Abstract
The inositol polyphosphate 5-phosphatase INPP5B hydrolyzes the 5-phosphate group from water- and lipid-soluble signaling messengers. Two synthetic benzene and biphenyl polyphosphates (BzP/BiPhPs), simplified surrogates of inositol phosphates and phospholipid headgroups, were identified by thermodynamic studies as potent INPP5B ligands. The X-ray structure of the complex between INPP5B and biphenyl 3,3′,4,4′,5,5′-hexakisphosphate [BiPh(3,3′,4,4′,5,5′)P6, IC50 5.5 μM] was determined at 2.89 Å resolution. One inhibitor pole locates in the phospholipid headgroup binding site and the second solvent-exposed ring binds to the His-Tag of another INPP5B molecule, while a molecule of inorganic phosphate is also present in the active site. Benzene 1,2,3-trisphosphate [Bz(1,2,3)P3] [one ring of BiPh(3,3′,4,4′,5,5′)P6] inhibits INPP5B ca. 6-fold less potently. Co-crystallization with benzene 1,2,4,5-tetrakisphosphate [Bz(1,2,4,5)P4, IC50 = 6.3 μM] yielded a structure refined at 2.9 Å resolution. Conserved residues among the 5-phosphatase family mediate interactions with Bz(1,2,4,5)P4 and BiPh(3,3′,4,4′,5,5′)P6 similar to those with the polar groups present in positions 1, 4, 5, and 6 on the inositol ring of the substrate. 5-Phosphatase specificity most likely resides in the variable zone located close to the 2- and 3-positions of the inositol ring, offering insights to inhibitor design. We propose that the inorganic phosphate present in the INPP5B–BiPh(3,3′,4,4′,5,5′)P6 complex mimics the postcleavage substrate 5-phosphate released by INPP5B in the catalytic site, allowing elucidation of two new key features in the catalytic mechanism proposed for the family of phosphoinositide 5-phosphatases: first, the involvement of the conserved Arg-451 in the interaction with the 5-phosphate and second, identification of the water molecule that initiates 5-phosphate hydrolysis. Our model also has implications for the proposed “moving metal” mechanism.
There are 10 human Mg2+-dependent inositol 5-phosphatase isoenzymes that cleave the 5-phosphate of some inositol phosphates and inositol phospholipid derivatives. Only type I inositol 5-phosphatase (INPP5A) is specific for inositol phosphates; the remaining nine enzymes can hydrolyze either inositol phospholipids or both inositol phospholipids and inositol phosphates.1
Some inositol 5-phosphatases are implicated in disorders including cancer, diabetes, obesity, and neurodegenerative diseases.1,2 Four 5-phosphatase crystal structures with bound ligands are known, namely, INPP5B in complex either with diC8PtdIns(4)P or diC8PtdIns(3,4)P2 (PDB, 3MTC and 4CML, respectively), the polyphosphate 5-phosphatase domain of SPsynaptojanin (PDB, 1I9Z) (from yeast Schizosaccharomyces pombe, hence SPsynaptojanin) in complex with Ins(1,4)P23 and a SHIP2–BiPh(2,3′,4,5′,6)P5 complex (PDB, 4A9C).4 The closest structural family member of INPP5B is OCRL-1 (INPP5F), also known as Lowe oculocerebrorenal syndrome protein, and is associated with the rare X-linked human genetic disorder.5 In the structure of OCRL (PDB, 4CMN),6 an inorganic phosphate molecule was identified in the active site and suggested to mimic either the 5-phosphate of the substrate or the scissile phosphate obtained after its hydrolysis. The coordinates of another inositol 5-phosphatase, the phospholipid-specific INPP5E (PDB, 2XSW), in its apo-form, are also available in the PDB.7
Type II inositol 5-phosphatase (INPP5B) is a 5-phosphatase that hydrolyzes both inositol phosphates and inositol phospholipids.8 INPP5B was first identified in platelets9 and has similar affinities for both PtdIns(4,5)P2 and Ins(1,4,5)P3.10 INPP5B plays a role in the endocytic pathway11 and the early secretory pathways,12 binding to the small GTPase protein Rab5 (that also binds OCRL). The Gram-negative bacterium, Yersinia pseudotuberculosis, causes Far East scarlet-like fever in humans. The organism invades host cells in a process similar to phagocytosis13 and hijacks INPP5B and OCRL, which are then recruited to hydrolyze phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2] from the host organism. Enzymatic hydrolysis of PtdIns(4,5)P2 together with the association of the GTPase Rab5 with prevacuoles completes the multistep invasion process.13 INPP5B and OCRL may be potential targets for inhibition by specific 5-phosphatase ligands. However, to date there is no specific human disease associated with INPP5B.
Because of the therapeutic importance of these enzymes, attempts to identify lead ligands for drug discovery have been initiated. A high throughput screening methodology for small molecule inhibitors of inositol 5-phosphatase enzymes was recently developed by the De Camilli group.14 Compound YU142670 (1, Figure 1) was identified as an inhibitor of two enzymes INPP5B (IC50 = 1.78 μM) and OCRL (IC50 = 0.71 μM). YU142670 became the lead compound for evaluating binding by isothermal titration calorimetry and several other assays, providing a platform for the development of new specific and potent inhibitors.
Figure 1.
Structures of YU142670 (1), d-myo-inositol 1,3,4,5-tetrakisphosphate (2), biphenyl 3,3′,4,4′,5,5′-hexakisphosphate (3), and benzene 1,2,4,5-tetrakisphosphate (4).
Several methods are currently available for characterizing protein binding partners without monitoring enzyme activity.15,16 One such method developed by the Structural Genomics Consortium (SGC)15 uses light scattering-based approaches to measure the variation of the thermal stability of proteins (thermal-shift) when screened against a library of compounds. The principle of this light scattering-based approach is that ligand binding stabilizes a protein structure to thermal unfolding and subsequent aggregation. Thus, the temperature at which the protein begins to aggregate, where light scattering is dramatically enhanced, increases in the presence of ligand. Those ligands which provide the highest temperature increase under optimum conditions are good candidates for X-ray crystal structure studies. The ligands that were selected were from the benzene and biphenyl phosphate class.
Previously, we demonstrated the potential of benzene- and biphenyl phosphates as inositol phosphate surrogates for structural studies.4,17 Benzene 1,2,3,4-tetrakisphosphate, Bz(1,2,3,4)P4, cocrystallized with the pleckstrin homology (PH) domain of protein kinase Bα (PKBα)17 and is a good surrogate for the phosphatidylinositol 3,4,5-trisphosphate [PtdIns(3,4,5)P3] headgroup inositol 1,3,4,5-tetrakisphosphate, Ins(1,3,4,5)P4 (2, Figure 1). These studies led by others generated drug-like inhibitors of the PKB PH domain by virtual screening.18 We have also cocrystallized SHIP2 in complex with biphenyl 2,3′,4,5′,6-pentakisphosphate, BiPh(2,3′,4,5′,6)P5; the resulting structure facilitated identification of a mobile loop located above the binding site.4 Synthetic benzene- and biphenyl phosphates (BzPs/BiPhPs) are compounds that could be used generically in the early stages of structure-based drug design targeting phosphoinositide-related pathways.
Here, we present the results of the thermal-shift screen that we used to select candidates from 40 ligands when assayed against 8 of the 10 catalytic domains of inositol phosphate 5-phosphatases. We report crystal structures for two INPP5B 5-phosphatase domain–ligand complexes formed in the presence of small molecule benzene phosphate (BzP and BiPhP) inhibitors that emerged from screening: first, INPP5B and biphenyl 3,3′,4,4′,5,5′-hexakisphosphate, BiPh(3,3′,4,4′,5,5′)P6 (3, Figure 1), and second, INPP5B in complex with benzene 1,2,4,5-tetrakisphosphate, Bz(1,2,4,5)P4 (4, Figure 1). The results further extend the potential of the BzP/BiPhP substrate surrogate approach, allowing us to identify the region of the inhibitors that is important to modulate their selectivity toward specific 5-phosphatase members and to offer new insights into the mechanism of INPP5B-catalyzed phosphate group cleavage (Supporting Information Scheme S1, Table S1, and Figure S1A–C).
Experimental Section
The general methods were followed according to a previous report,19 and the synthesis was performed according to Scheme 1.
Scheme 1. Synthesis of Biphenyl 3,3′,4,4′,5,5′-Hexakisphosphate (3).
(i) K2CO3, (2 equiv), 35°C, acetone/water (3:3.5), 0.5% Pd(OAc)2, 67%; (ii) BBr3, CH2Cl2, dry ice/acetone cooling, 94%; (iii) (EtO)2P–Cl, CH2Cl2, diisopropylethylamine, mCPBA (80%); (iv) TMSBr, CH2Cl2, H2O, TEAB (91%).
3,3′,4,4′,5,5′-Hexamethoxybiphenyl (7)
A mixture of K2CO3 (1.11 g, 8 mmol), Pd(OAc)2 (4 mg, 0.5%), 1,2,3-trimethoxy-5-bromobenzene (5) (988 mg, 4 mmol), and 3,4,5-trimethoxyphenylboronic acid (6) (1.27 g, 6 mmol), in distilled water (15 mL) and acetone (12 mL), was stirred for 1 h at 35 °C. The reaction mixture was partitioned between water and CH2Cl2 (50 mL of each), and the remaining solid was purified by flash chromatography (CH2Cl2 alone) and was pure enough without recrystallization. Yield (902 mg, 67%). Rf in CH2Cl2–ether (10:1) = 0.35; mp = 129–130 °C (Lit.,20 129–130 °C). 1H NMR (400 MHz, CDCl3) 3.89 (s, 6 H, 2 × ArOMe), 3.93 (2 s, 12 H, 4 × ArOMe), 6.72 (s, 4 H, 4 × ArH), 13C NMR (100 MHz, CDCl3) 56.24, (q, 4 × ArOMe), 60.91 (q, 2 × ArOMe), 104.58 (d, 4 × ArH), 137.54, 137.67 (s, Cq, Ar-Ar), 153.37 (s, Cq, ArOMe). m/z calcd. C18H23O6 [M + H]+ 335.1489; found, 335.1484.
