Abstract
The molecular mechanisms controlling genome packaging by single-stranded RNA viruses are still largely unknown. It is necessary in most cases for the protein to adopt different conformations at different positions on the capsid lattice in order to form a viral capsid from multiple copies of a single protein. We showed previously that such quasi-equivalent conformers of RNA bacteriophage MS2 coat protein dimers (CP2) can be switched by sequence-specific interaction with a short RNA stem–loop (TR) that occurs only once in the wild-type phage genome. In principle, multiple switching events are required to generate the phage T=3 capsid. We have therefore investigated the sequence dependency of this event using two RNA aptamer sequences selected to bind the phage coat protein and an analogous packaging signal from phage Qβ known to be discriminated against by MS2 coat protein both in vivo and in vitro. All three non-cognate stem–loops support T=3 shell formation, but none shows the kinetic-trapping effect seen when TR is mixed with equimolar CP2. We show that this reflects the fact that they are poor ligands compared with TR, failing to saturate the coat protein under the assay conditions, ensuring that sufficient amounts of both types of dimer required for efficient assembly are present in these reactions. Increasing the non-cognate RNA concentration restores the kinetic trap, confirming this interpretation. We have also assessed the effects of extending the TR stem–loop at the 5′ or 3′ end with short genomic sequences. These longer RNAs all show evidence of the kinetic trap, reflecting the fact that they all contain the TR sequence and are more efficient at promoting capsid formation than TR. Mass spectrometry has shown that at least two pathways toward the T=3 shell occur in TR-induced assembly reactions: one via formation of a 3-fold axis and another that creates an extended 5-fold complex. The longer genomic RNAs suppress the 5-fold pathway, presumably as a consequence of steric clashes between multiply bound RNAs. Reversing the orientation of the extension sequences with respect to the TR stem–loop produces RNAs that are poor assembly initiators. The data support the idea that RNA-induced protein conformer switching occurs throughout assembly of the T=3 shell and show that both positional and sequence-specific effects outside the TR stem–loop can have significant impacts on the precise assembly pathway followed.
Keywords: virus assembly, RNA–protein interaction, mass spectrometry
Introduction
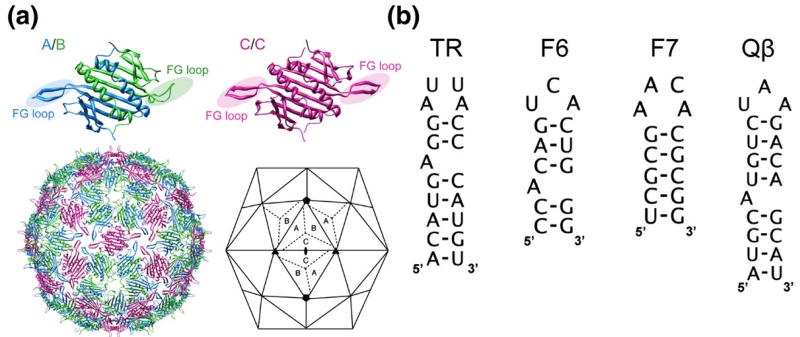
The mechanism used by simple viruses (i.e., those consisting of a nucleic acid genome enclosed in a protein shell) to regulate their self-assembly has been a puzzle since the structure of the first virus (tomato bushy stunt virus) was determined at atomic resolution.1,2 Bacteriophage MS2 conforms to a T=3 shell in the Caspar–Klug nomenclature,3 having an icosahedral surface lattice of 180 coat proteins packaged as 90 non-covalent dimers4,5 (Fig. 1a). We recently proposed that the quasi-equivalent conformers, A, B and C, are specified by sequence-specific RNA stem–loop binding to the coat protein dimers (CP2).6 This hypothesis is based on the results of in vitro assembly reactions starting with disassembled CP2, triggering the reaction by addition of an RNA stem–loop (TR) of just 19 nt (Fig. 1b). This interaction in vivo leads to translational repression of the replicase cistron and has long been a paradigm for sequence-specific RNA–protein recognition.7–13 The complex formed was also believed to constitute the minimal assembly-competent species.14,15
Fig. 1.
The molecular components used in the assembly assays. (a) Coat protein dimer quasi-equivalent conformers are shown as ribbon diagrams, with the FG loops highlighted. Their relationships within a capsid are also shown [Protein Data Bank (PDB) code 2MS24]; the A and C subunits have extended loops, while the B subunit loops fold back toward the globular core of the protein. In the complete capsid, A/B dimers surround the particle 5-fold axes with a ring of B-type loops, while the A- and C-type loops alternate around the 3-fold axes. (b) Sequences and secondary structures of the TR RNA, the aptamer consensus sequences F6 and F7 and the Qβ stem–loop.
Using non-covalent mass spectrometry,6 we demonstrated that the CP2:TR complex is kinetically trapped, i.e., that it is unable to assemble beyond this point in the absence of other molecular species. The effect of the trap can be relieved by subsequent addition of RNA-free CP2 to this complex, resulting in the rapid appearance of higher-order assembly intermediates and broad peaks with an m/z > 14,000, equivalent to those seen when recombinant T=3 shells are analysed. The speed of the assembly process under these conditions implies that the two types of CP2 present represent all the building blocks required for efficient assembly of the capsid. The CP2:TR complex is in equilibrium with the free components, although this lies heavily on the side of the complex under the reaction conditions (8 μM reactants and pH greater than 5.2). As a result, free CP2 is present in limiting concentrations. Such mixtures do make T=3 shells, but very slowly, with trace amounts appearing only after 90 min of incubation.
This result suggested that the RNA-free and RNA-bound forms of CP2 are different conformers, in particular that they mimicked the symmetric (C/C) and asymmetric (A/B) conformers needed to define the T=3 shell (Fig. 1).4,5 This assertion was supported by NMR data showing that addition of TR RNA to an isolated CP2 resulted in conformational changes from a symmetric dimer (assumed to be C/C-like) to an asymmetric one (A/B-like) and that these changes occurred primarily at the loop of polypeptide connecting the F and G β-strands within the CP monomer. Crystal structures of the wild-type virion and recombinant virus-like particles show that the FG loop is the site of the major conformational differences between quasi-conformers (Fig. 1a).
While this hypothesis is a satisfactory description of the assembly reaction using multiple copies of the TR stem–loop, it is clearly not an accurate description of assembly with genomic-length RNAs since there is only one copy of the TR sequence in the ~3500-nt genome.16 If such allosteric switching were the prerequisite for creation of the A/B conformers throughout the T=3 capsid, it would have to be caused by many other non-TR sequences. Some support for this idea comes from cryo-electron microscopy (EM) reconstructions of the wild-type phages of MS2, Qβ, fr and AP205.17–19 The MS2 virion structure, which is at the highest resolution (<10 Å), shows that the genomic RNA is organised as a double shell connected by columns of density at the particle 5-fold axes.17 The outer shell of RNA lies directly below the coat protein layer, i.e., it is positioned such that it could act as an allosteric conformer switch throughout the assembly process.
In order to test this idea further, we have studied MS2 capsid assembly reactions triggered by non-TR stem–loops. Three stem–loops (Fig. 1b), corresponding to two RNA aptamers (F6 and F7)20–22 that are known to bind to the same site on a CP2 as TR and the functionally equivalent stem–loop from phage Qβ, were used. The latter is known to be discriminated against by MS2 coat protein in vivo and is a very poor ligand in vitro.23,24 In addition, we examined the roles of non-TR genomic sequences directly with a series of 5′ and 3′ extensions to the core TR sequence. These either match the wild-type genomic sequences or reverse their order with respect to the TR, allowing both sequence-specific and positional effects to be tested. The data confirm that non-TR sequences can trigger the conformational change in CP2 required to initiate efficient assembly and show that sequences adjacent to TR in the genome play roles in directing the assembly pathway toward the T=3 shell. The results are consistent with an assembly pathway in which the genomic RNA contributes actively to its own encapsidation.
