Abstract
Sensitivity to MDM2 inhibitors is widely different among responsive TP53 wild-type cell lines and tumours. Understanding the determinants of MDM2 inhibitor sensitivity is pertinent for their optimal clinical application. Wild-type p53-inducible phosphatase-1 (WIP1) encoded by PPM1D, is activated, gained/amplified in a range of TP53 wild-type malignancies and is involved in p53 stress response homeostasis.
We investigated cellular growth/proliferation of TP53 wild-type and matched mutant/null cell line pairs, differing in PPM1D genetic status, in response to Nutlin-3/RG7388 ± a highly selective WIP1 inhibitor GSK2830371. We also assessed the effects of GSK2830371 on MDM2 inhibitor induced p53Ser15 phosphorylation, p53-mediated global transcriptional activity and apoptosis.
The investigated cell line pairs were relatively insensitive to single agent GSK2830371. However, a non-growth inhibitory dose of GSK2830371 markedly potentiated the response to MDM2 inhibitors in TP53 wild-type cell lines, most notably in those harbouring PPM1D activating mutations or copy number gain (up to 5.8-fold decrease in GI50). Potentiation also correlated with significant increase in MDM2 inhibitor induced cell death endpoints which were preceded by a marked increase in a WIP1 negatively regulated substrate, phosphorylated p53Ser15, known to increase p53 transcriptional activity. Microarray-based gene expression analysis showed that the combination treatment increases the subset of early RG7388 induced p53-transcriptional target genes.
These findings demonstrate that potent and selective WIP1 inhibition potentiates the response to MDM2 inhibitors in TP53 wild-type cells, particularly those with PPM1D activation or gain, while highlighting the mechanistic importance of p53Ser15 and its potential use as a biomarker for response to this combination regimen.
Keywords: MDM2 inhibitor, Potentiation, TP53, PPM1D/WIP1, GSK2830371
Introduction
Mutational inactivation of the p53 tumour suppressor protein, encoded by the TP53 gene, occurs in ≈50% of malignancies overall (1, 2). Non-genotoxic activation of p53 in the remaining TP53 wild-type malignancies has attracted attention as a therapeutic strategy (3-5). In unstressed cells p53 is rapidly turned over by binding to one of its transcriptional target gene products, MDM2, which inhibits p53-mediated transcription, promotes p53 ubiquitin-mediated nuclear export and its proteasomal degradation (3). Cellular stress can activate effector molecules (e.g. DNA-PK, ATM and ATR) which post-translationally modify MDM2 and or p53, leading to their dissociation followed by p53-mediated reversible cell cycle arrest, senescence or programmed cell death (6). Proof of concept that pharmacological inhibition of the MDM2-p53 interaction by small molecular weight MDM2 inhibitors can be successfully used for non-genotoxic activation of p53 has been established in preclinical and clinical settings with encouraging anti-tumour activity (4, 7). Although, It is firmly established that the most important determinant of response to MDM2 inhibitors is wild-type TP53 genetic status (Supplementary figure S1A and (8)), multiple independent studies using various classes of MDM2 inhibitors, and drug sensitivity data generated by the Sanger Institute, have shown that there is a wide range of sensitivity to MDM2 inhibitors among TP53 wild-type cell lines (Supplementary Figure S1B (8-10)). These highlight that the determinants of sensitivity to MDM2 inhibitors in a TP53 wild-type background are poorly understood. The use of combination regimens and patient stratification strategies are therefore being investigated to optimise tumour specific response in TP53 wild-type malignancies (11-13).
Another strategy for non-genotoxic activation of p53 currently in pre-clinical development is inhibition of Wild-type p53-inducible phosphatase-1 (WIP1/PPM1D) which is involved in homeostatic regulation of p53 function and stability following cellular stress (14-16). PPM1D is a bona fide oncogene which is activated, gained or amplified mostly in TP53 wild-type malignancies (17-19). Notably, PPM1D gain-of-function mutations (activation) and TP53 inactivating-mutations are mutually exclusive in brainstem gliomas, consistent with the role of WIP1 (PPM1D gene product) in negative regulation of p53 (18). Following cellular stress, p53 transcriptionally induces WIP1, which forms a negative auto-modulatory loop with the p53 network by dephosphorylating p53 and other signalling components involved in p53 post-translational regulation (15). In spite of selectivity and bioavailability challenges associated with pharmacological targeting of phosphatases (20), recently a highly selective allosteric WIP1 inhibitor, GSK2830371, which targets the unique flap-subdomain on WIP1 was identified and characterised (16). Although, the response to GSK2830371 was contingent on wild-type TP53, some TP53 wild-type cell lines did not respond in the dose range associated with on-target activity (16). In a subsequent publication which highlighted WIP1 as a potential target in neuroblastoma, GSK2830371 was shown to effectively inhibit the growth of TP53 wild-type cell lines with PPM1D copy number gain. However, there was greater than 52-fold range in sensitivity, with NGP cells (PPM1D copy number gain) showing no response at all within the dose range tested (10μM cut-off) (21).
MDM2 blocks the p53 transactivation domain by interacting with three key p53 amino acids (Phe19, Trp23 and Leu26) which are proximal to a WIP1 substrate, phosphorylated p53Ser15 (pp53Ser15) (22, 23). Unlike the strong influence of p53Ser20 phosphorylation on binding to MDM2, the phosphorylation of p53Ser15 has been reported to either have no or a modest effect on binding of p53 to MDM2 (24, 25), it has nevertheless been shown to contribute to increased p53 pro-apoptotic transcriptional transactivation activity (25-27). After MDM2 inhibitor mediated dissociation of p53 from MDM2, phosphorylated-p53Ser15 (pp53Ser15) is generated by the basal unstimulated activity of effector kinases which normally phosphorylate p53Ser15 following genotoxic stress (26). This suggests that a dynamic equilibrium exists between kinases and phosphatases in regulating pp53Ser15 following MDM2 inhibitor induced p53 stabilisation, which can be tilted in favour of the p53 activating kinases by inhibiting WIP1. Therefore in this study we have investigated whether a selective WIP1 inhibitor GSK2830371 can potentiate the response to MDM2 inhibitors Nutlin-3 and RG7388 in a panel of cell line pairs differing in their TP53 and PPM1D genetic status. Our findings show that a non-growth inhibitory dose of GSK2830371 can substantially increase sensitivity to MDM2 inhibitors in TP53 wild-type cell lines, particularly in those with PPM1D copy number gain or gain-of-function mutation. Furthermore, global gene expression analysis showed that RG7388 in the presence of GSK2830371 induces additional early p53 transcriptional target genes involved in apoptosis in TP53 wild-type cell lines which are not responsive to the WIP1 inhibitor alone. We propose that the combination of WIP1 and MDM2 inhibitors can selectively accentuate the sensitivity to MDM2 inhibitors in TP53 wild-type tumours with increased WIP1 expression or activity, with elevated pp53Ser15 as a potential mechanistic biomarker for response to this combination.
Materials and methods
Cell lines and growth conditions
All cell lines used were obtained from Northern Institute for Cancer Research cell line bank which only includes cell lines that have been authenticated using short tandem repeat (STR) DNA profiling (LGC Standards, Teddington, Middx, UK). Post-authentication passages were limited to 30 for experimental procedures (<6 Months) before replacing with lower passage number stocks. Cell line pairs used and their TP53 and PPM1D genetic status are described in Table 1. MCF-7 cells were used as a positive control for WIP1 protein expression and response to the WIP1 inhibitor GSK2830371. The S_N40R2 (SN40R2) and N_N20R1 (N20R1) cell lines were TP53 mutant, otherwise isogenic, Nutlin-3 resistant clones derived from SJSA-1 osteosarcoma and NGP neuroblastoma cells respectively and have been cited in pre-clinical studies of MDM2 inhibitors (28) (See Table 1 for mutation details). U2OS-DN cells overexpress the R175H variant of p53 which is reported to have a dominant negative effect (29).
Table 1.