3,3′,4,4′,5,5′-Hexahydroxybiphenyl (8)
3,3′,4,4′,5,5′-Hexamethoxybiphenyl (7) (865 mg, 2.58 mmol) was partially dissolved in dry CH2Cl2 (10 mL), and the solution was cooled using a dry ice acetone mixture. A solution of BBr3 in CH2Cl2 (1.0 M, 25 mL) was added over 5 min to the cooled solution that turned yellow and was allowed to warm to ambient temperature over a period of 19 h. An aqueous solution of 1 M HCl (50 mL) was added to the cooled mixture (dry ice–acetone), which resulted in a white and brick red precipitate. Water (100 mL) was then added and the layers separated. The aqueous layer was extracted with ethyl acetate (4 × 100 mL) and dried (MgSO4), and the solvent was evaporated. The remaining solid was suspended in ether (40 mL) to dissolve any of the impurities and filtered to give the title compound (8) as a salmon pink-colored solid (609 mg, 94%). 1H NMR (400 MHz, (d6-DMSO) 6.19, 6.35, 6.38 (4 H, 3 s, 4 × ArH), after D2O exch, 6.39 (s, 4 × ArH), 7.04 (v br s, 6 × OH, ArOH, D2O ex), (100 MHz, d6-DMSO) 104.83 (d, 4 × ArH), 131.38, 132.03, 146.11, 146.19 (s, Cq, Ar-Ar, 6 × ArOH). m/z calcd. C12H11O6 [M + H]+ 251.0550; found, 251.0540.
3,3′,4,4′,5,5′-Hexakis(diethoxyphosphoryloxy)biphenyl (9)
A mixture of diethyl chlorophosphite (1.33 mL, 7.8 mmol) and N,N-diisopropylethylamine (1.75 mL, 10.0 mmol) was stirred at room temperature in dry CH2Cl2 (10 mL) to give a yellow solution. 3,3′,4,4′,5,5′-Hexahydroxybiphenyl (8) (250 mg, 1 mmol) was added in small portions and the solid dissolved with the aid of ultrasound within 5–10 min. The solution remained a yellow color and was stirred for a further 30 min. The mixture was cooled using dry ice in acetone, mCPBA (2.58 g, 15 mmol) in CH2Cl2 (25 mL) was added in one portion, the color turned a dark olive green, and the solution was stirred for a further 30 min. The mixture was washed with 0.5 M aqueous phosphate buffer at pH 7.4 (2 × 100 mL), dried, and purified by flash chromatography, eluting with EtOAc then EtOAc–EtOH (5:1) to give the title compound as a pale yellow syrup (780 mg, 80%), Rf = 0.24 (EtOAc–EtOH, 5:1). 1H NMR (400 MHz, CDCl3) 1.35–1.41 (m, 36 H, 6 × ArOP(O) (OCH2CH3)2), 4.21–4.38 (m, 24 H, 6 × ArOP(O) (OCH2CH3)2). 13C NMR (100 MHz, CDCl3) 16.03, 16.10 (q, ArOP(O) (OCH2CH3)2), 64.94, 64.99, 65.05 (t, ArOP(O)(OCH2CH3)2), 115.53 (d, 4 × ArH), 133.03 (app q, Cq, J = 8.1 Hz,), 136.54 (s, Cq, Ar-Ar), 143.88 (dd, Cq, J = 3.7, 5.9 Hz, Cq, ArO(O)P(OCH2CH3)2). 31P (162 MHz, CDCl3) – 4.99 (s, 2P, ArO(O)P(OCH2CH3)2), – 5.79 (s, 2P, ArO(O)P(OCH2CH3)2), – 5.80 (s, 2P, ArO(O)P(OCH2CH3)2). m/z calcd. C36H65O24P6 [M + H]+ 1067.2286; found, 1067.2273. Calcd for C36H64O24P6 C 40.53, H 6.05; found, C 40.1, H 6.06.
3,3′,4,4,′5,5′-Biphenylhexakisphosphate (3)
3,3′,4,4′,5,5′-Hexakis-(diethoxyphosphoryloxy)biphenyl (9) (106.6 mg, 100 μmol), was dissolved in dry CH2Cl2 (5 mL). Bromotrimethylsilane (1.0 mL, 7.57 mmol) was added, and the solution was stirred for 3 days after monitoring the disappearance of the ethyl groups from the compound. The solvents were evaporated, and the remaining syrup was stirred in a mixed solvent of TEAB (1 mL) and water (2 mL) for 30 min. The title compound was purified over Q-Sepharose Fast Flow using a linear gradient of 0 → 2.0 M TEAB, eluting at 2.0 M buffer and the title compound obtained as a glassy triethylammonium salt (90.66 μmol, 91%). Compound 3 was reported surprisingly19 to stimulate the release of intracellular Ca2+, possibly via Ins(1,4,5)P3 receptors or via another mechanism. 1H NMR (400 MHz, D2O) 7.34 (s, 4 H, 4 × ArH). 13C NMR (100 MHz, D2O) 114.87 (d, 4 × ArH), 134.66 (s, Cq, Ar-Ar), 135.39 (app q, Cq, C-P coupling, J = 8.5 Hz, ArOPO32−), 145.77 (dd, Cq, J = 3.1, 6.1 Hz, C-P coupling, ArOPO32−). 31P (162 MHz, D2O) − 5.82 (6 P, 6 × ArOPO32–). m/z calcd. C12H15O24P6 [M – H]− 728.8384; found, 728.8369.
5-Phosphatase Inhibition Assay
The enzyme activity was monitored based on the method previously used4 that incorporated the established malachite green phosphate assay (BioAssay Systems) to measure the inorganic phosphate released by the reaction. A typical assay was carried out in a total volume of 50 μL with a buffer containing 20 mM HEPES at pH 7.5, 5% glycerol, 300 mM NaCl, 2 mM TCEP, and 2 mM MgCl2. The enzymes, 0.01 μM for INPP5B and 0.1 μM for untagged and His-tagged SHIP2, were incubated at 30 °C for 5 min with 50 μM Ins(1,4,5)P3 for INPP5B and 100 μM Ins(1,3,4,5)P4 for SHIP2. For IC50 determination, serial dilutions of the inhibitors were added. The assay was stopped by the addition of 12 μL of 10% TCA. The free phosphate was measured by the addition of 40 μL of malachite green working reagent and 98 μL of water, followed by incubation at room temperature for 30 min. The absorbance was measured at 620 nm.
Protein Expression and Purification
The sequences encoding INPP5B residues 259–563 were subcloned into the vector pNIC–CH2 adding a C-terminal 6 × His-Tag. The proteins were expressed in Escherichia coli strain BL21(DE3) R3 pRARE. Cultures were grown in TB medium supplemented with 8 g/L glycerol, 100 μg/mL kanamycin, and 34 μg/mL chloramphenicol in a LEX bioreactor system (Harbinger Biotechnology) at 37 °C until OD600 reached ∼2. The culture temperature was reduced to 18 °C over a period of 1 h before target expression was induced by the addition of 0.5 mM IPTG. Expression was allowed to continue overnight. Cells were harvested by centrifugation (4,430g, 10 min, 4 °C), and the pellet was resuspended in lysis buffer (100 mM HEPES, 500 mM NaCl, 10% glycerol, 10 mM imidazole, 0.5 mM TCEP, 2000 U Benzonase Merck, and Complete EDTA-free protease inhibitor, pH 8.0). Cells were disrupted by sonication (Vibra-Cell, Sonics) at 80% amplitude for 3 min effective time (pulsed 4 s on, 4 s off), and cell debris was removed by centrifugation (49,000g, 20 min, 4 °C). The supernatant was decanted and filtered through a 0.45 μm flask filter. The filtered lysate was loaded onto a Ni-charged HiTrap Chelating HP (GE Healthcare) column and washed with IMAC wash1 buffer (20 mM HEPES, 500 mM NaCl, 10% glycerol, 10 mM imidazole, and 0.5 mM TCEP, pH 7.5) followed by IMAC wash2 buffer (20 mM HEPES, 500 mM NaCl, 10% glycerol, 25 mM imidazole, and 0.5 mM TCEP, pH 7.5). Bound protein was eluted from the IMAC column with IMAC elution buffer (20 mM HEPES, 500 mM NaCl, 10% glycerol, 500 mM imidazole, and 0.5 mM TCEP, pH 7.5) and subsequently loaded onto a HiLoad 16/60 Superdex 200 Prep grade column (GE Healthcare) equilibrated in gel filtration buffer (20 mM HEPES, 300 mM NaCl, 10% glycerol, and 0.5 mM TCEP, pH 7.5).
Stability Studies by Differential Static Light Scattering (DSLS)
The effect of compounds and substrate/inhibitor analogues on thermal stability of 5-phosphatases was studied by differential static light scattering (Stargazer384, Harbinger). Protein samples at 0.2 mg/mL were heated from 25 to 80 °C at a rate of 1 °C/min in clear bottomed 384-well plates (Nunc) in 50 μL of buffer (20 mM Hepes, 300 mM NaCl, 20% glycerol, 2 mM MgCl2, and 2 mM TCEP, pH 7.5). Protein aggregation was measured by recording the scattered light using a CCD camera taking images of the plate every 0.5 °C. Aggregation temperature was calculated as previously described.21,22 ΔTagg was calculated as the difference in aggregation temperature between samples with no compound added and the sample with addition of 0.5 mM compound. The plot (Supporting Information, Scheme S1) shows the average ΔTagg values for duplicate samples.
Crystallization and Data Collection
Crystallization experiments were performed using the sitting-drop method. Prior to setting-up the crystallization trials, INPP5B was incubated on ice with either 2 mM BiPh(3,3′,4,4,′5,5′)P6 and 2 mM MgSO4 or 2 mM Bz(1,2,4,5)P4 and 2 mM MgSO4. Protein concentration used in the crystallization trials was 18 and 22 mg/mL, respectively. Crystals were obtained by mixing either 0.2 μL of protein solution with 0.1 μL of precipitant (complex with BiPh(3,3′,4,4′,5,5′)P6) or 0.1 μL of protein solution with 0.2 μL of precipitant (complex with Bz(1,2,4,5)P4). The precipitant was composed of 10% glycerol, 25% propanediol, and 0.1 M sodium/potassium phosphate at pH 6.2 (BiPh(3,3′,4,4′,5,5′)P6) or of 0.2 M Li2SO4, 0.1 M Bis-Tris at pH 5.5, and 25% PEG3350 (Bz(1,2,4,5)P4). Crystallization trials were incubated at 4 °C. Crystals were then harvested and briefly transferred into a cryoprotectant solution whose composition was similar to the precipitant supplemented with either 25% final concentration glycerol and 2 mM MgSO4 (complex with BiPh(3,3′,4,4,′5,5′)P6) or 21.7% glycerol, 2 mM MgSO4, and 2 mM Bz(1,2,4,5)P4 (complex with Bz(1,2,4,5)P4). Crystals were flash-frozen into liquid nitrogen. Data were recorded at BESSY light source on beamlines BL14.2 and BL14.1 for complexes with BiPh(3,3′,4,4′,5,5′)P6 and Bz(1,2,4,5)P4, respectively.
Data Processing, Structure Solution, Model Building, and Refinement
Data were integrated with XDS23 and scaled with Scala.24,25 Structures were solved by Molecular Replacement with Phaser26 using the structure of apo-INPP5B (PDB code 3N9V) as a probe. Model-building and refinement were performed by using iterative cycles of manual model building with Coot27 and maximum-likelihood refinement with Refmac 5.524,28 (complex with BiPh(3,3′,4,4′,5,5′)P6) or Buster 2.1129 (complex with Bz(1,2,4,5)P4). The geometric quality of the final models was assessed using Molprobity.30 Crystallographic statistics are summarized in Table 1.