Results and Discussion
The sequence specificity of quasi-equivalent conformer switching
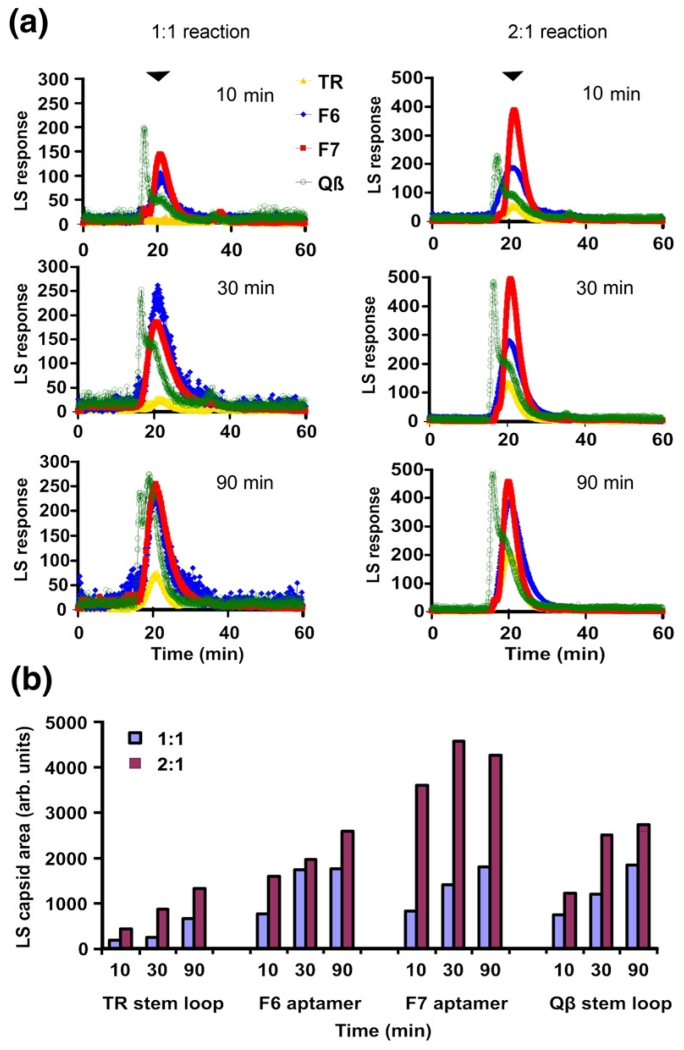
In order to test the sequence requirements for efficient RNA-mediated allosteric CP2 switching, we prepared the minimal TR RNA and three additional RNA stem–loops (Fig. 1) using solid-phase RNA synthesis.25,26 These RNAs include two aptamer family consensus sequences (F6 and F7) that can mimic the interactions at the TR stem–loop binding site of CP2 despite significant sequence differences. The X-ray structures of these RNAs bound to the CP2 have been determined.20,21 In addition, we tested the equivalent translational operator site from phage Qβ, which binds very poorly to the MS2 CP2 both in vivo and in vitro.12,23,24 The coat protein affinities of these variant stem–loops have been determined either in vitro27 or in in vivo repression assays. The results suggest normalised affinities for MS2 coat protein dimers of 1:0.5:0.06 and 0.04 for TR:F6:F7 and Qβ stem–loops, respectively. In order to facilitate simple comparison of kinetics and yield of T=3 capsid, we analysed assembly reactions by gel-filtration chromatography. The column outflow was monitored by both UV absorption and light-scattering to detect particulate species.6 The results of these experiments are shown in Fig. 2.
Fig. 2.
Assembly kinetics of TR and non-cognate stem–loops. (a) Gel filtration–light-scattering assays of capsid assembly. MS2 CP2 and RNA stem–loops (coloured as indicated) were mixed in 40 mM ammonium acetate, pH 5.2–5.7, to form 1:1 reactions, incubated at 4 °C for 10, 30 or 90 min and then loaded onto a Superose 6 gel-filtration column equilibrated in 50 mM Tris–acetate, pH 7.4, and eluted at 0.4 mL/min. The outflow from the column was analysed simultaneously via UV absorbance and light-scattering, but only the latter traces are shown for clarity. Similar reactions were set up and pre-incubated at a molar ratio of 1:1 for 10 min, and then an additional aliquot of protein was added to create 2:1 reactions. The position at which T=3 capsids eluted is marked with an arrowhead. (b) The extent of capsid formation at each point was estimated using the fitting procedure described in Materials and Methods and is shown here as a histogram. Note that the assembled peak for the Qβ stem–loop includes material eluting in front of the position for T=3 capsid and the capsid peak itself.
When the non-TR sequences were used at a molar ratio of 1:1 CP2:RNA, they rapidly triggered assembly of T=3 shells, while the equivalent TR reactions were kinetically trapped, as expected (cf., yellow traces in Fig. 2a, with data for the other RNAs). Addition of RNA-free protein to these reactions (to give 2:1 CP2:RNA) released the kinetic trap for TR and led to further assembly in the non-TR reactions, resulting in increased yields of the T=3 product in the assay period. All the reactions generated T=3 shells as judged by negative staining and transmission EM (data not shown). Therefore, it appears that assembly can be triggered by CP2: nonTR RNA interactions, consistent with conformer switching throughout the genome, and surprisingly that the non-cognate sequences are better triggers than the natural sequence (Fig. 2b). Note that for the Qβ sequence, a proportion of the assembled species runs faster than the T=3 shell on the gel-filtration column and perhaps represents aggregated material that can be difficult to distinguish by EM in assembling mixtures.
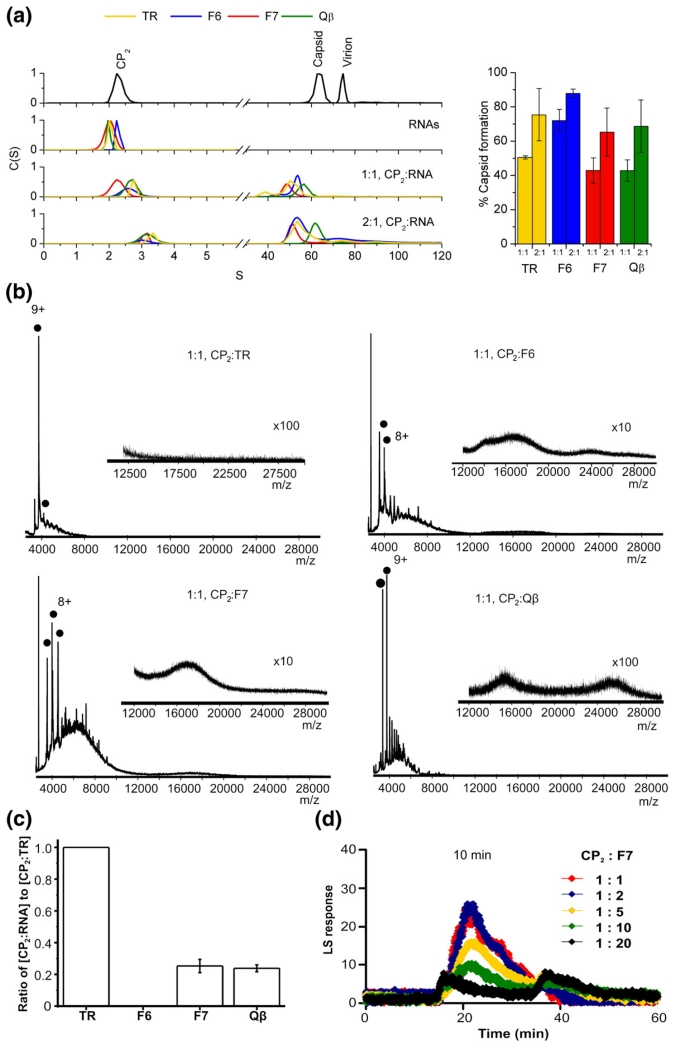
To probe these results in more detail, we then carried out sedimentation velocity analysis of the assembly reactions after these had equilibrated for ~4.5 h. The results (Fig. 3a) confirm that the high mass peaks from the gel-filtration column correspond to species with sedimentation coefficients close to those expected for the recombinant T=3 shell. However, the data also suggest that after the extended incubation, there are few, if any, differences between the four stem–loops, even at a 1:1 stoichiometry. If the effects seen for TR at this stoichiometry are indeed due to initial formation of a kinetically trapped CP2:TR species, it is clear that the trap has been overcome by 4.5 h (see below).