TP53 Wild-type and mutant/Null cell line pairs with different PPM1D genetic status. PPM1D status of SJSA-1 cells were obtained from sanger.ac.uk (51). TP53 mutant daughter cell lines of NGP and SJSA-1 cell lines were derived as described in Materials and Methods. Wt: wild-type; Mt: Mutant.
| TP53 Wild-type parental cell lines | TP53 mutant/Null daughter lines | Tumour of origin | PPM1D genetic alteration (citation) |
|---|---|---|---|
| SJSA-1 | SN40R2 (E285K) | Osteosarcoma | Wild-type (51) |
| HCT116+/+ | HCT116−/−(Null) | Colorectal carcinoma | c.1344delT/Wt (L450X) Gain-of-function (39) |
| U2OS | U2OS-DN (R175H) | Osteosarcoma | c.1372C>T/Wt (R458X) Gain-of-function (39) |
| NGP | N20R1 (P152T & P98H) | Neuroblastoma | Copy number gain (21) |
| MCF-7 | – | Breast adenocarcinoma | Amplified (38) |
Growth inhibition assay
Cells were seeded in 96-well plates 24 hours before treatment. Cells were then fixed with Carnoy’s fixative and Sulphorhodamine B assay was carried out as described in (30). A spectrophotometer (Biorad Model 680) was used for densitometry at 570nm.
Immunoblotting
Western blotting was carried out as described in (31). Antibodies used were MDM2 (Ab-1) 1:300 (#: OP46-100UG, Merck Millipore), MDMX (#: A300287A-2 Bethyl laboratories), WIP1 (F-10) 1:200 (#: sc-376257, Santa Cruz Biotechnology), p53 1:500 (#: NCL-L-p53-DO7, Leica Microsystems Ltd.), phospho-p53Ser-15 1:1000 (#: 9284 Cell Signalling), p21WAF1 1:100 (#: OP64, Calbiochem), BAX 1:1000 (#: 2772S, Cell Signalling), cleaved caspase-3 1:1000 (#: 9664S, New England Biolabs Ltd.), actin 1:3000 (#: A4700, Sigma-Aldrich). Secondary goat anti-mouse/rabbit HRP-conjugated antibodies (#: P0447/P0448, Dako) were used at 1:1000. All antibodies were diluted in 5% milk/1 × TBS-tween (w/v). Proteins were visualised using enhanced chemiluminescence (GE Life Sciences) and X-ray film (Fujifilm). Densitometry was carried out using ImageJ software.
Denaturing immunoprecipitation
Treated cells were lysed (50mM Tris, 150mM NaCl, 0.2mM Na3VO4, 1% NP40 v/v, 1mM PMSF, Roche cOmplete protease inhibitor tablet, 1mM DTT, 2% SDS) then aliquots were conserved as input. Non-denaturing (No SDS) lysis buffer was used to dilute the remainder of the lysates (<0.1% SDS). 1-2μg of rabbit anti-ubiquitin antibody (#: FL-76, Santa Cruz) or rabbit IgG control (#: X0903, Dako) was added to each appropriate vessel and incubated overnight at 4°C. Sepharose beads (#: 17-0618-01, GE Healthcare) were then added and incubated for a further 4Hrs at 4°C. Beads were washed first with 0.5M KCl then with 0.1M KCl, then treated as lysates in the immunoblotting protocol above.
Caspase3/7 assay
2 × 104 cells/well (≈60-70% confluence) were seeded in white 96-well plates (CELLSTAR®, Greiner Bio-One international) and treated after 24 hours. Caspase-3/7 enzymatic activities were quantified by adding a 1:1 ratio of CaspaseGlo® 3/7 reagent (Promega) to growth media 30 min before measuring the luminescence signal using a FLUOstar Omega plate reader (BMG Labtech) and all values were expressed as a ratio of signal relative to solvent control.
Expression array
NGP cells were seeded at 6 × 105 cells per well of a 6-well plate and treated with either DMSO or 75nM of RG7388 (≈GI50) ± 2.5μM GSK2830371 for 4 hours before RNA extraction using RNeasy Plus Mini Kit (Qiagen). Concentration and quality of mRNA were determined using Agilent RNA 6000 nano kit on an Agilent 2100 Bioanalyzer (RNA integrity numbers > 9). RNA samples were sent to AROS Applied Biotechnology (Aarhus, Denmark) for gene expression analysis using Illumina Beadchip expression arrays (HumanHT-12v4.0). Array data processing, background correction, normalisation and quality control checks were performed using the R package ‘Lumi’ (bioconductor.org). Probe intensity values were converted to VSD (variance stabilized data). Robust spline normalization (RSN) was used as an array normalization method. Poor quality probes (detection threshold < 0.01), and probes that are not detected at all in the remaining arrays were removed prior to downstream analysis. The remaining probe normalized intensity values (18,634) were used in the differential expression analysis. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (32) and are accessible through GEO Series accession number GSE75197 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75197).
RNA extraction and Quantitative real-time polymerase chain reaction (qRT-PCR)
Complementary DNA was generated using the Promega Reverse Transcription System (A3500, Promega) as described by the manufacturer. qRT-PCR was carried out using SYBR® green RT-PCR master mix (Life technologies) as per the manufacturer’s guidelines and the following primers (5’-3’): CDKN1A (Forward (F)-TGTCCGTCAGAACCCATGC, Reverse (R)-AAAGTCGAAGTTCCATCGCTC), TP53INP1 (F-TCTTGAGTGCTTGGCTGATACA, R-GGTGGGGTGATAAACCAGCTC), BTG2 (F-CCTGTGGGTGGACCCCTAT, R-GGCCTCCTCGTACAAGACG), MDM2 (F-CAGTAGCAGTGAATCTACAGGGA, R- CTGATCCAACCAATCACCTGAAT) and GAPDH (F-CAATGACCCCTTCATTGACC, R-GATCTCGCTCCTGGAAGAT). 50ng/μl of the cDNA samples per 10μl final reaction volume, with the standard cycling parameters (Stage 1: 50°C for 2min, Stage 2: 95°C for 10min then 40 cycles of 95°C for 15 Sec and 60°C for 1 min), were set and carried out on an ABI 7900HT sequence detection system. Data were presented as mean ± standard error of mean (SEM) relative quantities (RQ) of four independent repeats where GAPDH was used as endogenous control and DMSO used as the calibrator for each independent repeat with the formula 2−ΔΔCT. Analysis was carried out using SDS 2.2 software (Applied Biosystems).
Site-directed mutagenesis and p53 overexpression
The plasmid vector used in this study was pcDNA3.1 (+/−) (Life Technologies, #V790-20) and full-length human TP53 cDNA was cloned into this backbone. The Gozani lab protocol for site directed mutagenesis (33) was used to generate the p53Ser15 mutants. Primers used: p53S15A (F-GTCGAGCCCCCTCTGGCTCAGGAAACATTTTCA, R-TGAAAATGTTTCCTGAGCCAGAGGGGGCTCGAC) and p53S15D (F-GTCGAGCCCCCTCTGGACCAGGAAACATTTTCA, R-TGAAAATGTTTCCTGGTCCAGAGGGGGCTCGAC). HCT116−/− cells were transfected with Lipofectamine 2000 and plasmid DNA 12 hours before lysates were collected at different time intervals.
Flow Cytometry
After treatment, floating and adhered cells were pooled and incubated in propidium iodide solution (150μM propidium iodide (Calbiochem, San Diego, CA), 1.46μM DNase free-RNase A (Sigma, St. Louis, MO), 3.88mM sodium citrate (Sigma, St. Louis, MO) and 0.3% Triton-X 100 (Sigma, St. Louis, MO)) for 10 min at 25°C and then fluourescence-activated cell sorting (FACS) was carried out using a FACSCalibur™ (BD Biosciences). CellQuest software was used to establish cell cycle distribution and gated histograms.
Statistical analysis
Statistical tests were carried out in GraphPad Prism 6 software and all p-values represent two-tailed paired t-tests of 3 or more independent repeats unless otherwise stated. For microarray differential expression analysis R-statistical software was used. Microarray data was processed using R Bioconductor package lumi (34) Probes intensity values were transformed using variance stabilising transformation implemented in the lumi package before data normalisation. The robust spline normalization was used as a normalization method. Poor quality probes (detection threshold < 0.01), and probes that are not detected at all in the remaining arrays were removed. Differential expression analysis was performed using R Bioconductor package limma (35).
