. 11, 77–92. [DOI] [PubMed] [Google Scholar]
- 36.Sheth, B., Moran, B., Anderson, J. M. & Fleming, T. P. (2000) Development (Cambridge, U.K.) 127, 831–840. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki, N., Zara, J., Sato, T., Ong, E., Bakhiet, N., Oshima, R. G., Watson, K. L. & Fukuda, M. N. (1998) Proc. Natl. Acad. Sci. USA 95, 5027–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffman, L. M., Nix, D. A., Benson, B., Boot-Hanford, R., Gustafsson, E., Jamora, C., Menzies, A. S., Goh, K. L., Jensen, C. C., Gertler, F. B., et al. (2003) Mol. Cell. Biol. 23, 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colnot, C., Fowlis, D., Ripoche, M. A., Bouchaert, I. & Poirier, F. (1998) Dev. Dyn. 211, 306–313. [DOI] [PubMed] [Google Scholar]
- 40.Shimono, A. & Behringer, R. R. (2003) Curr. Biol. 13, 613–617. [DOI] [PubMed] [Google Scholar]
- 41.Paria, B. C., Lim, H., Wang, X. N., Liehr, J., Das, S. K. & Dey, S. K. (1998) Endocrinology 139, 5235–5246. [DOI] [PubMed] [Google Scholar]
- 42.Paria, B. C., Elenius, K., Klagsbrun, M. & Dey, S. K. (1999) Development (Cambridge, U.K.) 126, 1997–2005. [DOI] [PubMed] [Google Scholar]
- 43.Raab, G., Kover, K., Paria, B. C., Dey, S. K., Ezzell, R. M. & Klagsbrun, M. (1996) Development (Cambridge, U.K.) 122, 637–645. [DOI] [PubMed] [Google Scholar]
- 44.Das, S. K., Tsukamura, H., Paria, B. C., Andrews, G. K. & Dey, S. K. (1994) Endocrinology 134, 971–981. [DOI] [PubMed] [Google Scholar]
- 45.Paria, B. C., Das, S. K., Andrews, G. K. & Dey, S. K. (1993) Proc. Natl. Acad. Sci. USA 90, 55–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, Z. & Armant, D. R. (2004) Exp. Cell Res. 296, 317–326. [DOI] [PubMed] [Google Scholar]
- 47.Barnard, J. A., Graves-Deal, R., Pittelkow, M. R., DuBois, R., Cook, P., Ramsey, G. W., Bishop, P. R., Damstrup, L. & Coffey, R. J. (1994) J. Biol. Chem. 269, 22817–22822. [PubMed] [Google Scholar]
- 48.Iwamoto, R. & Mekada, E. (2000) Cytokine Growth Factor Rev. 11, 335–344. [DOI] [PubMed] [Google Scholar]
- 49.Kanehisa, M., Goto, S., Kawashima, S. & Nakaya, A. (2002) Nucleic Acids Res. 30, 42–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.