Fig. 3.
Examining the mechanism leading to the kinetic trap. (a) Normalised sedimentation coefficient distributions, C (S), derived from sedimentation velocity analysis of the MS2 capsid re-assembly reactions with TR, Qβ, F6 and F7 RNAs. The two uppermost traces show normalised C(S) plots of individual MS2 capsid re-assembly components. The two remaining C(S) plots show MS2 capsid re-assembly reactions at 1:1 and 2:1 CP2:RNA molar ratios. The bar chart (alongside) shows the percentage of capsid-like product formed in each reaction at the two reaction ratios. The colours used follow the scheme in Fig. 2 for the different RNAs. All samples were analysed after incubation in 40 mM ammonium acetate at 4 °C for 4 h, and the resulting sedimentation distribution data were analysed using SEDFIT as described in Materials and Methods. (b) Electrospray ionisation mass spectrometry assays of stem–loop induced re-assembly. The spectra were acquired over the m/z range 500–30,000 in ammonium acetate (40 mM). Filled circles represent the initiation complex [CP2:RNA]. The charge states for selected ions are also labelled. The spectra are for re-assembly reactions at a 1:1 stoichiometry of CP2:RNA (8 μM:8 μM), t=30 min, for CP2:TR, CP2:F6, CP2:F7 and CP2:Qβ, respectively. The insets highlight the m/z range 12,000–30,000 and show the broad unresolved higher mass-to-charge signals assigned to the T=3 capsid. Bars below the spectra indicate the magnification factor used for all ions above m/z 12,000 to enhance the clarity. Note the absence of high m/z peaks in the TR reaction even at the highest magnification. (c) Graphs of the competitive binding experiments. CP2:TR or competitor RNA (F6, F7 or Qβ) was added in a 1:1:1 ratio at a concentration of 10 μM. The reaction was allowed to equilibrate for 5 min, and then spectra were acquired for 1 min. The peak areas of [CP2:TR] and [CP2:RNA] were calculated assuming that the initiation complexes with the different RNAs have similar ionisation efficiencies. The peak area of [CP2:TR] was set at 100%, and the other initiation complexes were normalised to this. The competition experiments were done in triplicate, and error bars shown in the plots represent the ±SD. (d) Light-scattering peaks, as in Fig. 2, for the 10 -min time points of reactions set up with molar excesses of F7 RNA over CP2, as indicated in the colour key.
The implication of these data is that the kinetic trap is less of a problem for the non-TR stem–loops. To investigate this in more detail, we used mass spectrometry to examine the species present in these reactions. For the 1:1 assembly reactions, the CP2:RNA complex was formed as expected, but there were also higher-order species present for the non-TR RNAs not seen at equivalent time points for TR-induced assembly. For example, the broad unresolved higher mass-to-charge signals (greater than an m/z of 14,000) assigned to the MS2 T=3 capsid6 are visible in the 1:1 CP2:nonTR sequence assembly reactions after just 30 min (Fig. 3b), consistent with the gel-filtration data. Interestingly, the capsid signal observed for assembly with Qβ RNA was less intense than that seen with F6 and F7 RNAs. Since the light-scattering data (Fig. 2a and b) suggest that assembly with Qβ RNA may also result in aggregates that might not ionise under the same conditions in the mass spectrometer, this could account for the reduced signal observed. The major higher-order intermediates identified during assembly with TR6 (Morton et al., unpublished), corresponding either to 6 dimers bound to three TRs (the stoichiometry expected at a 3-fold axis) or to 10 dimers bound to five TRs (consistent with an extended 5-fold axis), were, however, not detected with the aptamers or Qβ RNA. One explanation could be that the more rapid formation of the T=3 capsid with these RNAs prevents build-up of these transient intermediates.
The mass spectrometry and gel-filtration data are consistent with the non-TR stem–loops showing no evidence of the initial kinetic assembly trap seen for TR. In order to understand why this is so, we then compared assembly competition between TR and non-TR RNAs. Assembly reactions were set up at a total CP2:RNA ratio of 1:2, in which the RNA component was a 50:50 mixture of TR and a non-TR stem–loop (Fig. 3c). Remarkably, although the affinities of the individual aptamers for CP2 are high as judged by separate binding assays,8 in the ammonium acetate buffer used for mass spectrometry, TR outcompeted the non-TR species in formation of the 1:1 CP2:RNA complex. This competition was complete in the case of aptamer F6.
If all stem–loops trigger the conformational change in the CP2, then when the protein is fully saturated with bound RNA, asymmetric A/B-like species would be expected to be present. However, if the non-TR stem–loops are non-saturating under these conditions, then a mixture of A/B-like and symmetrical C/C-like species would be present. This would be the case even in the 1:1 reactions leading to conditions equivalent to the 2:1 reactions for TR (i.e., entirely consistent with the apparently increased ability of non-TR RNAs to promote capsid formation). A prediction of this model is that increasing the concentration of a non-TR RNA would lead to reduced assembly because more of the CP2 present would be in the A/B-like conformation. The results of such an experiment for F7 are shown in Fig. 3d. As the RNA concentration is raised to 20-fold molar excess over the CP2, the amount of capsid formed at early time points, 10 min in this case, is progressively reduced, entirely consistent with this hypothesis.
There remains the issue of how all the assembly reactions escape the kinetic trap so that by 4.5 h there is no difference between them, except overall yield as the coat protein concentration is increased (Fig. 3a). Such a result would arise if there were some mechanism that steadily reduced the “available” concentration of RNA operating. One such mechanism would be binding of TR stem–loops to C/C dimers within assembling complexes. Such interactions are known to occur both in wild-type phage (see above) and in crystals of recombinant shells soaked with RNA.10,13,20,21 In each case, RNA binds to both A/B and C/C dimers and the protein shells are stable, implying that the binding to dimers that have been incorporated into the shell does not cause the conformational change to A/B. In larger assembly intermediates, therefore, TR could dissociate from A/B positions and then be recaptured at either A/B positions or C/C positions, the latter freeing a new site for higher-affinity TR binding. Such re-equilibration would have the effect of reducing the free RNA concentration, resulting in even the TR-mediated 1:1 reaction containing both sorts of quasi-equivalent conformer required for efficient assembly. The concentration of such larger assembly intermediates increases with time, consistent with the difference in results between gel filtration/mass spectrometry of early time points and the sedimentation velocity measurements after 4.5 h.