Results
GSK2830371 potentiates the response to MDM2 inhibitors Nutlin-3 and RG7388 in a p53-dependent manner
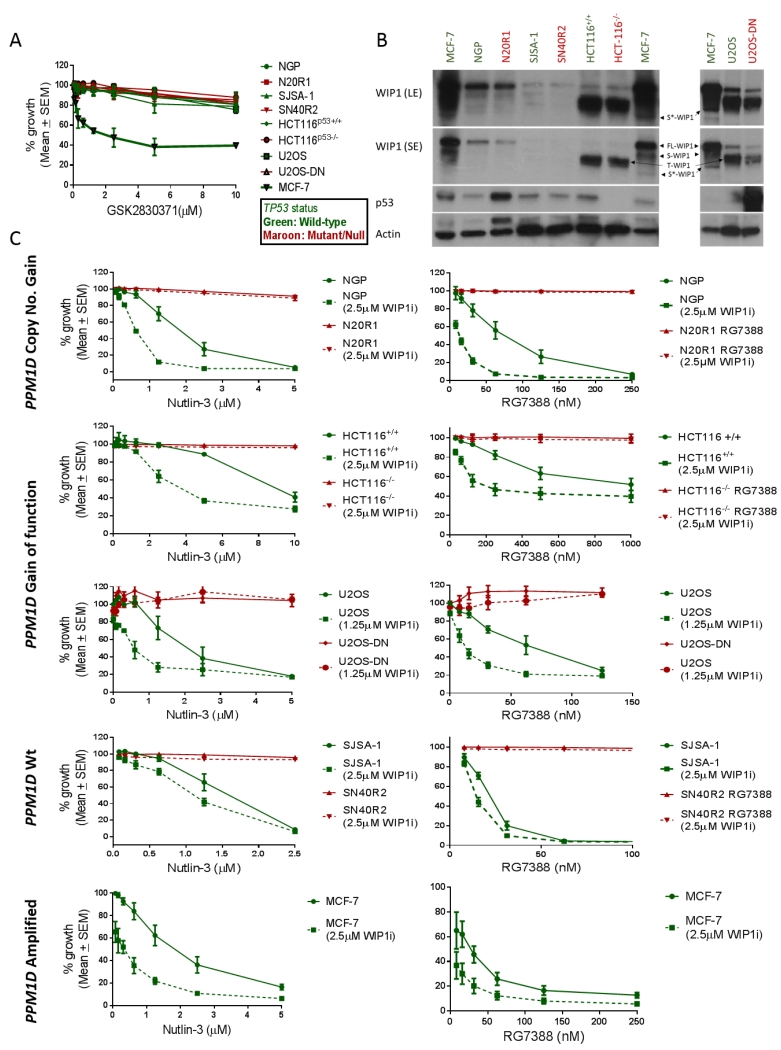
Growth inhibition assays were carried out on a panel of TP53 wild-type and mutant/null cell line pairs differing in their PPM1D genetic status, to assess their sensitivity to the selective allosteric WIP1 inhibitor GSK2830371 and its ability in turn to sensitise cells to MDM2 inhibitors (Figure 1A). GSK2830371 sensitive MCF-7 cells were used as a positive control for the growth inhibitory activity and biochemical effect of this compound. GSK2830371 had a 50% growth inhibitory concentration (GI50) of 2.65μM ± 0.54 (SEM) in MCF-7 cells. The growth inhibition curve for GSK2830371 plateaued in MCF-7 cells at doses 2.5μM-10μM suggesting that a subpopulation of MCF-7 cells are resistant to growth inhibition and apoptosis in response to maximal WIP1 inhibition (Figure 1A). All other cell line pairs were not sensitive to growth inhibition by GSK2830371 alone, with GI50 > 10μM irrespective of their PPM1D or TP53 genetic status (Figure 1A). Basal expression of WIP1 across the panel of cell line pairs was assessed by immunoblotting with an antibody that detects WIP1 (FL-WIP1), its previously described shorter isoform (S-WIP1) (36), and the two WIP1 gain-of-function mutant proteins in HCT116 and U2OS cell line pairs (Figure 1B). Transient knockdown of WIP1 using four different anti-PPM1D siRNA constructs resulted in a reduction in the intensity of all the bands detected by the WIP1 antibody (F-10) which suggests that all of the bands detected in these conditions correspond to WIP1 (Supplementary figure S2A). This includes a further WIP1 isoform detected by F-10 (≈55kDa), which is referred to here as S*-WIP1. PPM1D-amplified MCF-7 cells (37) showed the highest basal expression of WIP1 and its isoforms, consistent with their sensitivity to GSK2830371 single treatment (Figure 1B) (See (38)). NGP cells, with PPM1D copy number gain (21), and its otherwise isogenic TP53 mutant daughter cell line, N20R1, were insensitive to <10μM GSK2830371 despite showing the second highest expression of full-length WIP1 after MCF-7 cells. The SJSA-1 and SN40R2 TP53 wild-type and mutant pair, with wild-type PPM1D, showed the least WIP1 protein expression among all cell line pairs in the panel. Shorter bands corresponding to the previously reported truncated and activating WIP1 mutant proteins WIP1 L450X and WIP1 R458X were detected in lysates derived from HCT116 and U2OS cell line pairs respectively (Figure 1B) (39).
Figure 1.
A) The effect on growth of a panel of p53 wild-type (Green) and Mutant/null (Maroon) cell line pairs with different PPM1D genetic status to 0.08-10μM GSK2830371 exposure for 168hr, using sulforhodamine (SRB) growth inhibition assays. B) Basal expression of WIP1 and p53 in cell lines (SE-short film exposure, LE-long exposure). C) The sensitivity of a panel of p53 Wt (Green) and Mt/null (Maroon) cell line pairs with different PPM1D genetic status to 0.08-10μM Nutlin-3 and 0.008-1μM RG7388 in 168hr SRB growth inhibition assays in the presence and absence of the highest non-growth inhibitory dose of GSK2830371 (2.5μM). The U2OS cell line pair was treated with MDM2 inhibitors ± 1.25μM GSK2830371. WIP1i: GSK2830371; FL-WIP1: Full length WIP1; S-WIP1: WIP1 shorter isoform; T-WIP1: Truncated WIP1 mutants, WIP1 L450X in HCT116 cells or WIP1 R458X in U2OS cells; S*-WIP1 the shortest band detected by the F-10 antibody.
Due to the role of WIP1 in homeostatic feedback regulation of the p53 network we aimed to assess whether GSK2830371 (WIP1i) can potentiate the response to MDM2 inhibitors. Treatment with a combination of the highest non-growth inhibitory dose of GSK2830371 (2.5μM), potentiated the response to MDM2 inhibitors Nutlin-3 and RG7388 in a p53-dependent manner in cell lines that were not sensitive to growth inhibition by GSK2830371 alone (Figure 1C). TP53 wild-type parental cell lines HCT116+/+, NGP and SJSA-1 showed a 2.4-fold (p=0.007), 2.1-fold (p=0.039) and 1.3-fold (p =0.017) decrease respectively in their Nutlin-3 GI50 values in the presence of 2.5μM GSK2830371. In contrast Nutlin-3 GI50 did not change for theirTP53 Null/Mutant matched pairs HCT116−/−, N20R1 and SN40R2. However, pertinent to the widening of RG7388 therapeutic index in the clinic, the same dose of GSK2830371 resulted in a much greater potentiation of RG7388 in TP53 wild-type cell lines with either PPM1D gain-of-function or copy number gain: NGP 5.8-fold (p=0.049), and HCT116+/+ 4.8-fold (p=0.018) compared to PPM1D wild-type SJSA-1 cells 1.4-fold (p=0.020) (Figure 1C). U2OS TP53 wild-type cells showed a similar trend towards potentiation of Nutlin-3 in combination with GSK2830371 at 1.25μM as Nutlin-3 GI50 was reduced by 3.2-fold (p=0.08), however the same dose of the WIP1 inhibitor resulted in a 5.3-fold (p=0.039) decrease in RG7388 GI50. None of the TP53 mutant daughter cell lines showed increased sensitivity to RG7388 in the presence of the WIP1 inhibitor. Interestingly, the combination of Nutlin-3 or RG7388 with 2.5μM GSK2830371 also augmented the growth inhibitory effect in MCF-7 cells compared to each drug alone. Therefore, the most marked fold-change in sensitivity to both MDM2 inhibitors was observed in TP53 wild-type cell lines that have increased WIP1 expression or activity.