Table 1. Data Collection and Refinement Statistics.
| INPP5B | ||
|---|---|---|
| BiPh(3,3′,4,4′,5,5′)P6 | Bz(1,2,4,5)P4 | |
| PDB ID | 5A7I | 5A7J |
| Data Collection | ||
| synchrotron | BESSY | BESSY |
| beamline | BL14.1 | BL14.1 |
| wavelength (Å) | 0.91841 | 0.91841 |
| resolution range (Å) | 34.29–2.89 | 48.48–2.90 |
| (outest shell) | (3.05–2.89) | (3.06–2.9) |
| space group | P41212 | P21 |
| unit-cell dimensions (Å) | a = 96.97 | a = 52.26 |
| b = 96.97 | b = 94.44 | |
| c = 152.14 | c = 78.31 | |
| unit-cell angles (deg) | α = β = γ = 90 | α = γ = 90, β = 106.32 |
| completeness (%) | 99.9 (100.0) | 99.7 (99.7) |
| unique reflections | 16922 | 16223 |
| mean (I)/SD (I)a | 21.2 (2.2) | 6.9 (2.3) |
| redundancy | 7.0 (7.2) | 3.6 (3.6) |
| Rmeas (%)b | 7.5 (90.7) | 19.4 (67.9) |
| Refinement | ||
| resolution range (Å) | 34.29–2.89 | 48.48–2.90 |
| (outest shell) | (2.97–2.89) | (3.10–2.90) |
| Rcryst (%)c | 19.8 (33.8) | 20.2 (21.9) |
| Rfree (%)d | 22.3 (34.5) | 25.0 (31.0) |
| Model Content | ||
| protein atoms | 2545 | 4971 |
| ligand atoms | 42 | 52 |
| metal atoms | 1 | 0 |
| water molecules | 28 | 76 |
| others | 16 | 26 |
| average B factors (Å2) | ||
| protein atoms | 83.59 | 39.10 |
| ligand atoms | 76.44 | 65.84 |
| metal atoms | 66.37 | n.d. |
| water molecules | 71.94 | 19.15 |
| other atoms | 111.59 | 63.89 |
| r.m.s.d. bonds (Å) | 0.007 | 0.008 |
| r.m.s.d. angles (deg) | 1.191 | 1.000 |
| Ramachandran plot (%)e | ||
| (favored, outliers) | 95.82, 0.32 | 94.95, 0 |
Mean (I)/SD is the mean ratio for all reflections of ⟨Ih⟩/SD ⟨Ih⟩, where for each unique reflection h, ⟨Ih⟩ is the weighted mean of measured Ih, and SD ⟨Ih⟩ is the mean of estimated error SD(I).
Rmeas = 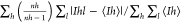
Rcryst =  , where Fobs and Fcalc are the observed and calculated
structure factors, respectively.
, where Fobs and Fcalc are the observed and calculated
structure factors, respectively.
Rfree is the same as Rcryst but based on a subset of 5% (5.1% and 5.0% for INPP5B-BiPh(3,3′,4,4′,5,5′)P6 and INPP5B-Bz(1,2,4,5)P4, respectively) of reflections omitted during refinement.
Values were computed by Molprobity (ref (26)).
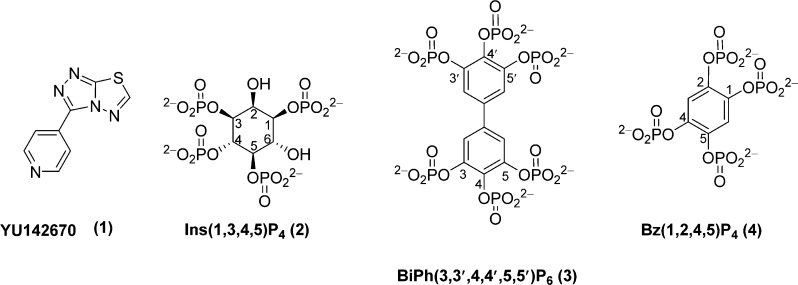
Production of the INPP5B 5-Phosphatase Mechanism
Figure 8A was produced by overlaying INPP5B–BiPh(3,3′,4,4′,5,5′)P6 and INPP5B–diC8PtdIns4P complex structures, and an Ins(1,4,5)P3 molecule was then overlaid onto the PtdIns4P. The Ins(1,4,5)P3 molecule and the dihedral angles of the 5-phosphate of Ins(1,4,5)P3 were rotated so that the 5-phosphate group points into the expected binding site, with the 5-phosphate in an equivalent orientation to the inorganic phosphate and minimal difference between the inositol rings of Ins(1,4,5)P3 and diC8PtdIns4P. Finally, the 5-phosphate of a second identically orientated Ins(1,4,5)P3 was replaced with a trigonal bipyramidal intermediate with 3 planar oxygen atoms in an equivalent orientation to the inorganic phosphate. For each stage of the mechanism, the residues and Mg2+ from the INPP5B–BiPh(3,3′,4,4′,5,5′)P6 complex are shown, and the other relevant parts of the overlay are added so the starting point adds the water molecule from the INPP5B– diC8PtdIns4P complex and the Ins(1,4,5)P3. The potential intermediate shows the trigonal bipyramidal-modified Ins(1,4,5)P3 or PtdIns(4,5)P2 at the 5-position, and the final stage shows the release of products.
Figure 8.
(A) Stick diagram showing participants in a pentacoordinate intermediate based on the AP endonuclease mechanism. After nucleophilic attack of an activated water molecule the trigonal bipyramidal intermediate is stabilized by a series of residues and the active site Mg2+ ion. Arg-451 (colored green) was not originally suggested as part of the mechanism, but is located such that it may help stabilize the intermediate and is conserved in all 5-phosphatases. Only schematic involvement of the various residues is depicted. (B) Mechanism for the hydrolysis of the 5-phosphate of Ins(1,4,5)P3 by INPP5B, based upon the stick diagram and interactions found in (A) together with additional contributing amino acid residues and the movement of the Mg2+ ion from site B towards site A. Attack by a water molecule produces a trigonal bipyramidal intermediate that collapses releasing the phosphate anion. The amino acids are color-coded for easier recognition. The anion that forms at the 5-position is probably quenched with a proton originating from a nearby water molecule, possibly Mg2+-bound, or from the adjacent protonated 4-phosphate group. This has not been shown on the diagram for the sake of clarity. Key: R = H for Ins(1,4,5)P3, R = diacylglycerol for PtdIns(4,5)P2. Only schematic involvement of the various residues is depicted. Negative charges during phosphoryl transfer are not shown.
Results
Identification of 5-Phosphatase Ligands Suitable for Structural Studies
Forty, predominantly synthetic ligands including inositol polyphosphates, inositol phospholipids, benzene-, and biphenyl polyphosphates, were screened against eight inositol polyphosphate 5-phosphatases using a differential static light scattering method in a multiwell-format.15 The stability of the protein was monitored by following temperature-induced protein aggregation using light scattering.15 The variation of the temperature of aggregation (ΔTagg) for all the compounds screened against the 5-phosphatases is shown in Supporting Information, Scheme S1. The compound names and structures are shown in Supporting Information, Table S1, and the structures of all the compounds used in the crystallization trials are found in the Supporting Information, Figure S1A, B, and C. The compounds that were considered as hits generated a ΔTagg above 2 °C, and these were followed up by crystallographic studies. Negative controls using dimethyl sulfoxide and inositol were used in the assay and did not reveal any effect on the stability of the protein.
By applying 2 °C in the ΔTagg cutoff, we identified 16 potential cocrystallization ligands for INPP5B. The highest ΔTagg were observed when the reaction media contained product or substrate analogues (between 11 to 13 °C) such as the three lipid derivatives, diC8-phosphatidylinositol 4,5-bisphosphate (STO1429), diC8-phosphatidylinositol 4-phosphate (STO1430), and diC8-phosphatidylinositol 3,4,5-trisphosphate (STO927).6 Noteworthy is that none of the product or substrate analogues lacking a phosphate in position 4 of the inositol group led to a ΔTagg exceeding the 2 °C cutoff. This observation can be related with previous studies that revealed that INPP5B is specific to PtdIns(4,5)P2 and PtdIns(3,4,5)P3 but is unable to mediate the hydrolysis of PtdIns(5)P and PtdIns(3,5)P2.10 Besides substrates and product analogues, compounds that led to the highest ΔTagg belonged to the BiPhP series. All three BiPhP derivatives (labeled STO1415, STO1416, and STO951 Supporting Information, Figure S1A and C) have ΔTagg values in the region of 8–10 °C. INPP5B cocrystallized in the presence of the biphenyl derivative BiPh(3,3′,4,4′,5,5′)P6 (STO1415) and the single-ringed ligand, Bz(1,2,4,5)P4 (STO949), leading to the highest ΔTagg in this family of compounds. While crystallization with STO1429 and STO927 led to structures containing the product of the reaction,6 the aromatic phosphates are resistant to hydrolysis when compared to their inositol-containing counterpart.
Surprisingly, ΔTagg for OCRL was <2 °C for inositol phospholipid derivatives, suggesting that either binding to the substrate does not generate a high thermal stabilization or a low affinity for the product of the reaction (likely produced during the course of the assay). However, the most promising ligands for OCRL, ranked according to the amplitude of the thermal-shift induced, are the three biphenylpolyphosphates. BiPh(2,3′,4,5′,6)P5 (STO951) gave the highest ΔTagg value ≈8 °C with BiPh(3,3′,4,4′,5,5′)P6 (STO1415) and BiPh(2,2′,4,4′,5,5′)P6 (STO1416) both having ΔTagg ≈ 6 °C. Because of the apparent stability of the complex between ligand and protein, these BiPhP derivatives may be suitable candidates for crystallization studies than the inositol phospholipid derivatives.