The pathway to T=3 shell formation
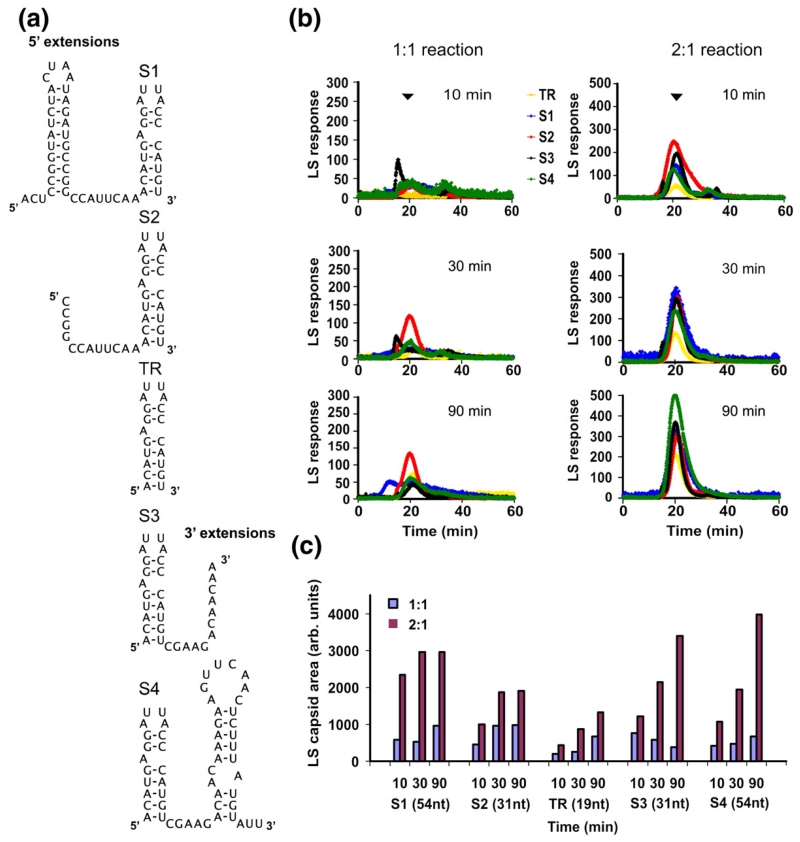
The results described above are consistent with RNA-induced protein conformer switching occurring throughout the capsid assembly pathway. We therefore examined the effects of increasing the sizes of RNAs used to initiate assembly. We produced a series of extended TR stem–loops, termed the S-series (S1–S4) (Fig. 4a), containing 12- or 35-nt extensions to the TR core sequence at either the 5′ end (S1 and S2) or the 3′ end (S3 and S4), respectively. Mfold26 suggests that these would all fold into secondary structures containing the TR stem–loop, with the two longer RNAs also encompassing an additional stem–loop. We have previously shown that both S2 and S3 separately trigger assembly of T=3 shells, and cryo-EM reconstruction of the S2 product confirms that it binds in essentially the same site as TR below each CP2.17,19
Fig. 4.
Assembly with extended TR RNAs. (a) Sequences and secondary structures of the extended TR stem–loops. (b) Gel filtration–light-scattering profiles for 1:1 and 2:1 re-assembly reactions with the RNAs shown in (a). Experimental details are as those given in Fig. 2. (c) Quantitation of the extent of capsid formation in each case.
The gel-filtration time courses of assembly of these species are shown in Fig. 4b for both CP2:TR 1:1 and 2:1 reactions, and the quantitative comparison of the extent of capsid formation is shown in Fig. 4c. In marked contrast to assembly with non-TR RNAs, each of the extended TR RNAs shows evidence of being at least partially kinetically trapped in 1:1 reactions (cf., the heights of the light-scattering peaks at the different time points in Fig. 4b). The trap was removed by addition of another aliquot of CP2, implying that all the longer RNAs are TR-like in their behaviour. This suggests that the kinetic trap is caused by high-affinity binding of the CP2 to the TR stem–loop within these longer RNAs. The apparent kinetics and yield of capsid assembly are, however, different from TR, with the longer RNAs leading to both faster and more efficient assembly during the time of the experiment.
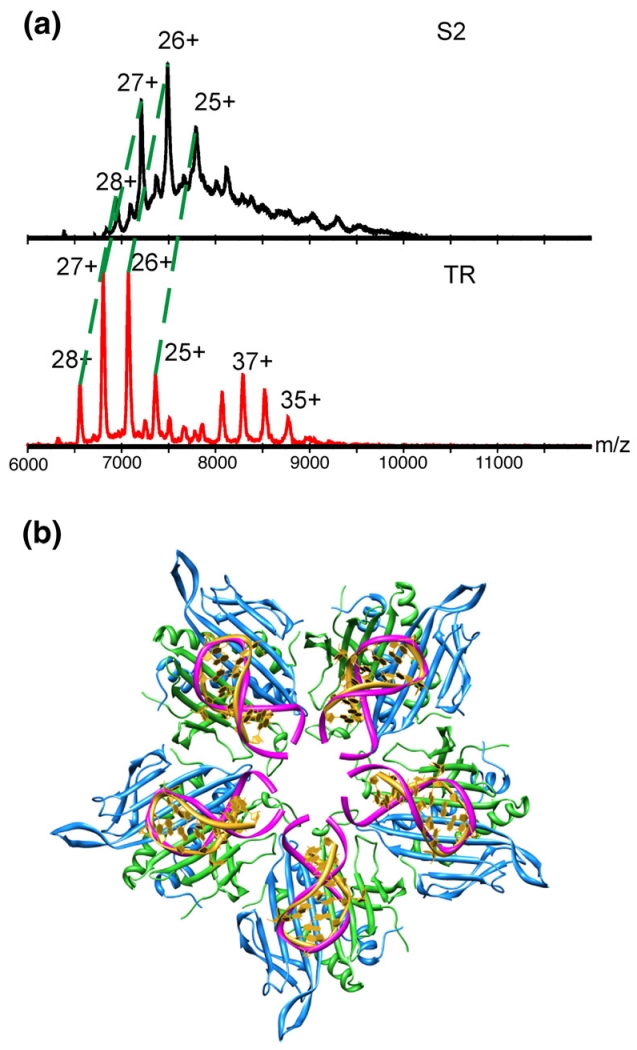
The implication is that the longer RNAs influence the efficiency of the assembly pathway. To test this idea, we then examined the S2 assembly reaction by mass spectrometry. Previously, we have shown that between the 1:1 complex of CP2:TR and the broad peak representative of the T=3 capsid, there are two dominant assembly intermediates. These have stoichiometries consistent with representing a fully formed capsid 3-fold axis [3(CP2:TR)+3CP2] and a 5-fold axis decorated with an additional layer of dimers [5(CP2:TR)+5CP2]6,28 (Morton et al., unpublished). The kinetic-trapping mechanism relies on the hypothesis that the interaction between CP2:TR complexes is weak and insufficient to create the minimal 5-fold axis, which could lead to assembly of a T=1 shell as well as a T=3 shell. No minimal 5-fold species has been detected by mass spectrometry, nor have we seen T=1 particles in cryo-EM samples, consistent with this idea. By implication, the interaction between CP2:TR and CP2 must be significantly stronger. Although in the crystal structures of the capsid there are more inter-dimer contacts between adjacent A/B and C/C dimers (e.g., at 3-fold axes) than between A/B and A/B dimers at 5-fold axes,5 it is not obvious why there should be a large difference in interaction energy. Both the 3-fold complex and the extended 5-fold (decameric) complex chase into capsid-related higher-order species at the same rate when the free CP2 concentration is raised, implying that they are both on assembly pathways that lead to T=3 shell formation.6
The mass spectrum of the S2 assembly reaction shows markedly reduced levels of the decameric complex but approximately normal levels of the complex corresponding to the 3-fold axis (Fig. 5a). Since we have shown that S2 supports T=3 assembly, this confirms that the 3-fold complex is on a capsid assembly pathway and that there are at least two such routes for a TR-mediated reaction. Furthermore, it shows that it is possible to change the flux through these pathways by altering the RNAs used to trigger assembly. Examination of the structure created by modelling stem–loops with extended stems into the TR binding sites at a 5-fold axis (Fig. 5b) suggests that the longer RNA would create steric/electrostatic clashes that would reduce the stability of a decameric complex containing A/B dimers fully bound by RNA stem–loops. This would not be expected to be a problem in the 3-fold complex. Unfortunately, poor ionisation of the CP2 complexes with the longer RNAs prevented extension of these mass spectrometry assays to the other extended TR RNAs. Figure 6 summarises the assembly pathways consistent with these data.