Inhibition of WIP1 catalytic activity by GSK2830371 is separable from its induction of ubiquitin-mediated WIP1 degradation
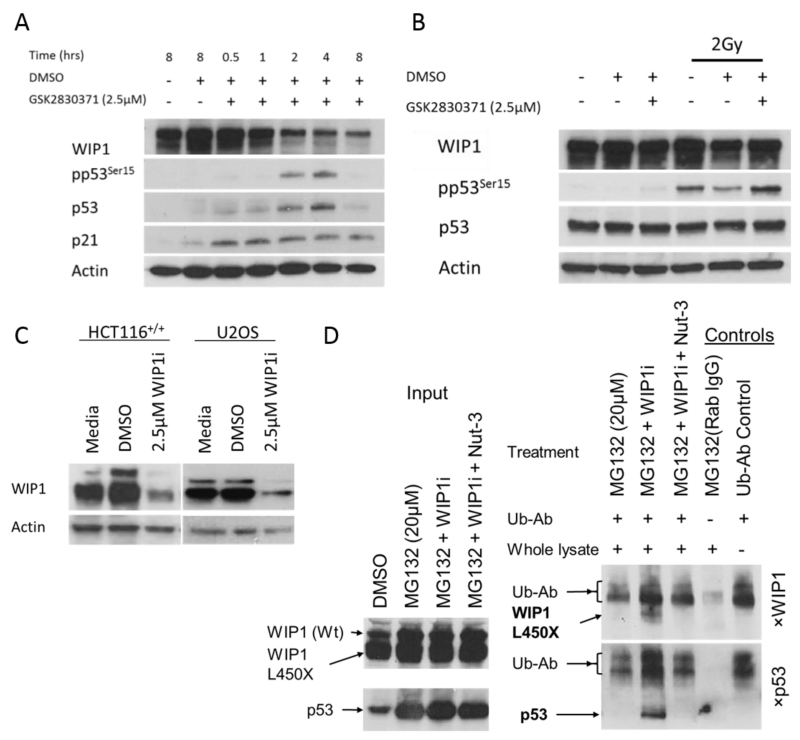
Treatment of MCF-7 cells with 2.5μM GSK2830371 resulted in marked time-dependent degradation of both isoforms of WIP1 over 8 hours which correlated with p53 stabilisation and phospho-p53Ser15 (pp53Ser15) consistent with previous reports by Gilmartin et al., (Figure 2A) (16). Quantification of WIP1 signal intensity is presented in Supplementary figure S2B.
Figure 2.
A) GSK2830371 (2.5μM) treatment of MCF-7 cells over 8 hours shows WIP1 degradation over time, p53 stabilisation and Phospho-p53Ser15 (pp53Ser15) accumulation. B) p53Ser15 phosphorylation in MCF-7 cells 30min post 2Gy ionising radiation in the presence or absence of 2.5μM. GSK2831371 inhibits WIP1 catalytic activity independent of WIP1 protein levels. C) 4 hours treatment with 2.5μM GSK2830371 results in degradation of full-length and truncated WIP1 in HCT116+/+ and U2OS cells. D) Lysates obtained from HCT116+/+ cells treated with 20μM proteasome inhibitor MG132 and 2.5μM GSK2830371 ± 3.0μM Nutlin-3 overnight underwent immunoprecipitation (IP) with anti-ubiquitin antibody (Ub-Ab) and the precipitates probed for WIP1 and p53 by western blot. Input samples (left panel) are western blots of total lysate before IP for comparison with the IP results on the right hand panel. An aliquot of MG132 treated lysate was precipitated with Rabbit IgG instead of Ub-Ab as a negative control. WIP1i: GSK2830371; Ub-Ab: anti-ubiquitin antibody; WIP1 (Wt): Wild-type WIP1, WIP1 L450X: Truncated WIP1.
To test the effect of the GSK2830371 inhibitor on WIP1 phosphatase activity separate from degradation of WIP1, its effect on the phosphorylation of p53Ser15 30 min following exposure of MCF7 cells to ionising radiation was assessed. GSK2830371 was seen to inhibit pp53Ser15 dephosphorylation at a time point when the WIP1 protein expression had not yet been affected by this compound (Compare pp53Ser15 on the last two tracks in Figure 2B). These data show that inhibition of the catalytic activity of WIP1 by GSK2830371 is separable from its ubiquitin-mediated degradation.
It was noteworthy that GSK2830371 also lead to the degradation of truncated WIP1 mutants within 4 hours (Figure 2C). We carried out a denaturing immunoprecipitation (IP) experiment probing for all ubiquitinated species in HCT116+/+ cells treated with the proteasome inhibitor MG132 and either GSK2830371 alone or in combination with Nutlin-3, to assess whether wild-type WIP1 and WIP1 L450X are both degraded by ubiquitin mediated processes (Figure 2C). The anti-ubiquitin antibody (Ub-Ab) migrated to a similar molecular weight as full-length WIP1 and it was detected by the goat anti mouse antibody (Figure 2D, last lane Ub-Ab Control); therefore, the ubiquitination of full-length WIP1 could not be determined. However, ubiquitinated WIP1 L450X was observed to be increased by GSK2830371 (Figure 2D). Interestingly this ubiquitination event was also reduced in the presence of Nutlin-3. Increased ubiquitination of p53 in the presence of MG132 + GSK2830371 was reversed by Nutlin-3 as expected because inhibiting MDM2-p53 interaction prevents MDM2 mediated p53-ubiquitination.
GSK2830371 significantly increases MDM2 inhibitor mediated apoptosis and reduces clonogenic cell survival in TP53 wild-type cell lines
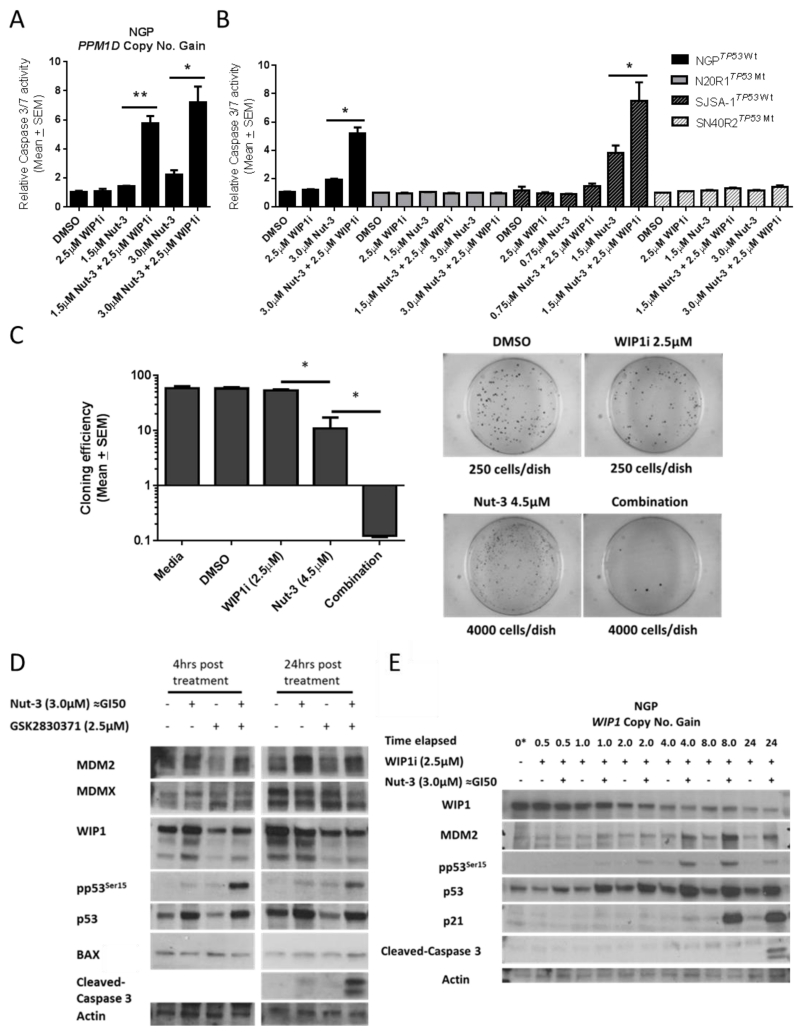
The combination of GSK2830371 and multiples of Nutlin-3 GI50 dose resulted in a marked increase in caspase-3/7 activity in both NGP and SJSA-1 cells compared to treatment with either drug alone (Figure 3A & 3B). For NGP cells, 24 hour treatment with 2.5μM GSK2830371 did not lead to detectable caspase-3/7 activity, whereas Nutlin-3 at 0.5 × and 1 × GI50 resulted in a dose-dependent increase in caspase 3/7 signal which was significantly enhanced (≈4-fold p=0.005 and ≈3-fold p=0.02 respectively) in the presence of 2.5μM GSK2830371 (Figure 3A). No increased caspase-3/7 activity was observed in SJSA-1 cells after 24 hours of exposure to 2.5μM GSK2830371 alone or Nutlin-3 ± 2.5μM GSK2830371 (data not shown). Similarly in both cell line pairs, 48 hours treatment with 2.5μM GSK2830371 alone resulted in no increased caspase-3/7 activity, whereas its presence significantly increased response to Nutlin-3 (≈2.7-fold p=0.01 in NGP, ≈2- fold p=0.04 in SJSA-1) in a p53-dependent manner (Figure 3B).
Figure 3.