Structures of the INPP5B–BiPh(3,3′,4,4′,5,5′)P6 and INPP5B–Bz(1,2,4,5)P4 Complexes
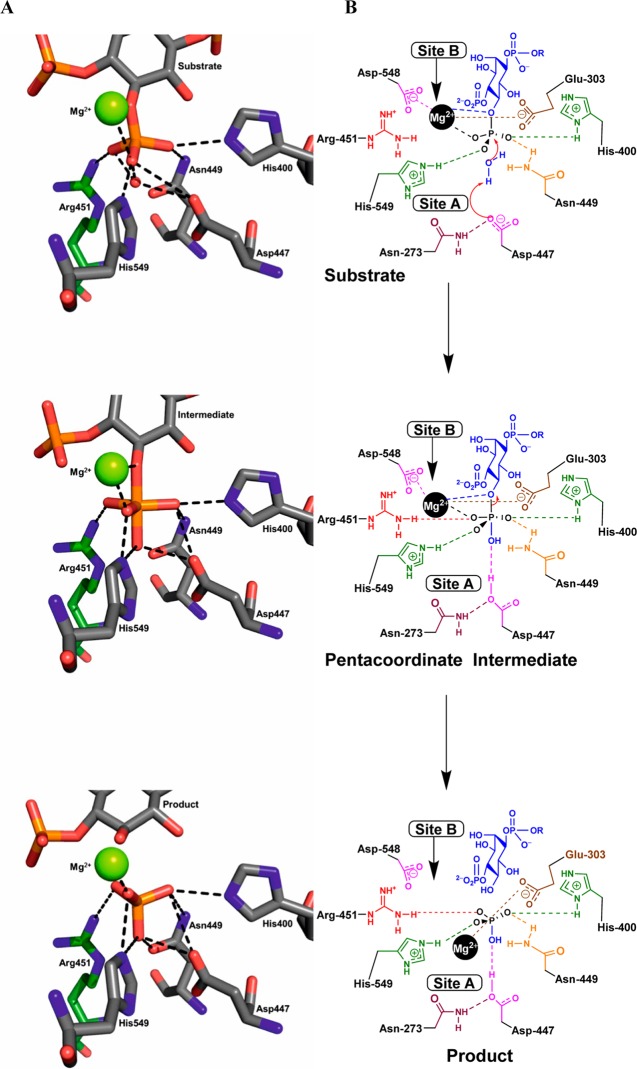
The crystal structure of the BiPh(3,3′,4,4′,5,5′)P6–INPP5B complex contains a single molecule of INPP5B in the asymmetric unit with BiPh(3,3′,4,4′,5,5′)P6 bound in a shallow pocket. The base of the pocket also contains one Mg2+ ion and a molecule of inorganic phosphate (Figure 2, panel A). Examination of the bound BiPh(3,3′,4,4′,5,5′)P6 reveals multiple interactions among the 3-, 4-, 5-, 4′-, and 5′-phosphates with side chain amino acids of INPP5B (Figure 3A and Supporting Information, Figure S2A). Most of the protein–ligand interactions are focused around the 3-, 4-, and 5-phosphates on one pole of the ligand inserted into the pocket of the active site. The aromatic ring of this pole has an edge to face π–π stacking interaction with His-404. The 3-phosphate interacts with Lys-380 and Asn-379 (although this is longer range, 3.2 Å). The side chain of Lys-380 is located between the 3- and 4-phosphates of BiPh(3,3′,4,4′,5,5′)P6, although the interactive bond length with the 4-phosphate is slightly longer, 3.2 Å. In addition to its interaction with Lys-380, the 4-phosphate also interacts with Glu-303 and forms a longer range interaction with His-400 (3.2 Å). The 5-phosphate interacts with the side chain hydroxyl of Tyr-502 and has a longer range interaction with Arg-518 (3.2 Å). The other pole of the ligand is orientated toward one side at the top of the active site pocket. The 5′-phosphate interacts with Arg-518, and Lys-516 is positioned between, and interacts with, both the 4′- and 5′-phosphates. There are additional interactions between phosphates 5-, 3′-, and 4′-phosphates and three water molecules. The Mg2+ ion and inorganic phosphate are positioned between the 4- and 5-phosphates within a network of potential interactions from the phosphate groups (Figure 3A).
Figure 2.
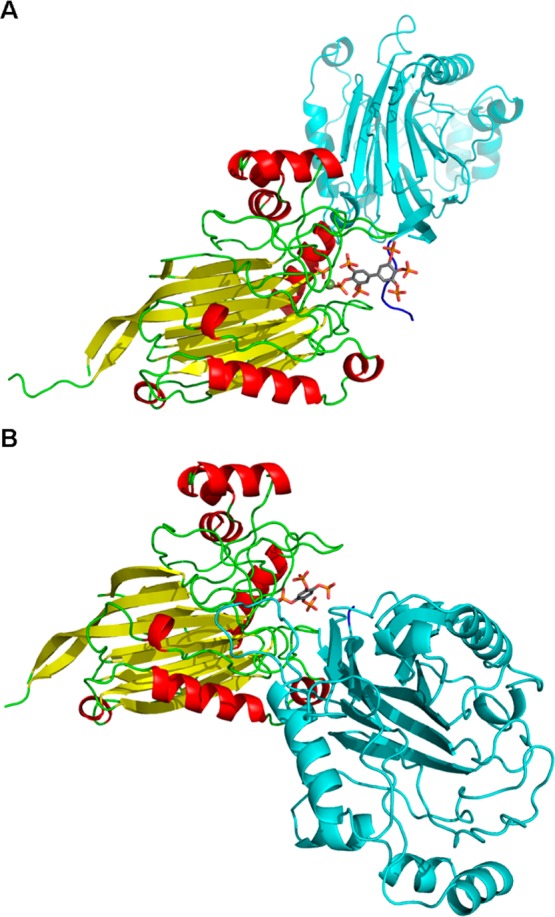
Cartoon representation of the crystal structure of INPP5B–BiPh(3,3′,4,4′,5,5′)P6 (panel A) and INPP5B–Bz(1,2,4,5)P4 (panel B) complexes. BiPh(3,3′,4,4′,5,5′)P6, inorganic phosphate, and Bz(1,2,4,5)P4 are shown as sticks, the Mg2+ ion is shown as a green sphere, β-sheets are in yellow, α-helices in red, and loop regions in green. For both complexes, the symmetry related molecule that provides additional interactions to the phosphorylated ligand is shown in cyan with the His-Tag highlighted in dark blue. Only chain A of the INPP5B–Bz(1,2,4,5)P4 complex is shown for clarity.
Figure 3.
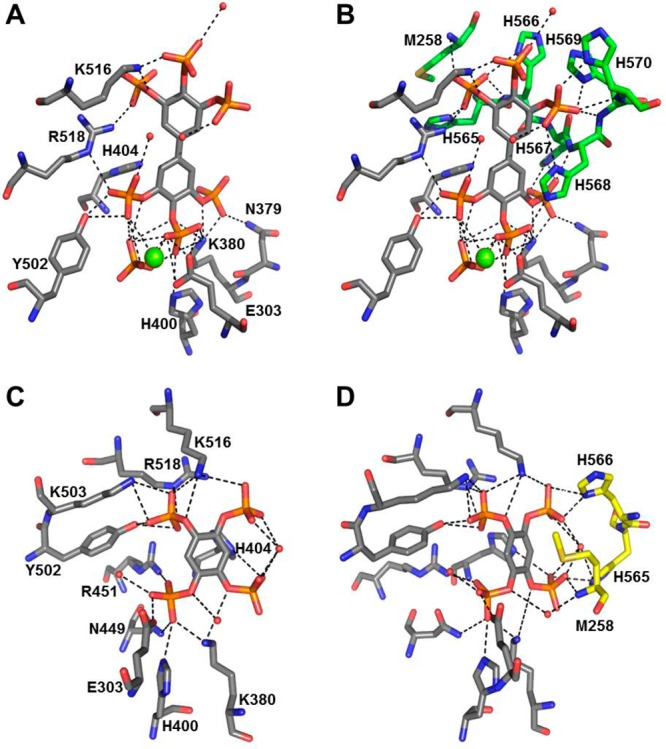
Panel A shows BiPh(3,3′,4,4′,5,5′)P6 bound in the catalytic site of INPP5B, with a molecule of inorganic phosphate positioned between the 3- and 4-phosphates. The green sphere is a magnesium ion (Mg2+), and the red spheres are water molecules in the absence of the His-Tag. Panel B shows the BiPh(3,3′,4,4′,5,5′)P6 bound in the catalytic site of INPP5B, in complex with the His-Tag. The carbon atoms of the two INPP5B molecules interacting with BiPh(3,3′,4,4′,5,5′)P6 are colored gray (catalytic site) and green (neighboring INPP5B molecule). Panel C shows Bz(1,2,4,5)P4 bound in the catalytic site of INPP5B. The red sphere indicates the presence of a molecule of water and in the absence of the His-Tag. Panel D shows the presence of Bz(1,2,4,5)P4 in the catalytic site of INPP5B (carbon atoms colored in gray) bound to the His-Tag of a neighboring INPP5B molecule (carbon atoms are colored yellow).
In contrast to the BiPh(3,3′,4,4′,5,5′)P6–INPP5B structure, the Bz(1,2,4,5)P4–INPP5B structure (Figure 2B, Figure 3C and D, and Supporting Information Figure S2B and C) has two molecules of INPP5B in the asymmetric unit. Each molecule has one Bz(1,2,4,5)P4 ligand bound in the same shallow pocket as BiPh(3,3′,4,4′,5,5′)P6 but lacking both the active site metal ion or inorganic phosphate (Figure 2B). An overlay of the two molecules in the asymmetric unit shows a shift ca. 0.4 Å of the aromatic core of Bz(1,2,4,5)P4, indicating a small variation in the position and orientation of the phosphate groups between the two molecules. However, for both INPP5B structures (Supporting Information, Figure S2B and C) all phosphates of Bz(1,2,4,5)P4 produce similar interactions with side chains of INPP5B. The only noticeable difference between the two structures involves the 1-phosphate, where chain A has a longer range interaction (3.6 Å) with His-404. However, for chain B the small shift of Bz(1,2,4,5)P4 allows the 1-phosphate to move away from His-404 and toward Lys-380, which itself shifts to form a bidentate interaction with the 1-phosphate. The 2-phosphate interacts with Glu-303, Lys-380, and His-400 side chains of chain A as well as showing longer range interactions with Asn-449 (3.3 Å) and Arg-451 (3.5 Å). In chain B, the shift in Lys-380 means it no longer interacts with the 2-phosphate, and the longer range interaction with Arg-451 increases to 3.7 Å. The 4-phosphate shows some variation in rotation between the two molecules, but this phosphate still interacts with the same residues (Tyr-502, Lys-503, Lys-516, and Arg-518), albeit with differences in bond length interactions. The 5-phosphate interacts with the side chain of Lys-516 in the asymmetric unit of both INPP5B molecules. In both subunits, there is a T-shaped π–π stacking interaction between the aromatic ring of Bz(1,2,4,5)P4 and His-404 (Supporting Information, Figure S2B and C). The aromatic ring also has a π-cation interaction with Arg-451 similar to the complex formed between INPP5B and BiPh(3,3′,4,4′,5,5′)P6. There are two equivalent waters in each INPP5B molecule with one located between, and interacting with, the 1- and 5-phosphates of Bz(1,2,4,5)P4 and the other binding to the 2-phosphate (Supporting Information, Figure S2B and C). There is one extra water molecule in the active site of chain A in the asymmetric unit that also interacts with the 2-phosphate.