Fig. 5.
Mass spectrometry of assembly with S2 RNA. (a) Stoichiometry of the 3-fold intermediate [3(CP2:RNA)+3CP2] using TR and S2 to initiate re-assembly in the 2:1 CP2:RNA stoichiometry at t=30 min. The charge-state distributions corresponding to the intermediates formed during TR and S2 assembly, in the m/z range 6000–12,000 are indicated. The difference in mass between TR and S2 is 4 kDa. The observed masses for the 3-fold intermediates are 182.7 and 194.7 kDa, yielding a mass difference of 12 kDa, as expected. The peaks corresponding to the extended 5-fold intermediate with TR are clearly visible in the lower spectrum with TR (35+ to 38+) but are essentially undetectable in the upper one for S2. (b) Modelling the effect of RNA stem–loop binding to all dimers in a particle 5-fold axis. The yellow RNA stem–loop represents TR (PDB code 1AQ3),30 and that shown in pink is a 38-nt stem–loop (PDB code 1TXS)31 that has been truncated to 23 nt and manually positioned over the TR stem–loop.
Fig. 6.
The inferred assembly pathway. The cartoon illustrates the inferred assembly pathways consistent with the mass spectrometry, gel-filtration and ultracentrifugation data. A C/C-like (red) coat protein dimer can be switched to an A/B-like dimer (blue/green) by binding an RNA stem–loop (yellow). In principle, such A/B-like species could self-associate, generating initially a pentameric complex that could create a closed shell with T=1 symmetry. This does not occur, ensuring formation of a larger capsid capable of packaging the phage genome. As a result, tightly binding RNA ligands “trap” the coat protein dimer in just one of the two quasi-equivalent conformers required for efficient T=3 shell formation. Addition of RNA-free (C/C-like) coat protein dimers provides the missing assembly component, leading to formation of two higher-order intermediates, corresponding to the 3- and 5-fold assemblies on the pathway to the T=3 shell. A similar outcome is observed with the weakly binding stem–loops because both sorts of coat protein dimer are always present, preventing the kinetic trap. Extended TR oligos provide additional binding sites for incoming protein subunits in the appropriate places to promote conformer switching. From the structures of the 3- and 5-fold intermediates, it is difficult to imagine their assembly pathways intersecting until the final T=3 capsid is completed.
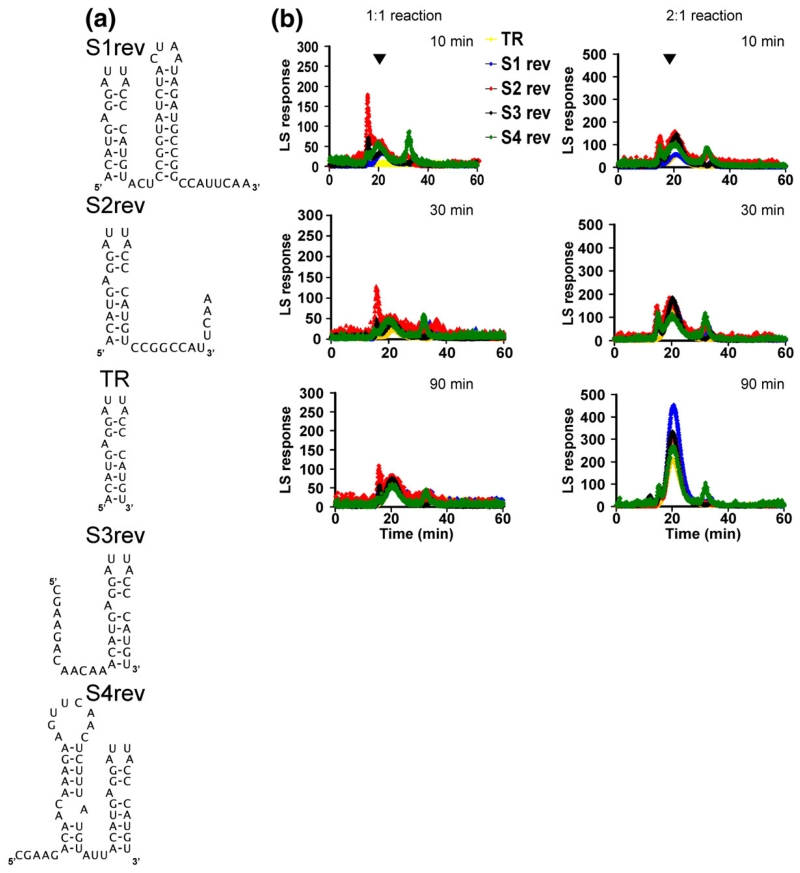
The faster assembly kinetics seen for the extended TR oligonucleotides could arise from the positions of the extensions, their sequences or a combination of both. To test these possibilities, we produced an additional set of extended TR oligonucleotides having the same genomic sequence extensions as S1–S4 but in the reverse orientation with respect to TR—that is, the natural 5′ extension sequences were added to the 3′ end of TR and vice versa, creating the stem–loops S1rev–S4rev (Fig. 7a). The results of using these new RNAs in assembly reactions are striking. The Srev stem–loops, which contain TR, show evidence of the kinetic trap in 1:1 reactions, but this is much less efficiently relieved in 2:1 reactions compared with the S-series (cf., the 30 -min time points in Figs. 4 and 7b). The reversed RNAs also show more evidence for formation of aggregates than the natural sequences. Clearly, the effects seen in the stem–loops are consequences of both the position and the sequence of the genomic extensions of TR.
Fig. 7.
Assembly with the scrambled extended TR RNAs. (a) Sequences and secondary structures of the Srev-series of RNAs. (b) Gel filtration–light-scattering profiles for 1:1 and 2:1 re-assembly reactions with these RNAs. Experimental details are as those given in Fig. 2.
Biological implications
In contrast to double-stranded RNA and DNA viruses that package their genomes into pre-formed containers, single-stranded RNA viruses, such as MS2, assemble their capsids around their genomic RNAs.29 In a number of examples, high-affinity genome packaging signals, such as the MS2 TR, have been reported,2,32–34 but in the majority of these viruses, no such signal has been detected. In addition, many studies have shown that single-stranded RNA viral coat proteins can package a variety of species (non-specific RNAs, DNAs and even polymers and nanoparticles35,36), leading to the impression that the proteins are the dominant component of the assembly reaction and that the principal role of the RNA is simply to localise such proteins, presumably in an electrostatically stabilised complex. The data described here and in our previous articles suggest that this view requires refinement to acknowledge the active, specific role(s) that cognate RNAs can play in creating an efficient assembly pathway.