A) Dose-dependent increased in caspase 3/7 activity of NGP cells after 24 hours treatment with Nutlin-3 (Nut-3 GI50 ≈ 3.0μM) alone or in combination with 2.5μM GSK2830371. B) Increase in caspase 3/7 activity in NGP and SJSA-1 cells and their TP53 mutant daughter cell lines 48 hours after treatment with Nutlin-3 ± 2.5μM GSK2830371. C) Reduction in clonogenic efficiency of HCT116+/+ cells following exposure to 0.5 × GI50 concentration of Nut-3 in the presence of 2.5μM GSK2830371 compared to either inhibitor alone over 10 days. D) Immunoblot of NGP cells showing Nut-3 dependent phosphorylation of p53 at Ser15 is markedly enhanced by GSK2830371 at 4 & 24hrs exposure and leads to increased caspase 3 cleavage at 48 hours. E) Time-course of NGP response to 3.0μM Nutlin-3 (≈GI50) ± 2.5μM GSK2830371 over 24 hours. WIP1i: GSK2830371; *: p ≤0.05; **: p≤0.005
Caspase-3/7 activity could not be detected in HCT116+/+ and for up to 48 hours following treatment (data not shown), so continuous exposure cloning efficiency experiments were carried out as described in Figure 3C (see caption) in order to assess clonogenic cell survival. There was no reduction in clonogenic efficiency in the presence of the GSK2830371 alone in comparison with untreated controls. Cloning efficiency of HCT116+/+ cells in the presence of 0.5 × Nutlin-3 GI50 significantly decreased (p=0.008) when GSK2830371 was present at 2.5μM (Figure 3C).
In NGP cells pp53Ser15 was not affected by the GI50 dose of Nutlin-3 or 2.5μM GSK2830371 alone, whereas in combination there was a marked increase in pp53Ser15 at 4 hours which persisted through to 24 hours and correlated with the detection of cleaved caspase-3 (Figure 3D & 3E). GSK2830371 alone resulted in modest p53 stabilisation in NGP cells, after 24 hours treatment, which did not result in detectable induction of p53 direct transcriptional targets p21WAF1 and MDM2 (Figure 3E). Interestingly, monotreatment with the same dose of GSK2830371 in MCF-7 cells was sufficient for WIP1 degradation, p53 stabilisation and increase pp53Ser15 in contrast to NGP cells (Figure 2A v Figure 3E). Consistently, WIP1 was also degraded in NGP cells in the presence of the WIP1 inhibitor (track 3 v’s track 1 in Figure 3D) even when WIP1 was induced by Nutlin-3 (track 4 v’s track 2 in Figure 3D). The lack of a p53 response of NGP cells to 2.5μM GSK2830371 may explain their insensitivtiy to GSK2830371 monotreatment compared to MCF-7 cells. Also the addition of 2.5μM GSK283037 did not affect MDM2 induction by Nutlin-3 (Figure 3D). This suggests that the reported role of WIP1 in downregulation of MDM2 (14) may be counterbalanced by the p53-dependent transcriptional induction of MDM2 in the presence of Nutlin-3. There was a reduction in MDMX expression 24 hours after combination treatment compared to Nutlin-3 treatment alone (Figure 3D). Given that MDMX increased expression has been proposed to contribute to reduced sensitivity to MDM2 inhibitors (40) it is likely that the role of WIP1 in negative regulation of MDMX stability (41) may be a factor in its ability to potentiate MDM2 inhibitors in MDMX overexpressing NGP cells. There was no change the expression of the p53 proapoptotic transcriptional target BAX at 4 and 24 hours following combination treatment compared to Nutlin-3 or WIP1 inhibitor monotreatments (Figure 3D). This suggests that BAX is likely not involved in potentiation of Nutlin-3 by the WIP1 inhibitors.
GSK2830371 increases RG7388 induced p53-dependent transcription of growth inhibitory and pro-apoptotic genes
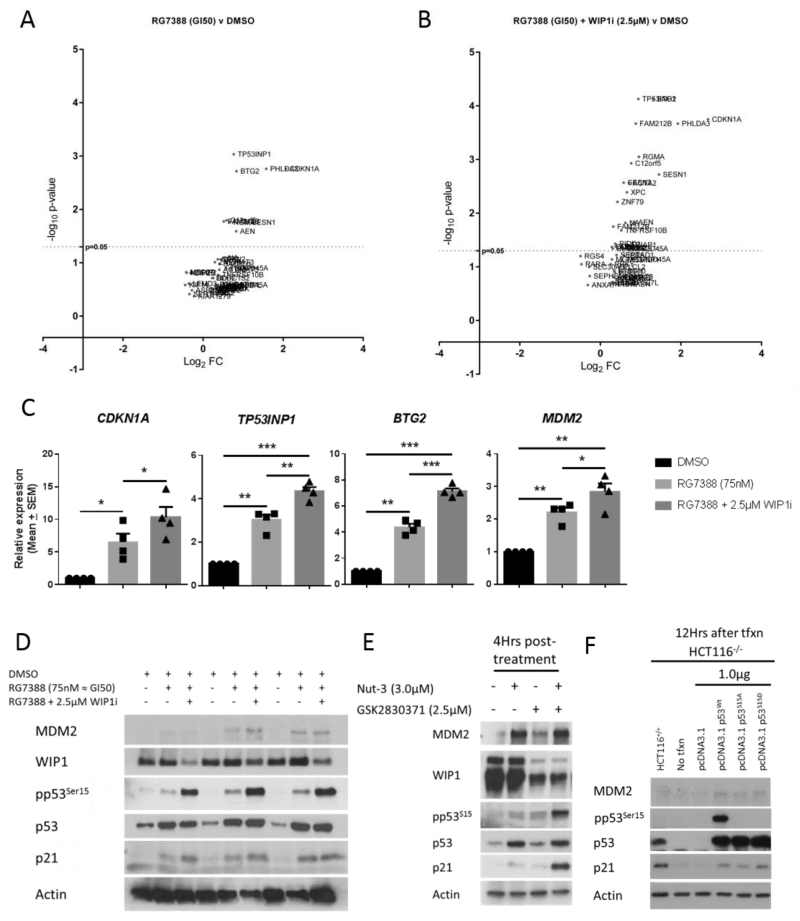
Phosphorylation of p53Ser15 has been reported to increase p53-mediated transcriptional transactivation but not to be necessary for dissociation of p53 from MDM2 in response to DNA damage (24, 26). Also reports in the literature have suggested that p53 post-translational modifications can behave as variable barcodes and induce transcription of alternate sets of p53 target genes which could lead to different cell fates after p53 activation (42). As the greatest potentiation of MDM2 inhibitors by GSK2830371 was observed in NGP cells (5.8-fold), we assessed whether the subset of early genes activated in response to RG7388 in this cell line differed in the presence of 2.5μM GSK2830371 which produced a marked increase in pp53Ser15. Peak p53 transcriptional target expression (e.g. p21WAF1 and MDM2) as detected by western blotting is reached by 6-8 hours after treatment (Figure 3E). Because later changes in transcription may be secondary effects and not directly p53-dependent, we assessed changes in global gene expression 4 hours following RG7388 (GI50 ≈ 75nM) ± 2.5μM GSK2830371 using the Illumina BeadChip expression array platform. A testament to the specificity of RG7388 in exclusively activating p53, 4 hours of exposure to a GI50 dose of RG7388 led to significantly increased mRNA expression of only 9 genes, all which were known p53 transcriptional targets (Figure 4A). The top 41 genes just below the statistical significance cut-off point (p>0.05 after correction for multiple testing) were also mostly genes that are well-established to be direct p53 transcriptional targets. Interestingly, in the presence of 2.5μM GSK2830371 the subset of statistically significant RG7388 mediated transcriptional changes increased from 9 to 24 genes, indicating that inhibition of WIP1 results in a significant increase of additional p53-mediated transcriptional activity at this early time point (Figure 4B) (For a list of genes refer to Supplementary table S1A and Supplementary table S1B).
Figure 4.