Unlike other costructures between 5-phosphatases and ligands, the binding of BiPh(3,3′,4,4′,5,5′)P6 and Bz(1,2,4,5)P4 by INPP5B (Figure 3B and D) and also that of BiPh(2,3′,4,5′,6)P5–SHIP24 results in additional interactions from a symmetry related molecule. Additional interactions with BiPh(3,3′,4,4′,5,5′)P6 and Bz(1,2,4,5)P4 are mediated by the N-terminal Met-258 and His-tag residues that are carried on the INPP5B construct we used for structural studies. There are interactions with the 3-, 3′, 4-, and 5′-phosphates of BiPh(3,3′,4,4′,5,5′)P6 and the 1- and 5-phosphates of Bz(1,2,4,5)P4. The electron density maps for BiPh(3,3′,4,4′,5,5′)P6 and Bz(1,2,4,5)P4 in complex with INPP5B are shown in Supporting Information, Figure S3).
Inhibition of INPP5B and SHIP2 by BiPh and BzP Ligands
Inhibition of INPP5B activity was measured using a small library of biphenyl and single ring benzene polyphosphates to investigate which structural components of BiPh(3,3′,4,4′,5,5′)P6 contribute to potent inhibition. Enzyme activity was assayed using the malachite green phosphate assay (BioAssay Systems) to measure the release of inorganic phosphate from the substrate. A comparison of IC50 values for these inhibitors (Table 2) shows that the biphenyl polyphosphates are the most potent inhibitors: IC50 = 2.4 ± 0.3 μM for BiPh(2,3′,4,5′,6)P5, 5.4 ± 0.7 μM for BiPh(2,2′,4,4′,5,5′)P6, and 5.5 ± 0.5 μM for BiPh(3,3′,4,4′,5,5′)P6. Bz(1,2,4,5)P4 was the most potent single ring benzene polyphosphate evaluated with an IC50 = 6.3 ± 0.8 μM and only slightly less potent than the biphenyl polyphosphate inhibitors. Bz(1,2,3)P3 is the weakest inhibitor; IC50 = 33.5 ± 6.8 μM. It is worth noting that the KM of INPP5B for Ins(1,4,5)P3, previously reported using the same malachite green phosphate assay, was 55 ± 22 μM.10
Table 2. IC50 Values of a Series of Compounds for His-Tagged-INNPP5B (0.01 μM enzyme, 50 μM Ins(1,4,5)P3), Compared to Untagged- and His-Tagged-SHIP2 (0.1 μM enzymes, 100 μM Ins(1,3,4,5)P4).
| IC50 (μM) |
|||
|---|---|---|---|
| His-Tag-INPP5B | SHIP2 (no tag) | His-Tag-SHIP2 | |
| Bz(1,2,3)P3 | 33.5 ± 6.8 | >1000 | >1000 |
| Bz(1,2,4,5)P4 | 6.3 ± 0.8 | 69.3 ± 15.4 | 108.3 ± 20.3 |
| BiPh(2,3′,4,5′,6)P5 | 2.4 ± 0.3 | 24.8 ± 3.0 | 28.4 ± 3.8 |
| BiPh(2,2′,4,4′,5,5′)P6 | 5.4 ± 0.7 | 15.5 ± 2.5 | 19.5 ± 3.1 |
| BiPh(3,3′,4,4′,5,5′)P6 | 5.5 ± 0.5 | 8.4 ± 1.6 | 12.3 ± 3.2 |
As described above, the crystal structures of INPP5B with Bz(1,2,4,5)P4 and BiPh(3,3′,4,4′,5,5′)P6, as well as the BiPh(2,3′,4,5′,6)P5–SHIP2 complex, all show interactions with the His-Tag from a symmetry-related molecule. To investigate the role of the His-tag in modulating the interaction with the ligands present in our library, we performed a comparative IC50 screen using both the His-tag and nontagged versions of SHIP2, the sole member of the 5-phosphatases we studied available under a nontagged form. One hundred micromolar Ins(1,3,4,5)P4 was used as the substrate concentration [Ins(1,4,5)P3 is not a substrate for SHIP2], with the same range of inhibitors as that used with INPP5B, and data are shown in Table 2. The IC50 values for Bz(1,2,3)P3, Bz(1,2,4,5)P4, BiPh(2,3′,4,5′,6)P5, BiPh(2,2′,4,4′,5,5′)P6, and BiPh(3,3′,4,4′,5,5′)P6 with His-Tagged-SHIP2 are >1000, 108.3 ± 20.3, 28.4 ± 3.8, 19.5 ± 3.1, and 12.3 ± 3.2 μM, respectively, and they are >1000, 69.3 ± 15.4, 24.8 ± 3.0, 15.5 ± 2.5, and 8.4 ± 1.6 μM, respectively, for untagged-SHIP2. These trends show the biphenyl polyphosphate inhibitors to be more potent than the single ring benzene polyphosphate compounds, and the most potent biphenyl derivative is BiPh(3,3′,4,4′,5,5′)P6. The results show that the His-Tag has little influence on the inhibition of SHIP2. Small inhibitory differences are more likely due to small intramolecular variations in folding or flexibility caused by the His-Tag, rather than any direct intermolecular interactions between the His-Tag and the bound inhibitor.
Discussion
Further analysis of the crystal structures provides interesting information on two areas related to these enzymes. First, comparison of all the 5-phosphatase structures that were crystallized as ligand complexes can be used to provide information for the design of future potent and specific inhibitors. Second, closer examination of the active site residues, metal ions, and the location of the inorganic phosphate ion in the BiPh(3,3′,4,4′,5,5′)P6/INPP5B structure can be used to further elucidate the mechanism of these enzymes.
Ligand–Enzyme Structure Comparison
Initial comparison of the two ligand-bound INPP5B complexes solved in this study shows there is little change in the overall protein structure (rmsd of 0.34 Å for 268 Cα that structurally align). In addition, the overlay shows the aromatic core of Bz(1,2,4,5)P4 occupying a similar position as one ring of BiPh(3,3′,4,4′,5,5′)P6 is located near the base of the active site pocket (Figure 4) but with some difference in their orientation. This results in the 4- and 5-phosphates of BiPh(3,3′,4,4′,5,5′)P6 overlaying well with the 2- and 4-phosphates of Bz(1,2,4,5)P4, respectively, and sharing many of the same interactions with INPP5B (Figure 3A and C). In the overlay, the 3-phosphate of BiPh(3,3′,4,4′,5,5′)P6 is close to the 1-phosphate of Bz(1,2,4,5)P4. The change in the orientation of the two ligands is due to the position of the cycles between Bz(1,2,4,5)P4 and BiPh(3,3,′4,4′,5,5′)P6 which have been rotated by 48° along the axis formed by carbons 1 and 4 (according to the BiPh(3,3′,4,4′,5,5′)P6 numbering) on the ring. The consequence is a displacement of phosphorus 3 and 5 of 1.19 and 1.44 Å, respectively, when comparing the two complexes. The rotation may be caused by interactions of the 1-phosphate of Bz(1,2,4,5)P4 with a symmetry related molecule (see above) that does not occur in solution or by a polar interaction between His-404 and the 5-phosphate of Bz(1,2,4,5)P4 that stabilizes a conformation where the ligand is more deeply buried in the active site. This comparison shows that the two ligands orientate in the active site largely in order to maximize interactions with their phosphates at the base of the site.
Figure 4.
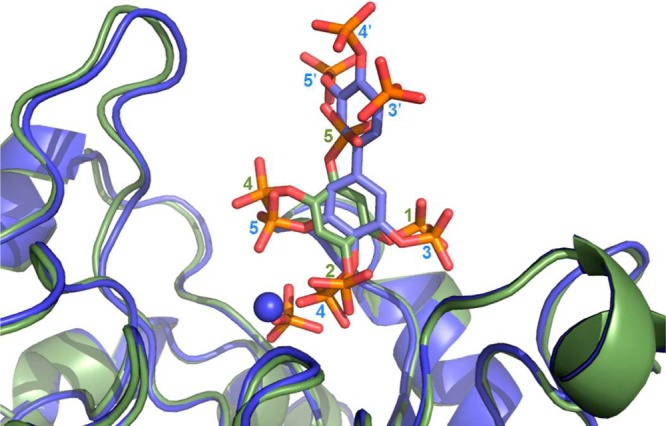
Overlay of Bz(1,2,4,5)P4–INPP5B (green) and BiPh(3,3′,4,4′,5,5′)P6–INPP5B (blue) complex structures showing good overlay between the two molecules and different phosphate groups. The Mg2+ (blue sphere) and inorganic phosphate (sticks) from the BiPh(3,3′,4,4′,5,5′)P6–INPP5B structure are shown, and the numbering of the ligand phosphates are included.
A comparison of these structures with the other structures of INPP5B available (apo, diC8–PtdIns4P- and diC8–PtdIns(3,4)P2-bound) revealed that the protein did not undergo major conformational changes upon binding the different ligands, with rmsd between the different structures in the range of 0.08–0.34 Å where nearly all of the Cα structurally align (Table 1). The influence of the ligand on the protein structure is only noticeable in small differences in the loop region surrounding it.
Two other 5-phosphatase proteins have also been crystallized in complex with ligands, namely, SHIP2 in complex with BiPh(2,3′,4,5′,6)P5, and SPsynaptojanin in complex with Ins(1,4)P2. A comparison of all the 5-phosphatase domains in complex with their ligands shows that there is little difference in the core structure of the proteins, with at least 66% structurally aligning giving rmsd values for these regions all less than 0.77 Å (Supporting Information, Table S2). The main differences between the different proteins are in their external loop regions.
An alignment of all ligand-bound 5-phosphatase structures could give insight into the regions of the enzyme that might be targeted to produce specific, high affinity inhibitors for different 5-phosphatases. When all 5-phosphatase/ligand complexes are superimposed (Figure 5), the overlay of the ligands shows three distinct regions where phosphates and/or hydroxyls align well and are likely to be good target regions for creating new high affinity inhibitors for all 5-phosphatases. The interactions between the ligands and proteins in the six structures are listed in Supporting Information, Table S3A and B.
Figure 5.