This realisation arises from the observation that the 1:1 complex between MS2 CP2 bound to the TR RNA fragment is kinetically trapped and that the resulting protein dimer is in a distinct conformation from its RNA-free equivalent. The TR binding site is at least 12 Å away from the nearest FG-loop atoms,6 where these conformational changes must take place, so it cannot be due to a direct effect of the RNA (Fig. 1). In an accompanying article by Dykeman et al.,37 a mechanism for this effect is suggested based on dynamic allostery due to the differential effects of binding an RNA stem–loop on the mobility of peptide loops within the CP2. This model is independent of the sequence of the stem–loop bound and accounts for the behaviour of an assembly mutant protein (W82R38), since TR binding in this case does not result in enhanced mobility in the FG loop. Such a mechanism is also consistent with NMR data on the unliganded C/C dimer showing considerable dynamic behaviour on both the nanosecond and millisecond timescales.38 This mobility is normally asymmetric distributed within the CP2, being highest in the FG loops but extending down the adjacent F and G β-strands sufficiently far that it overlaps the TR binding site.
Recruitment of RNA-free coat protein dimers by RNA-bound CP2 is then the next step on the pathway to T=3 capsid formation, and this is necessary to avoid a kinetic trap. We have shown that although multiple stem–loops can trigger the assembly reaction, these bind with lower affinity and therefore the reactions are not kinetically trapped. This is consistent with coat protein conformer switching occurring throughout assembly. It also raises a question about whether assembly with genomic RNA must always initiate with a coat protein dimer binding to TR. In principle, this is not required and the presence of the TR sequence in long RNAs has been shown to result in only marginal increases in assembly efficiency.14,15 These authors also noted the importance of non-TR sequences for the assembly reaction. However, in vivo there is a known preference for packaging cognate RNAs even in the presence of a competing RNA phage.24 Such specificity may be due to the influence of phage maturation proteins, which are known to make sequence-specific contacts to the RNA.39
The results with the longer RNAs provide additional evidence in support of active roles for the genomic RNA during assembly. Although the presence of a TR stem–loop confers a kinetic trap at low coat protein stoichiometries, as expected, even the addition of a single 5′ adenosine nucleotide to TR leads to a large increase in association rate constant with coat protein.14,15 This is consistent with the results reported here where small genomic sequence extensions to TR at both 5′ and 3′ ends lead to faster assembly during the time course of the experiments.
Since it seems impossible to envisage assembly sub-pathways based around the 3-fold axis or the 5-fold axis coalescing before the complete T=3 shell is formed, this result shows that separate pathways are used to assemble the capsid in TR-mediated reactions. In principle, a vast number of combinatorial pathways could exist for such a shell (R. Twarock, personal communication), although this total reduces significantly if the genomic RNA plays a role in assembly.18 From the data reported here, it is clear that there are at least two assembly pathways possible for MS2 (Fig. 6). Strikingly, for the one extended TR RNA where it was possible to obtain interpretable mass spectra of assembly reactions (S2), there is almost complete loss of the extended 5-fold complex. There appear to be simple steric/electrostatic repulsion arguments explaining why this is the case. The phage, however, must assemble its T=3 shell around a cognate genome of 3569 nt that contains just one copy of the TR site. If other stem–loop structures within the phage RNA result in conformational switching throughout the assembly pathway, then the increased assembly yields seen with the longer cognate RNAs make sense. The reversed S-series implies that the positions of defined sequences/secondary structures with respect to TR are also important. In summary, it appears that the RNA serves as a scaffold instructing the coat protein shell on which assembly pathway to take via controlled switching of quasi-equivalent dimers. Whether this restricts the assembly pathway to fewer possibilities remains unknown, but the net result of these features of the packaged RNA is greatly increased assembly efficiency.
The results do not mean that only MS2 RNA containing a TR site can be packaged into T=3 shells. Indeed, it has been shown with longer RNAs that TR confers only a slight packaging advantage,14,15 and our recombinant starting material before dissociation contains a significant amount of stochastically packaged Escherichia coli RNAs. It suggests that the cognate reactions will be more efficient, especially at lower concentrations and shorter times. The known preference for packaging cognate RNAs in vivo even in the presence of a competing RNA phage24 may also be due to the influence of phage maturation proteins, which are known to make sequence-specific contacts to the RNA.39 These results do not imply that RNA-free assembly is impossible. Presumably, the coat protein dynamics revealed by NMR sample both symmetrical and asymmetric conformers.38 The RNA stem–loops merely bias this equilibrium, extending the lifetimes of asymmetric species, thus facilitating the capsid assembly pathway. This is similar to the emerging view of the way enzymes sample conformational sub-states, one of which is then selected by substrate binding.40
Materials and Methods
Preparation of coat proteins and RNA stem–loops
Proteins
Wild-type recombinant coat proteins in the form of T=3 shells containing randomly incorporated cellular RNAs were prepared by overexpression in E. coli as described previously.41 These capsids were purified, and then disassembled coat protein dimers were isolated by treatment with glacial acetic acid followed by exchange into 20 mM acetic acid.8,42 This latter step removes the majority of the cellular RNA as judged from the absorbance at 260/280 nm.
RNAs
The MS2 RNA translational operator stem–loop (TR), aptamers F6 and F7 and the S-series (S1–S4) were synthesized and characterised using the protocols described previously.25,27 The Qβ stem–loop and Srev-series were supplied by Dharmacon (Thermo Scientific Inc., Belgium).
Gel filtration–light-scattering assays
Assays of re-assembly efficiency at differing stoichiometries and times were monitored by gel filtration using both UV absorbance and light-scattering to monitor the outflow with a UV VIS Detector LC 1200 (Polymer Laboratories, Varian, UK) at 260 nm and a Dynamic Light Scattering Detector PD2010 combined with correlator PD 2000 DLS (Precision Detectors, USA). The reactions were set up with slight variations on the protocol for mass spectrometry. For convenience, the concentrations of CP2 were 10 μM in the 1:1 reaction and 20 μM for the 2:1 sample, with the final TR concentration being 10 μM for both. The final 100-μL reaction volume was made up with assembly buffer (40 mM ammonium acetate, pH 6.9); the samples were incubated on ice for various times until injection onto a Tricorn high-performance column of Superose 6 10/300 GL (GE Healthcare Bio-Sciences AB, Sweden) thermostated at 12 °C. The eluant was 50 mM Tris acetate, pH 7.4, used to avoid problems due to degassing with ammonium acetate, at the flow rate of 0.4 mL/min, provided by HPLC pump LC 1120 (Polymer Laboratories, Varian). Samples were injected via a 100-μL loop. Samples of the dissociated MS2 coat protein were routinely checked by injecting onto the gel-filtration column prior to setting up the re-assembly reactions to confirm that they did not contain any undissociated/reassembled material. The instrument was calibrated prior to each experiment with bovine serum albumin (Sigma-Aldrich, UK) as a molecular mass protein standard (mass of 66,432 Da). The recombinant MS2 capsid protein (10 μM) was also injected to establish the expected elution time of the re-assembled capsid peak.