A & B) Volcano plots showing the significantly induced genes by RG7388 treatment of NGP cells for 4hrs in the presence or absence of 2.5μM GSK2830371 (RG7388 v DMSO and RG7388 + WIP1 (2.5μM) v DMSO). The results represent the mean of 4 independent biological repeats. y–axis: −Log10 of the p-value adjusted for false discovery rate using the Benjamini-Hochberg procedure; x-axis Log2 FC: Log2 fold-change in normalised mRNA expression. For a list of genes induced see Supplementary table S1A and Supplementary table S1B. Microarray data have been deposited in the NCBI GEO databank (Accession number: GSE75197). C) qRT-PCR validation of p53 target genes implicated in increased sensitivity to MDM2 inhibitors:TNFRSF10B, AEN , XPC and CDKN1A using the same RNA samples as used for the microarray analysis. D) Western blot for NGP cells showing changes in p53, pp53Ser15, MDM2, WIP1 and p21WAF1 protein expression in response to 75nM RG7388 (GI50) ± 2.5μM GSK2830371. Lysates were obtained from samples treated in parallel to the microarray experiment and represent the first 3 independent repeats. E) Western blot of HCT116+/+ cells 4 hours following treatment with the stated doses of Nutlin-3, GSK2830371 or their combination. F) Western blot showing changes in pp53Ser15, MDM2, and p21WAF1 detection 12 hours after ectopic expression of wild-type p53 (Wt), p53 mutants Ser15Ala (S15A) and Ser15Asp (S15D) in HCT116−/− cells. WIP1i: GSK2830371; *: p ≤0.05; **: p≤0.005; ***: p≤0.0005
To validate the results of the array we assessed the expression of the top three genes with the highest odds ratio difference between single and combination treatments (CDKN1A (p21WAF1), TP53INP1, and BTG2) and one of the genes that was exclusively induced in the combination treatment (MDM2) by qRT-PCR using the same mRNA samples used in the expression array experiment (Figure 4C). Consistent with the array data, all the genes tested showed significant increase in their mRNA expression in combination treatment compared to the RG7388 alone (Figure 4C). TP53INP1/P53DINP1 is a known pro-apoptotic p53 transcriptional target gene, the overexpression or induction of which following cellular stress has been associated with increased p53-mediated apoptosis (43). Among the 16 additional p53 regulated target genes induced exclusively in response to the combination treatment were tumour necrosis factor super family member 10B (TNFRSF10B) and p53-induced death domain protein 1 (PIDD1), two genes critical for extrinsic and intrinsic pro-apoptotic pathways respectively (44-47). Interestingly, despite neither of the agents being genotoxic, one of the other genes that showed differential expression in response to the combination treatment was the DNA base excision repair gene xeroderma pigmentosum complementation group C (XPC), the increased basal expression of which has been reported to correlate with increased sensitivity to MDM2 inhibitors in a large panel of TP53 wild-type cell lines and predict a better clinical response to RG7112 and RG7388 in acute myeloid leukaemia patients (13).
The increase in the subset of genes expressed correlated with a marked increase in the proportion of pp53Ser15 to total p53 in immunoblots of lysates prepared in parallel to the mRNA samples used in the expression array experiment (Figure 4D & see Supplementary figure S3A). These data suggest that the underlying mechanism for the observed potentiation of MDM2 inhibitors in combination with WIP1 phosphatase inhibition may be contributed to by the increased p53Ser15 phosphorylation which enhances p53-dependent pro-apoptotic gene transcription.
In line with this hypothesis, increased pp53Ser15 phosphorylation in HCT116+/+ following combination treatment resulted in an in increase in the p21WAF1 product of the CDKN1A gene in comparison to monotreatment with either drug (Figure 4E). In order to confirm earlier findings showing that phosphorylation of p53Ser15 increases p53-mediated transcription in HCT116 cells, we overexpressed wild-type (Wt) p53, mutant p53S15A or p53S15D in HCT116−/− cells and assessed p53-mediated expression of p21WAF1 protein encoded by the CDKN1A gene, which had showed the biggest fold-change in expression on the array following the combination treatment in NGP cells. Consistent with previous findings (24, 26) Wt p53 and phospho-mimetic p53S15D mutant constructs increased p53-mediated expression of p21WAF1 and MDM2 following transfection compared to the p53S15A mutant which could not be phosphorylated on that residue (Figure 4F). See Supplementary figure S3B & S3C for repeat and densitometry data.
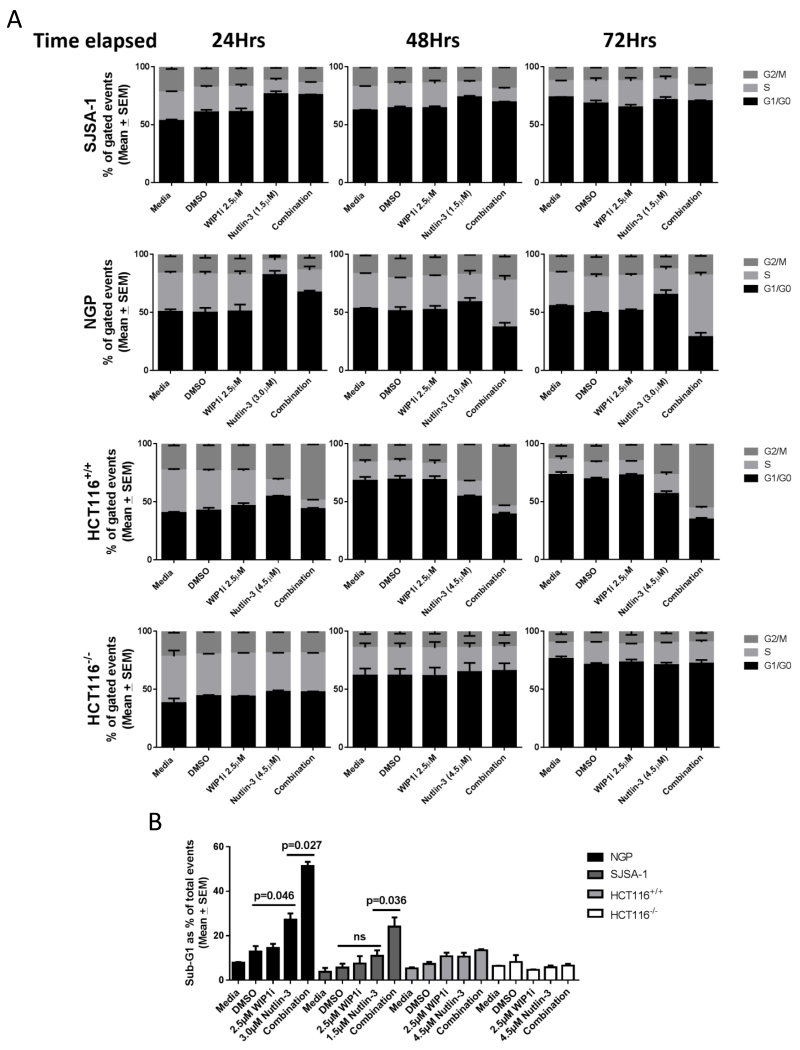
The effect of combined MDM2 and WIP1 inhibition on cell cycle distribution
Given that WIP1 inhibition potentiated the growth inhibitory and apoptotic response of TP53 wild-type cell lines to MDM2 inhibitors, and the highest fold increase in transcription was of the CDKN1A (p21WAF1) cyclin-dependent kinase inhibitor gene, we investigated changes in cell cycle distribution following this combination treatment. In all cell lines, 2.5μM GSK2830371 alone did not significantly affect cell cycle distribution throughout 72 hours of treatment (Figure 5A & Supplementary figure S4). Changes in cell cycle distribution after exposure to Nutlin-3 ± 2.5μM GSK2830371 were cell line-dependent. In SJSA-1 and NGP cell lines, 24 hours exposure to Nutlin-3 resulted in an increase in the proportion of cells in G1/G0 phases of the cell cycle. In SJSA-1 cells this effect of Nutlin-3 remained unchanged in the following 48 hours treatment with Nutlin-3 ± GSK2830371. However, in NGP cells the relative proportion of cells in G2/M and S-phase increased over the following 48 hours when Nutlin-3 and the WIP1 inhibitor were combined compared to Nutlin-3 alone. In HCT116+/+ cells Nutlin-3 resulted in an increase in the proportion of cells in G0/G1 and G2/M phases at 24 hours, which persisted to the 72 hours treatment time point (Figure 5A), consistent with the increase in CDKN1A (p21WAF1) expression in response to the combination treatment (Figure 4E). Cell cycle distribution was not affected in HCT116−/− cells regardless of the treatment condition, suggesting that the changes in cell cycle distribution observed in HCT116+/+ cells are p53-dependent (Figure 5A).
Figure 5.
A) Time-course of cell cycle distribution changes over 72Hrs of treatment using FACS analysis. B) % of Sub-G1 signals for NGP, SJSA-1, HCT116+/+ and HCT116−/− cells in response to the stated treatments at 72 hours. WIP1i: GSK2830371
Sub-G1 changes
In response to the combination treatment compared to Nutlin-3 alone, the increase in Sub-G1 FACS signal after exposure to Nutlin-3 was significantly augmented in the presence of 2.5μM GSK2830371 (WIP1i) in both SJSA-1 and NGP cell lines (Figure 5B and Supplementary figure S4B). This is in keeping with the increased cleaved caspase-3/7 activity in NGP and SJSA-1 cells (Figure 3A & 3B). Sub-G1 signals were not significantly changed in HCT116+/+cells throughout the 72 hours of Nutlin-3 ± GSK2830371 treatment (Figure 5B).