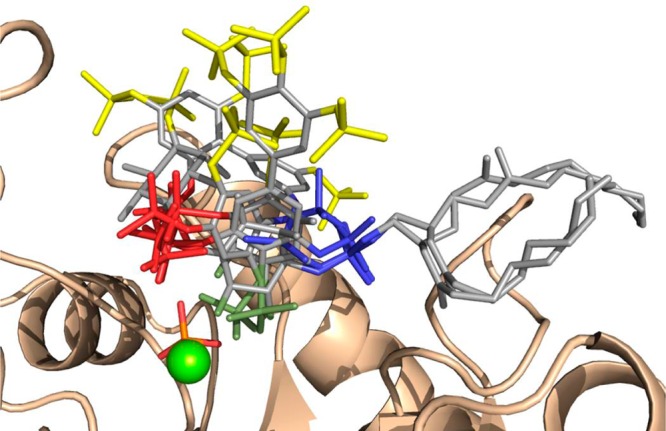
Overlay of the INPP5B–BiPh(3,3′,4,4′,5,5′)P6 crystal structure with the ligands from the other 5-phosphatase-ligand crystal structures (PtdIns4P, PtdIns(3,4)P2, and Bz(1,2,4,5)P4 from INPP5B complexes, SHIP2–BiPh(2,3′,4,5′,6)P5, and SPsynaptojanin–Ins(1,4)P2) highlighting the similarities and differences in the phosphate and hydroxyl positions. The 1-phosphate region is shown in blue, the 5- and 6-hydroxyl region in green, and the 4-phosphate region in red. Other phosphates that show a variety of different interactions with the protein are shown in yellow. The Mg2+ ion is shown as a green sphere and the inorganic phosphate as sticks.
Common Interactions between the Ligand and 5-Phosphatases
The first region where a phosphate group is located in all 5-phosphatase/ligand complexes is equivalent to the position of the 4-phosphate in PtdIns4P and PtdIns(3,4)P2. In all of the structures, the phosphorus atom of a phosphate either almost directly superimposes (Δ 0.54 Å for the 5-phosphate of BiPh(3,3′,4,4′,5,5′)P6 in INPP5B and Δ 0.76 Å for the 4-phosphate of Ins(1,4)P2 in SPsynaptojanin) or is at least close by (Δ 1.11 Å for the 4-phosphate of Bz(1,2,4,5)P4 in INPP5B and Δ 2.6 Å for the 2-phosphate of BiPh(2,3′,4,5′,6)P5 in SHIP2). It is worth noting that the orientation of Ins(1,4)P2 in SPsynaptojanin is very different from that of PtdIns4P and PtdIns(3,4)P2 in INPP5B and that the 5-hydroxyl of Ins(1,4)P2 is located some distance from the proposed active site residues. Therefore, it is unlikely that this orientation of Ins(1,4)P2 is correct for the substrate binding position in SPsynaptojanin and has been discussed previously.6 As shown in Supporting Information, Table S3A and B, some residues and interactions are conserved in all structures, while some interactions are provided by alternative residues in the different structures. Additionally, the only reported examples where a 5-phosphatase showed any activity toward a substrate without a 4-phosphate were SPsynaptojanin and SHIP2 that dephosphorylated PtdIns(3,5)P2.
The second region is centered around the 1-phosphate of PtdIns4P and PtdIns(3,4)P2 that overlays with the 3-phosphate from BiPh(3,3′,4,4′,5,5′)P6 and the 1-phosphate of Bz(1,2,4,5)P4 in the INPP5B structures. Although neither the 3′- or 5′-phosphate of BiPh(2,3′,4,5′,6)P5 from SHIP2 overlays exactly with the 1-phosphate of PtdIns4P, they are positioned on either side. As with the first region, this site has some conserved interactions, but also all of the phosphate groups have additional interactions with the protein, which indicates that this may be another important region to target for high affinity inhibitors.
Finally, in the overlay, the 4-phosphate of BiPh(3,3′,4,4′,5,5′)P6 and the 2-phosphate of Bz(1,2,4,5)P4 in the INPP5B complex structures are located in the region between the 5- and 6-hydroxyls of PtdIns4P and PtdIns(3,4)P2. Similar to the other two regions, this area has some largely conserved as well as additional specific interactions. Therefore, these three regions will be good areas to target in order to provide potential inhibitors with high affinity, but due to all 5-phosphatases providing many interactions in these regions, targeting them is unlikely to give specificity.
A potentially more effective inhibitor could be designed based upon structures similar to our aromatic phosphate compounds by extending a phosphate containing one or two carbon units, in order to provide a longer reach than the 4-phosphate of BiPh(3,3′,4,4′,5,5′)P6. The phosphate group of a new inhibitor should potentially locate in the same binding position as the 5-phosphate anion observed in the BiPh(3,3′,4,4′,5,5′)P6–INPP5B structure and increase the number of potential interactions from the catalytic residues described below.
Specific Interactions between the Ligand and the 5-Phosphatases
Specific inhibitors could be designed by considering the interactions that are not conserved in the ligand–enzyme overlay. Figure 5 shows that these are located away from the active site itself. The 3-phosphate of PtdIns(3,4)P2, the 3′-, 4′-, and 5′-phosphates of BiPh(3,3′,4,4′,5,5′)P6, and the 5-phosphate of Bz(1,2,4,5)P4 all have additional interactions with INPP5B. Likewise, the 4- and 6-phosphates of BiPh(2,3′,4,5′,6)P5 and the 1-phosphate of Ins(1,4)P2 have additional interactions with SHIP2 and SPsynaptojanin, respectively. These phosphates do not overlay with each other, have varied interactions, and are thus more likely to be specific. By targeting this region, a selective 5-phosphatase inhibitor is more likely to be produced. Another potential method to increase inhibitor specificity is to target differences in the loop regions at the entrance to the active site. For example, the flexible SHIP2 loop (residues 672–684)4 is elongated compared to equivalent loops in INPP5B and SPSynaptojanin. If BiPh(2,3′,4,5′,6)P5 bound in the same orientation as BiPh(3,3′,4,4′,5,5′)P6 in the INPP5B structure, steric clashes would occur with the loop. The apex of this loop is flexible and provides a potential target for designing potent ligands.4
Implications for the Catalytic Mechanism Model
The catalytic mechanism by which the 5-phosphatases hydrolyze their substrates is conserved within the apurinic/apyrimidic family of endonucleases (AP-endonucleases). Recently, structures of INPP5B bound to product analogues6 allowed detailed mapping of the catalytic residues when compared with mechanistic studies performed using members of the AP-endonuclease family. While the role of the general base is confirmed as residue Asp-447, there is still uncertainty as to the number and the location of the participating metal ions. The competing models involve either a single-metal, a dual-metal, or single “moving metal” mechanism.34 To date, all metal-bound structures of either 5-phosphatases or AP-endonucleases show the presence of a single metal located in the so-called A-site. The only structure comprising two metals has been observed in the case of an AP-endonuclease bound to two Pb atoms (one in the A-site, the second in a site named B). The size of the atoms and the pH used during the crystallization experiments has raised questions about the relevance of this structure and the plausibility of a dual-metal mechanism. Molecular dynamics studies have led to a hypothesis in which the metal would be first positioned in site B of the moving metal mechanism. Here, the metal ion would help to activate the attacking water molecule to move to site A where it would help to stabilize the product after the appearance of an additional negative charge. The most recent studies based upon alignment with distant homologues of AP-endonucleases favors the existence of a single metal in site A which helps to stabilize the intermediate and release the product.
In the structure of product analogue-bound INPP5B, a metal is present in site A. In an OCRL structure, an inorganic phosphate present has been attributed a role as an inositol 5-phosphate mimic. Therefore, superimposition of the OCRL structure with INPP5B product bound structures allowed modeling of an intact substrate and then to identify residues involved in the catalysis.6 Noteworthy, OCRL also contains a metal positioned in site A. To aid our understanding of the catalytic mechanism of 5-phosphatases, we performed a superimposition of these two structures together with the structure of INPP5B bound to BiPh(3,3′,4,4′,5,5′)P6 (Figure 7). The inorganic phosphate present in the complex with BiPh(3,3′,4,4′,5,5′)P6 appears to be located at 2.38 Å from the phosphate present in the OCRL structure. Moreover, its phosphorus center is located 3.7 Å from the 5-hydroxyl present in INPP5B bound to PtdIns(3,4)P2. The inorganic phosphate present in the complex with BiPh(3,3′,4,4′,5,5′)P6 is also interacting directly with Asp-447 which is expected to be the catalytic base. The distance between its phosphorus atom and Asp-447 is only 3.3 Å and thus would not allow the catalytic water molecule to be correctly oriented to attack the phosphorus atom. Therefore, our structure cannot represent the scissile 5-phosphate of an intact substrate due to these distances.
Figure 7.
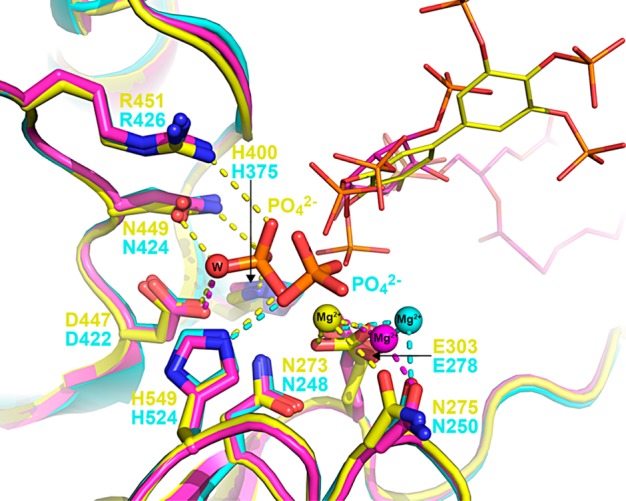
Superposition of INPP5B-BiPh(3,3′,4,4′,5,5′)P6 (yellow), INPP5B-PtdIns(3,4)P2 (magenta), and OCRL (cyan). Amino-acids whose side-chains are shown as sticks are labeled using the same color as the protein to which they correspond. BiPh(3,3′,4,4′,5,5′)P6 (yellow) and PtdIns(3,4)P2 (magenta) are represented as thin sticks. Mg2+ ions and the putative catalytic water molecule are displayed as spheres. Protein–metal, protein–water, or protein–phosphate interactions are represented by dashes. Spheres and dashes are both colored yellow, magenta, or cyan whether they belong to INPP5B-BiPh(3,3′,4,4′,5,5′)P6, INPP5B-PtdIns(3,4)P2, or OCRL structures, respectively. The water molecule (present in the structure of INPP5B-diC8–PtdIns(3,4)P2) that attacks the 5-phosphate upon deprotonation in the mechanism we propose is depicted by a red sphere, labeled with a “W”. Free phosphates are labeled using either a yellow or a cyan font whether they were present in INPP5B-BiPh(3,3′,4,4′,5,5′)P6 or the OCRL structure.
Although Arg-451 is conserved in all members of the 5-phosphatase family, no role could be attributed to this residue whether catalytic or substrate-binding. In addition to Asp-447, Arg-451 is one of the main coordinating residues to inorganic phosphate in the structure of INPP5B-BiPh(3,3′,4,4′,5,5′)P6. Therefore, we believe that the structure of INPP5B-BiPh(3,3′,4,4′,5,5′)P6 helps us to pick up an essential intermediate in the catalytic cycle of the 5-phosphatase in which the 5-phosphate is cleaved but is still retained in the active site.