Quantitation of gel-filtration data
The light-scattering and UV-absorption traces of the column outflow revealed the presence of a T=3 capsid peak, which overlaps with aggregates, and incomplete capsid fragments (CP2:RNA complex and free RNA). The area of the capsid was evaluated using multi-Gaussian peak fitting with the program Igor Pro 5.0.5.7 (Wave-Metrics Inc., Lake Oswego, OR).
Assembly reactions
Reaction conditions for the gel-filtration experiments
Assembly reactions were carried out in 40 mM ammonium acetate, pH 6.9, at 4 °C. Initially, CP2 was mixed with TR to a final concentration of 8 μM, and further aliquots of CP2 were added as indicated. The concentration of the CP2 solution was calculated from UV absorbance.
Electrospray ionisation mass spectrometry
Samples were analysed by positive ionisation nano-electrospray using an LCT Premier mass spectrometer (Waters Corporation, Manchester, UK) with collisional cooling capabilities equipped with a NanoMate (Advion Biosciences Inc., Ithaca, NY) temperature-controlled, automated sample handling and ionisation interface, which was maintained at 4 °C. A capillary voltage of 1.9 kV was set with a nitrogen gas flow of 0.5 psi for sample introduction and ionisation. The sampling cone voltage was optimised at 100 V, Ion Guide 1 was at 130 V and Aperture 1 was at 60 V. Data were acquired over the m/z range 500–30,000, and data processing was performed using the MassLynx software supplied with the mass spectrometer. An external calibration using CsI clusters was applied to the data.
For the competitive binding experiments, all mass spectra were recorded using 40 mM ammonium acetate at pH 6.9. Equimolar concentrations (10 μM) of coat protein dimer, TR RNA and any of the competing ligands (F6, F7 or Qβ RNA) were added in a 1:1:1 ratio. The reaction was allowed to equilibrate on ice for 5 min, and then spectra were acquired at 2.4 scan s−1 for 1 min. Averaged spectra were background subtracted and smoothed using the MassLynx software. Peak areas corresponding to the [CP2:TR] and [CP2:RNA] were summed, and peak areas were normalised to the signal of [CP2:TR]. It was assumed in all cases that the ionisation efficiency is the same for each protein–RNA complex. All RNAs were shown to form equivalents of the initiation complex in the stoichiometry [CP2:RNA], implying that all are able to bind CP2. The competition experiments were done in triplicate, and error bars shown in the plots represent the ±SD.
Sedimentation velocity assay of assembly
Capsid re-assembly reactions were performed at a 2 μM final concentration of RNA in 40 mM ammonium acetate at 4 °C. The reaction volume was 320 μL, which, following incubation for 4 h, was loaded along with 340 μL of buffer in the reference sector into a two-sector meniscus-matching Epon centrepiece cell. Sedimentation velocity analysis was carried out using a Beckman Optima XL-I ultracentrifuge using an An-50 Ti rotor. Sedimentation velocity profiles were collected at 17,000 and 40,000 rpm by absorbance at 260 nm every ~2 min (A260 of ~0.5). Due to the large difference in sedimentation rate of the substrates and products of MS2 capsid assembly reactions, the reactions were spun at both 17,000 and 40,000 rpm, collecting 50 and 100 scans, respectively. The resulting sedimentation profiles were analysed using the C(S) method in SEDFIT.43 The buffer viscosity and density were calculated with the program SEDNTERP† and were calculated to be 0.001008 poise and 0.99835 g/mL, respectively. A partial specific volume of 0.53 mL/g44 was used in fitting all acquired data.
Acknowledgements
This work was supported in part by the Leverhulme Trust, the Wellcome Trust and the UK Biotechnology and Biological Sciences Research Council. O.R. is grateful to the Wellcome Trust for a postgraduate studentship. We thank Dr. Abdul Rashid, Dr. Iain Manfield and Ms. Amy Magoolagan for production of some of the RNAs used in the experiments. We also thank Dr. Katerina Toropova for help with the figures and Prof. Reidun Twarock (York), Dr. Neil Ranson and our other colleagues for helpful discussions of the results and the manuscript.
Abbreviation used
- EM
electron microscopy
Footnotes
References
- 1.Harrison SC, Olson AJ, Schutt CE, Winkler FK, Bricogne G. Tomato bushy stunt virus at 2.9 Å resolution. Nature. 1978;276:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- 2.Sorger PK, Stockley PG, Harrison SC. Structure and assembly of turnip crinkle virus: II. Mechanism of reassembly in vitro. J. Mol. Biol. 1986;191:639–658. doi: 10.1016/0022-2836(86)90451-1. [DOI] [PubMed] [Google Scholar]
- 3.Caspar DL, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 4.Golmohammadi R, Valegard K, Fridborg K, Liljas L. The refined structure of bacteriophage MS2 at 2.8 Å resolution. J. Mol. Biol. 1993;234:620–639. doi: 10.1006/jmbi.1993.1616. [DOI] [PubMed] [Google Scholar]
- 5.Valegard K, Liljas L, Fridborg K, Unge T. The three-dimensional structure of the bacterial virus MS2. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- 6.Stockley PG, Rolfsson O, Thompson GS, Basnak G, Francese S, Stonehouse NJ, et al. A simple, RNA-mediated allosteric switch controls the pathway to formation of a T = 3 viral capsid. J. Mol. Biol. 2007;369:541–552. doi: 10.1016/j.jmb.2007.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carey J, Uhlenbeck OC. Kinetic and thermodynamic characterization of the R17 coat protein–ribonucleic acid interaction. Biochemistry. 1983;22:2610–2615. doi: 10.1021/bi00280a003. [DOI] [PubMed] [Google Scholar]
- 8.Lago H, Parrott AM, Moss T, Stonehouse NJ, Stockley PG. Probing the kinetics of formation of the bacteriophage MS2 translational operator complex: identification of a protein conformer unable to bind RNA. J. Mol. Biol. 2001;305:1131–1144. doi: 10.1006/jmbi.2000.4355. [DOI] [PubMed] [Google Scholar]
- 9.Stockley PG, Stonehouse NJ, Valegard K. Molecular mechanism of RNA phage morphogenesis. Int. J. Biochem. 1994;26:1249–1260. doi: 10.1016/0020-711x(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 10.Talbot SJ, Goodman S, Bates SR, Fishwick CW, Stockley PG. Use of synthetic oligoribonucleotides to probe RNA–protein interactions in the MS2 translational operator complex. Nucleic Acids Res. 1990;18:3521–3528. doi: 10.1093/nar/18.12.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Crystal structure of an RNA bacteriophage coat protein–operator complex. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 12.Valegard K, Murray JB, Stonehouse NJ, van den Worm S, Stockley PG, Liljas L. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein–RNA interactions. J. Mol. Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 13.Witherell GW, Gott JM, Uhlenbeck OC. Specific interaction between RNA phage coat proteins and RNA. Prog. Nucleic Acid Res. Mol. Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- 14.Beckett D, Uhlenbeck OC. Ribonucleo-protein complexes of R17 coat protein and a translational operator analog. J. Mol. Biol. 1988;204:927–938. doi: 10.1016/0022-2836(88)90052-6. [DOI] [PubMed] [Google Scholar]