Discussion
Although mutant TP53 status is a dominant mechanism of resistance to MDM2-p53 binding antagonists, there is nevertheless a clinically relevant wide range of sensitivity to MDM2 inhibitors among TP53 wild-type cancer cell lines. Importantly, this variation of response is not exclusive to one class of MDM2 inhibitors and is clearly seen in panels of cell lines with validated wild-type TP53 status. Amgen have recently reported a wide range of sensitivity to their MDM2 inhibitor AMGMDS3 among their carefully curated panel of TP53 wild-type and functional cell lines showing a 500-fold GI50 difference between the least to most sensitive (8). These observations suggest that there are a diverse set of underlying genetic variables that determine cell fate following a dose of activated/stabilised p53. Here we have shown a non-growth inhibitory dose of the selective orally bioavailable allosteric WIP1 phosphatase inhibitor, GSK2830371, can modulate the phosphorylation state of p53 and potentiate both the growth inhibitory and apoptotic response to MDM2 inhibitors in TP53 wild-type cell lines, especially those with increased WIP1 expression or activity. MDM2 inhibitor potentiation was at its greatest when the cell line harboured either PPM1D copy number gain/elevated expression or gain-of-function truncating mutations, thus providing a rationale for specific combination treatment targeting of tumours with this genotype.
The argument in support of p53Ser15 phosphorylation increasing p53-mediated transcription (24, 26, 27) is compelling and consistent with our findings. In contrast to what was originally reported by Vassilev et al., in 2004, phosphorylation of p53 following treatment with MDM2 inhibitors is observed, however it is not as intense and immediate compared to p53Ser15 phosphorylation following DNA damaging agents of equivalent growth inhibitory dose (7, 26). As shown by Loughery et al., the basal activity of kinases involved in phosphorylation of p53Ser15 (e.g. ATM and ATR) in response to DNA damage are also likely responsible for, or contribute to, this post-translational modification in response to MDM2 inhibitors (26). Our findings have shown that, in the presence of a selective WIP1 inhibitor, the minimal phosphorylation of p53Ser15 in response to MDM2 inhibitors is markedly accentuated, which correlates with potentiation of apoptotic and growth inhibitory response to MDM2 inhibitors in TP53 wild-type cells, particularly in those with high WIP1 expression or activity. This was also associated with significantly increased transcript levels from an increased number of immediate p53 transcriptional target genes as compared to those induced by single agent RG7388. Increased p53Ser15 phosphorylation in response to the combination treatment also resulted in increased p21WAF1 and MDM2 protein expression. Consistent with the reported role of p53Ser15 phosphorylation in increasing transcriptional activity of p53 we also confirmed that mutation of this residue influenced expression of p21WAF1 and MDM2 proteins. Thus our current working model includes evidence for the role of enhanced p53 transcriptional activity in response to the combination of MDM2 inhibitors and GSK2830371 (Figure 6). It is likely however, that direct and or indirect WIP1 mediated post-translational modifications that effect the stability and function of stress response proteins and their cross-talk with the p53 network (As reviewed in (15)) may also contribute to MDM2 inhibitor potentiation in the presence of GSK2830371. Regardless of this, our data strongly suggest that increased phosphorylated p53Ser15 can be considered a surrogate marker of p53 dissociation from MDM2 in response to single agent GSK2830371 treatment or its combination with MDM2 inhibitors, since this modification coincides with p53 transcriptional activation that precedes the subsequent enhanced p53-mediated growth inhibitory or apoptotic response to each of these treatments.
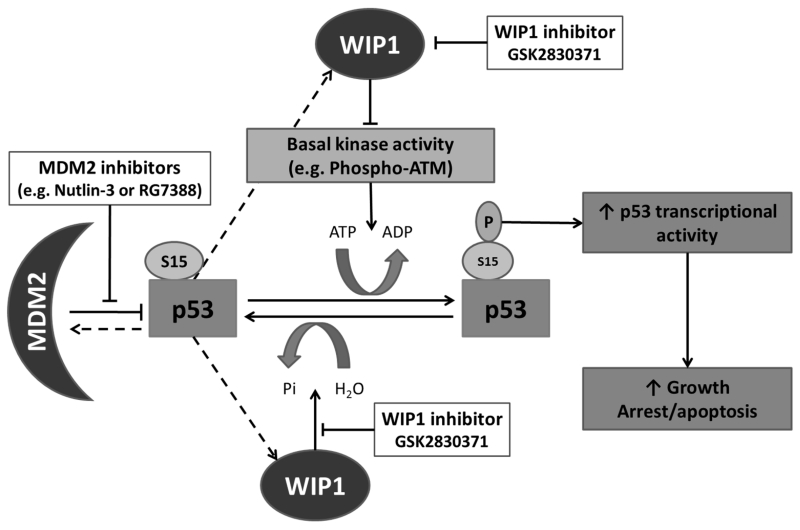
Figure 6.
Proposed model for potentiation of cellular response to MDM2 inhibitors by the selective allosteric WIP1 inhibitor GSK2830371. After activation of p53 by MDM2 inhibitors, p53Ser15 is unmasked and therefore available as a substrate for the basal level activity of multiple kinases and phosphatases normally involved in post-translational modification of this residue in response to cellular stress. In normal homeostatic control, phosphorylation of p53Ser15 is kept in check by an equilibrium between the kinase and phosphatase activities. Inhibition of WIP1 by GSK2830371 tilts this balance in favour of the activating kinases which in turn increases p53 transcriptional activity and is enhanced in combination with MDM2 inhibitors. Dashed lines indicate direct p53 transcriptional upregulation of the corresponding genes for MDM2 and WIP1.
Nutlin-3 mediated changes in cell cycle distribution were all enhanced in the presence of a dose of GSK2830371 which on its own did not affect cell cycle distribution. The observed increase in Sub-G1 FACS analysis signals with combination treatment of NGP cells is consistent with potentiation of apoptosis and growth inhibition in this cell line. Enhancement of Nutlin-3 mediated G1:S and G2:S ratios in the presence of GSK2830371 are also consistent with the increased CDKN1A (p21WAF1) expression observed at both transcript and protein levels and its importance in negative regulation of cell cycle progression (48, 49). Kleiblova et al., (39) had previously shown that transient knockdown of truncated PPM1D increases G1 checkpoint in response to ionising radiation (IR). Interestingly, in our current study WIP1 inhibition and depletion by GSK2830371 in HCT116+/+ cells harbouring a PPM1D L450X truncation mutation also resulted in an increase in p53-dependent G1 arrest following p53 activation by Nutlin-3, while this did not occur in NGP and SJSA-1 cell lines that do not have PPM1D gain of function mutations. Lindqvist A. et al., 2009 (50) also reported that WIP1 knockdown ablates the competence of cellular p53-dependent G2 checkpoint recovery following cellular stress, although the authors were not aware of the gain of function WIP1 R458X mutation in U2OS cells used in their study, as it had not yet been reported. These findings suggest that the increase in G2:S ratio observed in HCT116+/+ cells treated with the combination of MDM2 inhibitors and GSK2830371 is likely due to inhibition of WIP1 L450X which would otherwise be negatively regulating p53 transcriptional activity in these cell lines. Of note we have also shown that GSK2830371 increases the ubiquitin mediated degradation of truncated WIP1 as postulated by Gilmartin et al., (16). Interestingly, this increase in the ubiquitination of truncated WIP1 was reversed by inhibition of MDM2 which suggests that MDM2 is directly or indirectly involved in WIP1 ubiquitination following GSK2830371.
Wild-type TP53 genetic status is the most important determinant of response to MDM2 inhibitors, while being necessary but not sufficient for growth inhibitory response to WIP1 inhibition by GSK2830371. Following their promising clinical outcomes so far, MDM2 inhibitors will be explored in combination with other anticancer agents to optimise their therapeutic potential. Combination regimens of these non-genotoxic agents could minimise DNA damage to healthy tissue which does not express altered forms of PPM1D. Here we have shown that specific pharmacological inhibition of WIP1 combined with MDM2 inhibitors is a promising therapeutic strategy in TP53 wild-type tumours that show increased WIP1 function, and that phosphorylated p53Ser15 and PPM1D genotype are important both mechanistically and as predictive biomarkers for response to this combination treatment.