The mechanism suggested for 5-phosphatases based on the AP endonuclease proposes the activation of a water molecule by Asp-447 (INPP5B numbering), which removes a proton forming an attacking nucleophile. In INPP5B–PtdIns4P, SPsynaptojanin–Ins(1,4)P2 apo-SPsynaptojanin and SHIP2–BiPh(2,3′,4,5′,6)P5, a water molecule close to Asp-447 (or equivalent residue), overlays precisely with one of the oxygen atoms of inorganic phosphate in the crystal structure of complex between INPP5B and BiPh(3,3′,4,4′,5,5′)P6 (Figure 6B). It is located at 3.6 Å from the carboxylic moiety of catalytic Asp-447, 3 Å from the phosphorus of the inorganic phosphate present in OCRL (which has been postulated to mimic the prescissile position of the 5-phosphate), and is in-line between these two groups. Therefore, it appears as a suitable candidate for playing the role of the attacking water molecule (Figure 7).
Figure 6.
(A) Overlay of the proposed catalytic residues for INPP5B–BiPh(3,3′,4,4′,5,5′)P6 (green), INPP5B–Bz(1,2,4,5)P4 (yellow), INPP5B–PtdIns4P (dark blue) SHIP2–BiPh(2,3′,4,5′,6)P5 (light blue), and SPsynaptojanin–Ins(1,4)P2 (red) complexes. Numbering is for the INPP5B enzyme. (B) Overlay of inorganic phosphate from the INPP5B–BiPh(3,3′,4,4′,5,5′)P6 complex structure with water molecules from INPP5B–PtdIns4P, SPsynaptojanin-Ins(1,4)P2 apo-SPsynaptojanin, and SHIP2–BiPh(2,3,′4,5′,6)P5 structures. Only the catalytic residues surrounding the water molecule are shown for clarity. The Mg2+ from the INPP5B–BiPh(3,3′,4,4′,5,5′)P6 complex structure is shown as a green sphere, and the PtdIns4P from the INPP5B complex is shown in blue. (C) Overlay of the phosphate anion and co-ordinating residues from the INPP5B–BiPh(3,3′,4,4′,5,5′)P6 complex with the ligand and the INPP5B–PtdIns4P complex. (D) Overlay of the active site regions of INPP5B–BiPh(3,3′,4,4′,5,5′)P6 (gray carbons; Mg2+ as a green sphere; inorganic phosphate in green) AP endonuclease–DNA (pink carbons; Mn2+ as a purple sphere; DNA shown as a cartoon with the cleaved sugar–phosphate groups shown as sticks). Residue numbering is for INPP5B with AP endonuclease in parentheses.
The phosphate in this mechanism will go through a trigonal bipyramidal intermediate that will need to be stabilized by interactions with the protein. Direct evidence for such an intermediate has been previously observed in the crystal structure of phosphorylated β-phosphoglucomutase in the presence of the Mg2+ and either of the substrates glucose 1-phosphate or glucose 6-phosphate.35,36 In both structures of the enzyme–Mg2+–glucose 1,6-bisphosphate complex there is a stabilized trigonal bipyramidal intermediate of the 1-phosphate. Arg-451 is close to one side of the phosphate ion (3.2 Å) and is suitably orientated to help stabilize the intermediate. A further possible mechanism for the enzymatic hydrolysis of a phosphate monoester by type II inositol 5-phosphatase37 could involve a metaphosphate (PO3–) intermediate. Evidence from other previous experiments for this potential intermediate is derived from trifluoromagnesate (MgF3–), a metaphosphate surrogate and a discrete transition state analogue that has been cocrystallized in the presence of β-phosphoglucomutase together with glucose 6-phosphate or α-galactose 1-phosphate and Mg2+. Currently, there is no evidence for this metaphosphate involvement in the mechanism of INPP5B but this possibility is included for completeness (see ref (37) and references therein).
A comparison between the structure of INPP5B-PtdIns(3,4)P2 and INPP5B-BiPh(3,3′,4,4′,5,5′)P6 shows that only two residues are subjected to conformational changes; these are Asn-273 and the fully conserved Glu-303. Both of these residues are involved in the Mg2+ coordination in the two structures through their side chains. Similar to the position of the inorganic phosphate, the location of the Mg2+ also differs in the INPP5B-BiPh(3,3′,4,4′,5,5′)P6 and OCRL structures (by 3.15 Å) but is still coordinated by the inorganic phosphate in both cases.
Therefore, in order to complete the model previously proposed, we suggest that the water molecule previously described would act as the nucleophile after deprotonation by Asp-447. The resulting trigonal bipyramidal intermediate would then be stabilized by Arg-451, and after the cleavage of the phosphate monoester, the motion exerted by Asn-275 and Glu-303 would transport the Mg2+ and inorganic phosphate away from the other reaction product, potentially, also aided by Asn-273. This step is stabilized by the interaction between the released phosphate and conserved residues Arg-451 and Asp-447 that would then have a dual role during the catalytic cycle (Figure 8A and B).
The structure of INPP5B-BiPh(3,3′,4,4′,5,5′)P6 can be considered as a snapshot in the catalytic cycle of 5-phosphatases and provides, together with the other snapshot represented by the overlay of INPP5B-diC8–PtdIns(3,4)P2 with the inorganic phosphate of OCRL, atomic details about the transformations and conformational changes which occur during the reaction catalyzed by 5-phosphatases.
In this case, the Mg2+ would move from site A toward site B (but not such that it reaches it) (Figure 8B). Moreover, the region around site B is able to accommodate a metal (as suggested by crystallography or molecular dynamics), but the role of this site is not to activate the nucleophile but to stabilize the phosphate leaving group and help the release of the other product. Here, we propose that the metal is moving throughout catalysis but to a smaller extent than previously described in the controversial moving-metal mechanism. The structure of INPP5B-BiPhP is thus a key element in unifying the different theories developed around the number and the role of the metal ions in either 5-phosphatase- or AP-endonuclease-mediated catalysis.
Although both INPP5B and SHIP2 bind biphenyl polyphosphates more strongly than the single ring benzene polyphosphate compounds, there are some differences between the two proteins. BiPh(2,3′,4,5′,6)P5 is the most potent inhibitor of INPP5B, although only slightly more potent than the other biphenyl compounds, whereas BiPh(3,3′,4,4′,5,5′)P6 is clearly the most potent inhibitor of SHIP2. For single ring compounds, Bz(1,2,4,5)P4 is only slightly less potent than the biphenyl inhibitors for INPP5B, and Bz(1,2,3)P3 is the weakest INPP5B inhibitor but still in the low micromolar range. The single ring ligands have significantly lower affinity for SHIP2 compared to the biphenyl compounds with Bz(1,2,3)P3 showing very poor inhibition >1 mM.
Conclusions
The inositol 5-phosphatase family of proteins is of pharmacological importance.14 The orally active AQX-1125, a small molecule activator of the 5-phosphatase family member SHIP1, is currently in phase 2 trials.38 The structures of our INPP5B-ligand complexes presented here may further aid the development of new drug-like compounds for this family. We also propose some wider structural generalisations to this end. Additionally, benzene and biphenyl polyphosphates are useful emerging tools to modulate cell signaling processes, both through enzymes in the inositol 5-phosphatase family and at the receptor level,31,33 and via protein kinase Bα PH domain.33
Ligands potentially suitable for this INPP5B 5-phosphatase structural study were evaluated using a thermal shift assay, and two noninositol polyphosphate-based compounds were selected, resulting in two X-ray cocrystal structures. The high ΔTagg obtained for these compounds suggests that they might be prioritized over inositol polyphosphate derivatives, for example, for type I 5-phosphatase (INPP5A) or other inositol polyphosphate binding proteins whose crystal structure is as yet unavailable. These benzene and biphenyl polyphosphates are emerging more generally as useful inositol polyphosphate surrogates.4,17 Inhibition studies of INPP5B showed that all the biphenyl phosphates were more potent than the benzene phosphate derivatives. Although interactions between the inhibitors and the protein purification tag were observed in the crystal structures, solution data suggest that ligand binding and inhibition are not influenced by this extra sequence.
The mechanism of dephosphorylation by a 5-phosphatase is currently based upon the fact that it is related to an AP endonuclease and postulates that Mg2+, His-400, Asn-449, and His-549 (INPP5B numbering) should be close to, and interact with, the 5-phosphate from a suitable inositol polyphosphate substrate. The structure of the BiPh(3,3′,4,4′,5,5′)P6–INPP5B complex revealed a molecule of inorganic phosphate located close to Asp-447, proposed to activate the attacking water molecule. We propose that this inorganic phosphate occupies the location of the cleaved 5-phosphate after substrate hydrolysis by the enzyme. Interestingly, in our structure Arg-451 (conserved throughout the 5-phosphatase family) is located close to the inorganic phosphate, suggesting that it plays a role in the 5-phosphatase mechanism, although not previously identified as such. Arg-451 may help to stabilize the pentacoordinate intermediate and facilitate the hydrolysis of the 5-phosphate by aiding its release from the substrate. From our structure and the overlays in Figure 8, the identification of both Arg-451 and a candidate water molecule for the first step of the hydrolysis reaction refines the catalytic mechanism for INPP5B 5-phosphatase and with implications for the “moving metal” mechanism but still broadly in line with that generally proposed for the 5-phosphatase family.
Glossary
Abbreviations
- BiPhP
biphenyl polyphosphate
- BzP
benzene polyphosphate
- mCPBA
meta-chloroperbenzoic acid
- diC8–PtdIns(4)P
diC8–PtdIns(4)P
- HEPES
2-[4-(2-hydroxyethyl)piperazin-1-yl) ethanesulfonic acid
- IMAC
immobilized metal affinity chromatography
- INPP5A
type I inositol 5-phosphatase
- INPP5B
type II inositol 5-phosphatase
- Ins(1,4)P2
myo-inositol 1,4-bisphosphate
- Ins(1,4,5)P3
myo-inositol 1,4,5-trisphosphate
- Ins(1,3,4,5)P4
myo-inositol 1,3,4,5-tetrakisphosphate
- OCRL-1
Lowe oculocerebrorenal syndrome protein (INPP5F)
- PH-domain
pleckstrin homology domain
- PKBα
protein kinase Bα
- PtdIns(3,4,5)P3
phosphatidyinositol 3,4,5-trisphosphate
- SHIP1
SH2-domain containing inositol 5-phosphatase type 1
- SHIP2
SH2-domain containing Inositol 5-phosphatase, type 2
- TEAB
triethylammonium bicarbonate
- TCA
trichloroacetic acid
- TCEP
(tris(2-carboxethyl)phosphine)
- TMSBr
bromotrimethylsilane
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.5b00838.