- 15.Beckett D, Wu HN, Uhlenbeck OC. Roles of operator and non-operator RNA sequences in bacteriophage R17 capsid assembly. J. Mol. Biol. 1988;204:939–947. doi: 10.1016/0022-2836(88)90053-8. [DOI] [PubMed] [Google Scholar]
- 16.Fiers W, Contreras R, Duerinck F, Haegeman G, Iserentant D, Merregaert J, et al. Complete nucleotide sequence of bacteriophage MS2 RNA: primary and secondary structure of the replicase gene. Nature. 1976;260:500–507. doi: 10.1038/260500a0. [DOI] [PubMed] [Google Scholar]
- 17.Koning R, van den Worm S, Plaisier JR, van Duin J, Pieter Abrahams J, Koerten H. Visualization by cryo-electron microscopy of genomic RNA that binds to the protein capsid inside bacteriophage MS2. J. Mol. Biol. 2003;332:415–422. doi: 10.1016/s0022-2836(03)00846-5. [DOI] [PubMed] [Google Scholar]
- 18.Toropova K, Basnak G, Twarock R, Stockley PG, Ranson NA. The three-dimensional structure of genomic RNA in bacteriophage MS2: implications for assembly. J. Mol. Biol. 2008;375:824–836. doi: 10.1016/j.jmb.2007.08.067. [DOI] [PubMed] [Google Scholar]
- 19.van den Worm SH, Koning RI, Warmenhoven HJ, Koerten HK, van Duin J. Cryo electron microscopy reconstructions of the Leviviridae unveil the densest icosahedral RNA packing possible. J. Mol. Biol. 2006;363:858–865. doi: 10.1016/j.jmb.2006.08.053. [DOI] [PubMed] [Google Scholar]
- 20.Convery MA, Rowsell S, Stonehouse NJ, Ellington AD, Hirao I, Murray JB, et al. Crystal structure of an RNA aptamer–protein complex at 2.8 Å resolution. Nat. Struct. Biol. 1998;5:133–139. doi: 10.1038/nsb0298-133. [DOI] [PubMed] [Google Scholar]
- 21.Hirao I, Spingola M, Peabody D, Ellington AD. The limits of specificity: an experimental analysis with RNA aptamers to MS2 coat protein variants. Mol. Diversity. 1998;4:75–89. doi: 10.1023/a:1026401917416. [DOI] [PubMed] [Google Scholar]
- 22.Rowsell S, Stonehouse NJ, Convery MA, Adams CJ, Ellington AD, Hirao I, et al. Crystal structures of a series of RNA aptamers complexed to the same protein target. Nat. Struct. Biol. 1998;5:970–975. doi: 10.1038/2946. [DOI] [PubMed] [Google Scholar]
- 23.Horn WT, Tars K, Grahn E, Helgstrand C, Baron AJ, Lago H, et al. Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2. Structure. 2006;14:487–495. doi: 10.1016/j.str.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling CM, Hung PP, Overby LR. Independent assembly of Qbeta and MS2 phages in doubly infected Escherichia coli. Virology. 1970;40:920–929. doi: 10.1016/0042-6822(70)90138-8. [DOI] [PubMed] [Google Scholar]
- 25.Murray JB, Collier AK, Arnold JR. A general purification procedure for chemically synthesized oligoribonucleotides. Anal. Biochem. 1994;218:177–184. doi: 10.1006/abio.1994.1157. [DOI] [PubMed] [Google Scholar]
- 26.Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrott AM, Lago H, Adams CJ, Ashcroft AE, Stonehouse NJ, Stockley PG. RNA aptamers for the MS2 bacteriophage coat protein and the wild-type RNA operator have similar solution behaviour. Nucleic Acids Res. 2000;28:489–497. doi: 10.1093/nar/28.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rolfsson O, Toropova K, Morton VL, Francese S, Basnak G, Thompson GS, et al. RNA packing specificity and folding during assembly of the bacteriophage MS2. Comput. Math. Methods Med. 2008;9:339–349. doi: 10.1080/17486700802168445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hohn T. Role of RNA in the assembly process of bacteriophage fr. J. Mol. Biol. 1969;43:191–200. doi: 10.1016/0022-2836(69)90088-6. [DOI] [PubMed] [Google Scholar]
- 30.van den Worm SH, Stonehouse NJ, Valegard K, Murray JB, Walton C, Fridborg K, et al. Crystal structures of MS2 coat protein mutants in complex with wild-type RNA operator fragments. Nucleic Acids Res. 1998;26:1345–1351. doi: 10.1093/nar/26.5.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du Z, Yu J, Ulyanov NB, Andino R, James TL. Solution structure of a consensus stem–loop D RNA domain that plays important roles in regulating translation and replication in enteroviruses and rhinoviruses. Biochemistry. 2004;43:11959–11972. doi: 10.1021/bi048973p. [DOI] [PubMed] [Google Scholar]
- 32.Zimmern D. The nucleotide sequence at the origin for assembly on tobacco mosaic virus RNA. Cell. 1977;11:463–482. doi: 10.1016/0092-8674(77)90065-4. [DOI] [PubMed] [Google Scholar]
- 33.Qu F, Morris TJ. Encapsidation of turnip crinkle virus is defined by a specific packaging signal and RNA size. J. Virol. 1997;71:1428–1435. doi: 10.1128/jvi.71.2.1428-1435.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stoker K, Koperzwarthoff EC, Bol JF, Jaspars EMJ. Localization of a high-affinity binding site for coat protein on the 3′-terminal part of RNA-4 of alfalfa mosaic virus. FEBS Lett. 1980;121:123–126. [Google Scholar]
- 35.Sun J, DuFort C, Daniel MC, Murali A, Chen C, Gopinath K, et al. Core-controlled polymorphism in virus-like particles. Proc. Natl Acad. Sci. USA. 2007;104:1354–1359. doi: 10.1073/pnas.0610542104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aniagyei SE, Kennedy CJ, Stein B, Willits DA, Douglas T, Young MJ, et al. Synergistic effects of mutations and nanoparticles templating in the self-assembly of cowpea chlorotic mottle virus capsids. Nano Lett. 2008;9:393–398. doi: 10.1021/nl8032476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dykeman EC, Stockley PG, Twarock R. Dynamic allostery controls coat protein conformer switching during MS2 phage assembly. J. Mol. Biol. 2009;395:916–923. doi: 10.1016/j.jmb.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Stockley PG, Ashcroft AE, Francese S, Thompson GS, Ranson NA, Smith AM, et al. Dissecting the fine details of assembly of a T=3 phage capsid. Comput. Math. Methods Med. 2005;6:119–125. [Google Scholar]
- 39.Shiba T, Suzuki Y. Localization of A protein in the RNA–A protein complex of RNA phage MS2. Biochim. Biophys. Acta. 1981;654:249–255. doi: 10.1016/0005-2787(81)90179-9. [DOI] [PubMed] [Google Scholar]
- 40.Kern D, Zuiderweg ER. The role of dynamics in allosteric regulation. Curr. Opin. Struct. Biol. 2003;13:748–757. doi: 10.1016/j.sbi.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 41.Mastico RA, Talbot SJ, Stockley PG. Multiple presentation of foreign peptides on the surface of an RNA-free spherical bacteriophage capsid. J. Gen. Virol. 1993;74(Pt. 4):541–548. doi: 10.1099/0022-1317-74-4-541. [DOI] [PubMed] [Google Scholar]
- 42.Sugiyama T, Nakada D. Control of translation of MS2 RNA cistrons by MS2 coat protein. Proc. Natl. Acad. Sci. USA. 1967;57:1744–1750. doi: 10.1073/pnas.57.6.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuck P. Size-distribution analysis of macro-molecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Enger MD, Stubbs EA, Mitra S, Kaesberg P. Biophysical characteristics of the RNA-containing bacterial virus R17. Proc. Natl Acad. Sci. USA. 1963;49:857–860. doi: 10.1073/pnas.49.6.857. [DOI] [PMC free article] [PubMed] [Google Scholar]