A supplementary thermal shift scatter graph for all compounds evaluated and 5-phosphatase proteins, compound structures, electron density maps for INPP5B and ligand, tables of ligand protein interactions, and interaction diagrams between ligand and 5-phosphatases and protein residues are provided. (PDF)
Accession Codes
Coordinates and structure factors were deposited in the Protein Data Bank (PDB). The accession codes for the coordinate entries are 5A7I for biphenyl 3,3′,4,4′,5,5′-hexakisphosphate and 5A7J for benzene 1,2,4,5-tetrakisphosphate.
Author Present Address
# C.S.: Sprint Bioscience, Teknikringen 38A, 114 28 Stockholm, Sweden.
This research was supported by the Wellcome Trust (Programme Grant 082837 to BVLP), the Swedish Cancer Society, and Swedish Research Council (to P.N.). B.V.L.P. is a Wellcome Trust Senior Investigator (grant no. 101010). We gratefully acknowledge the staff of the BESSY beamline BL14.1 (Berlin, Germany) for assistance with X-ray data collection. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research. and the Wellcome Trust.
The authors declare no competing financial interest.
Supplementary Material
References
- Ooms L. M.; Horan K. A.; Rahman P.; Seaton G.; Gurung R.; Kethesparan D. S.; Mitchell C. A. (2009) The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 419, 29–49. 10.1042/BJ20081673. [DOI] [PubMed] [Google Scholar]
- Majerus P. M.; York J. D. (2009) Phosphoinositide phosphatases and disease. J. Lipid Res. 50, S249–254. 10.1194/jlr.R800072-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujishita Y.; Guo S.; Stolz L. E.; York J. D.; Hurley J. H. (2001) Specificity determinants in phosphoinositide dephosphorylation: crystal structure of an archetypal inositol polyphosphate 5-phosphatase. Cell 105, 379–389. 10.1016/S0092-8674(01)00326-9. [DOI] [PubMed] [Google Scholar]
- Mills S. J.; Persson C.; Cozier G.; Thomas M. P.; Trésaugues L.; Erneux C.; Riley A. M.; Nordlund P.; Potter B. V. L. (2012) A synthetic polyphosphoinositide headgroup surrogate in complex with SHIP2 provides a rationale for drug discovery. ACS Chem. Biol. 7, 822–828. 10.1021/cb200494d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attree O.; Olivos I. M.; Okabe I.; Bailey L. C.; Nelson D. L.; Lewis R. A.; McInnes R. R.; Nussbaum R. L. (1992) The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature 358, 239–242. 10.1038/358239a0. [DOI] [PubMed] [Google Scholar]
- Trésaugues L.; Silvander C.; Flodin S.; Welin M.; Nyman T.; Gräslund S.; Hammarström M.; Berglund H.; Nordlund P. (2014) Structural basis for phosphoinositide recognition, catalysis, and membrane interactions in human inositol polyphosphate 5-phosphatases. Structure 22, 744–755. 10.1016/j.str.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Protein Data Bank. http://www.rcsb.org.
- Chi Y.; Zhou B.; Wang W.-Q.; Chung S.-K.; Kwon Y.-U.; Ahn Y.-H.; Chang Y.-T.; Tsujishita Y.; Hurley J. H.; Zhang Z.-Y. (2004) Comparative mechanistic and substrate specificity study of inositol polyphosphate 5-phosphatase, Schizosaccharomyces pombe synaptojanin and SHIP2. J. Biol. Chem. 279, 44987–44995. 10.1074/jbc.M406416200. [DOI] [PubMed] [Google Scholar]
- Mitchell C. A.; Connolly T. M.; Majerus P. W. (1989) Identification and isolation of a 75 kDa inositol polyphosphate-5-phosphatase from human platelets. J. Biol. Chem. 264, 8873–8877. [PubMed] [Google Scholar]
- Schmid A. C.; Wise H. M.; Mitchell C. A.; Nussbaum R.; Woscholski R. (2004) Type II phosphoinositide 5-phosphatases have unique sensitivities towards fatty acid composition and head group phosphorylation. FEBS Lett. 576, 9–13. 10.1016/j.febslet.2004.08.052. [DOI] [PubMed] [Google Scholar]
- Shin H.-W.; Hayashi M.; Christofordid S.; Lacas-Gervais S.; Hoepfner S.; Wenk M. R.; Modregger J.; Uttenweiler-Joseph S.; Wilm M.; Nystuen A.; Frankel W. N.; Solimena M.; De Camilli P.; Zerial M. (2005) An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170, 607–618. 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.; Choudhury R.; McKenzie E.; Lowe M. (2007) Targeting of the type II inositol polyphosphate 5-phosphatase INPP5B to the early secretory pathway. J. Cell Sci. 120, 3941–3951. 10.1242/jcs.014423. [DOI] [PubMed] [Google Scholar]
- Sarantis H.; Balkin D. M.; De Camilli P.; Isberg R. R.; Brumell J. H.; Grinstein S. (2012) Yersinia entry Rab5-dependent dephosphorylation of PI(4,5)P2 and membrane scission. Cell Host Microbe 11, 117–128. 10.1016/j.chom.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirruccello M.; Nandez R.; Idevall-Hagren O.; Alcazar-Roman A.; Abriola L.; Berwick S. A.; Lucast L.; Morel D.; De Camilli P. (2014) Identification of inhibitors of inositol 5-phosphatases through multiple screening strategies. ACS Chem. Biol. 9, 1359–1368. 10.1021/cb500161z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedadi M.; Niesen F. H.; Allali-Hassani A.; Fedrov O. Y.; Finerty P. J. Jr.; Wasney G. A.; Yeung R.; Arrowsmith C.; Ball L. J.; Berglund H.; Hui R.; Marsden B. D.; Nordlund P.; Sundstrom M.; Weigelt J.; Edwards A. M. (2006) Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc. Natl. Acad. Sci. U. S. A. 103, 15835–15840. 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A.; Jansson A.; Åberg A.; Nordlund P. (2011) Efficiency of hit generation and structural characterization in fragment-based ligand discovery. Curr. Opin. Chem. Biol. 15, 482–488. 10.1016/j.cbpa.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Mills S. J.; Komander D.; Trusselle M. N.; Safrany S. T.; van Aalten D. M. F.; Potter B. V. L. (2007) Novel inositol phospholipid headgroup surrogate crystallized in the pleckstrin homology domain of protein kinase Bα. ACS Chem. Biol. 2, 242–246. 10.1021/cb700019r. [DOI] [PubMed] [Google Scholar]
- Du-Cuny L.; Song Z.; Moses S.; Powis G.; Mash E. A.; Meuillet E. J.; Zhang S. (2009) Computational modeling of novel inhibitors targeting the Akt pleckstrin homology domain. Bioorg. Med. Chem. 17, 6983–6992. 10.1016/j.bmc.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills S. J.; Luyten T.; Erneux C.; Parys J. B.; Potter B. V. L. (2012) Multivalent benzene polyphosphate derivatives are non-Ca2+-mobilizing Ins(1,4,5)P3 receptor antagonists. Messenger 1, 167–181. 10.1166/msr.2012.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stjernstrom N. E.; Tengedal J. E. (1970) Reactions between p-benzoquinone and pyragallol. Acta Chem. Scand. 24, 338–340. 10.3891/acta.chem.scand.24-0338. [DOI] [Google Scholar]
- Hong B. S.; Senisterra G.; Rabeh W. M.; Vedadi M.; Leonardi R.; Zhang Y. M.; Rock C. O.; Jackowski S.; Park H. W. (2007) Crystal structures of human pantothenate kinases. Insights into allosteric regulation and mutations linked to a neurodegeneration disorder. J. Biol. Chem. 282, 27984–27993. 10.1074/jbc.M701915200. [DOI] [PubMed] [Google Scholar]
- Senisterra G. A.; Ghanei H.; Khutoreskaya G.; Dobrovetsky E.; Edwards A. M.; Prive G. G.; Vedadi M. (2010) Assessing the stability of membrane proteins to detect ligand binding using differential static light scattering. J. Biomol. Screening 15, 314–320. 10.1177/1087057109357117. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800. 10.1107/S0021889893005588. [DOI] [Google Scholar]
- Bailey S. (1994) The Ccp4 suite - programs for protein crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 50, 760–763. 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr., Sect. D: Biol. Crystallogr. 62, 72–82. 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. (2010) Features and development of coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 53, 240–255. 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P, Sharff A., Smart O. S., Vonrhein C., and Womack T. O. (2011) BUSTER, version 2.11.1, Global Phasing Ltd., Cambridge, United Kingdom. [Google Scholar]
- Chen V. B.; Arendall W. B. 3rd; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa S. E.; Shin D. S.; Mol C. D.; Izumi T.; Arvai A. S.; Mantha A. K.; Szczesny B.; Ivanov I. N.; Hosfield D. J.; Maiti B.; Pique M. E.; Frankel K. A.; Hitomi; Cunningham R. P.; Mitra S.; Tainer J. A. (2013) Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J. Biol. Chem. 288, 8445–8455. 10.1074/jbc.M112.422774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri S. D.; Zhang G.; Dunaway-Mariano D.; Allen K. N. (2003) The pentacovalent phosphorus intermediate of a phosphoryl transfer reaction. Science 299, 2067–2071. 10.1126/science.1082710. [DOI] [PubMed] [Google Scholar]
- Tremblay L. W.; Zhang G.; Dunaway-Mariano D.; Allen K. N. (2005) Chemical confirmation of a pentavalent phosphorene in complex with β-phosphoglucomutase. J. Am. Chem. Soc. 127, 5298–5299. 10.1021/ja0509073. [DOI] [PubMed] [Google Scholar]
- Baxter N. J.; Bowler M. W.; Alizadeh T.; Cliff M. J.; Hounslow A. M.; Wu B.; Berkowitz D. B.; Williams N. H.; Blackburn G. M.; Waltho J. P. (2010) Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed MgF3– rather than by phosphoranes. Proc. Natl. Acad. Sci. U. S. A. 107, 4555–4560. 10.1073/pnas.0910333106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquinox. www.aqxpharma.com.
- Mills S. J.; Vandeput F.; Trusselle M. N.; Safrany S. T.; Erneux C.; Potter B. V. L. (2008) Benzene polyphosphates as tools for cell signalling: inhibition of inositol 1,4,5-trisphosphate 5-phosphatase and interaction with the PH domain of protein kinase Bα. ChemBioChem 9, 1757–1766. 10.1002/cbic.200800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeput F.; Combettes L.; Mills S. J.; Backers K.; Wohlkönig A.; Parys J. B.; De Smedt H.; Missiaen L.; Dupont G.; Potter B. V. L; Erneux C. (2007) Biphenyl 2,3′,4,5′,6-pentakisphosphate, a novel inositol polyphosphate surrogate, modulates Ca2+ responses in hepatocytes. FASEB J. 21, 1481–1491. 10.1096/fj.06-7691com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.