Supplementary Material
Acknowledgements
We would like to thank Professor Nicola Curtin for providing the U2OS cell line pair. We would also like to acknowledge Mrs Mahsa Azizyan, Drs Olivier Binda and Richard Heath for their provision of reagents and advice on experimental procedures. The encouragement and provision of RG7388 by Professor Newell and the NICR Drug Discovery team are gratefully acknowledged.
Financial support:
J. Lunec: Cancer Research UK (C240/A15751)
A. Esfandiari: Newcastle University & Northern Institute for Cancer Research (C0190R4011)
S. Nakjang: Newcastle University (C0190R4011)
T. Hawthorne: Newcastle University (C0190R4011)
Footnotes
Conflict of interest: The authors disclose no potential conflicts of interest.
References
- 1.Levine AJ, Momand J, Finlay CA. THE P53 TUMOR SUPPRESSOR GENE. Nature. 1991;351:453–6. doi: 10.1038/351453a0. [DOI] [PubMed] [Google Scholar]
- 2.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 3.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–73. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Y, Aguilar A, Bernard D, Wang S. Small-Molecule Inhibitors of the MDM2–p53 Protein–Protein Interaction (MDM2 Inhibitors) in Clinical Trials for Cancer Treatment. Journal of Medicinal Chemistry. 2015;58:1038–52. doi: 10.1021/jm501092z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q, Lozano G. Molecular pathways: targeting Mdm2 and Mdm4 in cancer therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:34–41. doi: 10.1158/1078-0432.CCR-12-0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meek DW, Hupp TR. The regulation of MDM2 by multisite phosphorylation—Opportunities for molecular-based intervention to target tumours? Seminars in Cancer Biology. 2010;20:19–28. doi: 10.1016/j.semcancer.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–8. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 8.Saiki AY, Caenepeel S, Cosgrove E, Su C, Boedigheimer M, Oliner JD. Identifying the determinants of response to MDM2 inhibition. Oncotarget. 2015;6:7701–12. doi: 10.18632/oncotarget.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–5. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–307. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Z, Shen J, He J, Hu C. Combination therapy with p53–MDM2 binding inhibitors for malignancies. Med Chem Res. 2015;24:1369–79. [Google Scholar]
- 12.Sullivan KD, Padilla-Just N, Henry RE, Porter CC, Kim J, Tentler JJ, et al. ATM and MET kinases are synthetic lethal with non-genotoxic activation of p53. Nature chemical biology. 2012;8:646–54. doi: 10.1038/nchembio.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong H, Chen G, Jukofsky L, Geho D, Han SW, Birzele F, et al. MDM2 antagonist clinical response association with a gene expression signature in acute myeloid leukaemia. British Journal of Haematology. 2015 doi: 10.1111/bjh.13411. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X, Ma O, Nguyen T-A, Joness SN, Oren M, Donehower LA. The Wip1 phosphatase acts as a gatekeeper in the p53-Mdm2 autoregulatory loop. Cancer Cell. 2007;12:342–54. doi: 10.1016/j.ccr.2007.08.033. [DOI] [PubMed] [Google Scholar]
- 15.Lowe J, Cha H, Lee M-O, Mazur SJ, Appella E, Fornace AJ. Regulation of the Wip1 phosphatase and its effects on the stress response. Frontiers in bioscience : a journal and virtual library. 2012;17:1480–98. doi: 10.2741/3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilmartin AG, Faitg TH, Richter M, Groy A, Seefeld MA, Darcy MG, et al. Allosteric Wip1 phosphatase inhibition through flap-subdomain interaction. Nat Chem Biol. 2014;10:181–7. doi: 10.1038/nchembio.1427. [DOI] [PubMed] [Google Scholar]
- 17.Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, et al. Amplification of PPM1D in human tumors abrogates p53 tumor-suppressor activity. Nature Genet. 2002;31:210–5. doi: 10.1038/ng894. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Chen LH, Wan H, Yang R, Wang Z, Feng J, et al. Exome sequencing identifies somatic gain-of-function PPM1D mutations in brainstem gliomas. Nat Genet. 2014;46:726–30. doi: 10.1038/ng.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–10. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vintonyak VV, Antonchick AP, Rauh D, Waldmann H. The therapeutic potential of phosphatase inhibitors. Current Opinion in Chemical Biology. 2009;13:272–83. doi: 10.1016/j.cbpa.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Richter M, Dayaram T, Gilmartin AG, Ganji G, Pemmasani SK, Van Der Key H, et al. WIP1 Phosphatase as a Potential Therapeutic Target in Neuroblastoma. PLoS One. 2015;10:e0115635. doi: 10.1371/journal.pone.0115635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Böttger A, Böttger V, Garcia-Echeverria C, Chène P, Hochkeppel H-K, Sampson W, et al. Molecular characterization of the hdm2-p53 interaction. Journal of Molecular Biology. 1997;269:744–56. doi: 10.1006/jmbi.1997.1078. [DOI] [PubMed] [Google Scholar]
- 23.Lu X, Nannenga B, Donehower LA. PPM1D dephosphorylates Chk1 and p53 and abrogates cell cycle checkpoints. Genes & Development. 2005;19:1162–74. doi: 10.1101/gad.1291305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. The EMBO Journal. 1999;18:7002–10. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chehab NH, Malikzay A, Stavridi ES, Halazonetis TD. Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proceedings of the National Academy of Sciences. 1999;96:13777–82. doi: 10.1073/pnas.96.24.13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loughery J, Cox M, Smith LM, Meek DW. Critical role for p53-serine 15 phosphorylation in stimulating transactivation at p53-responsive promoters. Nucleic Acids Research. 2014;42:7666–80. doi: 10.1093/nar/gku501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unger T, Sionov RV, Moallem E, Yee CL, Howley PM, Oren M, et al. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene. 1999;18:3205–12. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Rousseau RF, Middleton SA, Nichols GL, Newell DR, Lunec J, et al. Pre-clinical evaluation of the MDM2-p53 antagonist RG7388 alone and in combination with chemotherapy in neuroblastoma. Oncotarget. 2015;6:10207–21. doi: 10.18632/oncotarget.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Allan LA, Fried M. p53-dependent apoptosis or growth arrest induced by different forms of radiation in U2OS cells: p21WAF1/CIP1 repression in UV induced apoptosis. Oncogene. 1999;18:5403–12. doi: 10.1038/sj.onc.1202931. [DOI] [PubMed] [Google Scholar]
- 30.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, et al. New Colorimetric Cytotoxicity Assay for Anticancer-Drug Screening. Journal of the National Cancer Institute. 1990;82:1107–12. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 31.Chen L, Zhao Y, Halliday GC, Berry P, Rousseau RF, Middleton SA, et al. Structurally diverse MDM2-p53 antagonists act as modulators of MDR-1 function in neuroblastoma. Br J Cancer. 2014;111:716–25. doi: 10.1038/bjc.2014.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gozani O. Site Directed Mutagenesis with Stratagene Pfu Turbo. Available from: http://web.stanford.edu/group/gozani/cgi-bin/gozanilab/wp-content/uploads/2014/01/Site-Directed-Mutagenesis-with-Stratagene-Pfu-Turbo.pdf.
- 34.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–8. doi: 10.1093/bioinformatics/btn224. (Oxford, England) [DOI] [PubMed] [Google Scholar]
- 35.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chuman Y, Kurihashi W, Mizukami Y, Nashimoto T, Yagi H, Sakaguchi K. PPM1D430, a Novel Alternative Splicing Variant of the Human PPM1D, can Dephosphorylate p53 and Exhibits Specific Tissue Expression. Journal of Biochemistry. 2009;145:1–12. doi: 10.1093/jb/mvn135. [DOI] [PubMed] [Google Scholar]
- 37.Castellino R, De Bortoli M, Lu X, Moon S-H, Nguyen T-A, Shepard M, et al. Medulloblastomas overexpress the p53-inactivating oncogene WIP1/PPM1D. J Neurooncol. 2008;86:245–56. doi: 10.1007/s11060-007-9470-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KCQ, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–4. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 39.Kleiblova P, Shaltiel IA, Benada J, Ševčík J, Pecháčková S, Pohlreich P, et al. Gain-of-function mutations of PPM1D/Wip1 impair the p53-dependent G1 checkpoint. The Journal of Cell Biology. 2013;201:511–21. doi: 10.1083/jcb.201210031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marine J-CW, Dyer MA, Jochemsen AG. MDMX: from bench to bedside. Journal of Cell Science. 2007;120:371–8. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 41.Zhang X, Lin L, Guo H, Yang J, Jones SN, Jochemsen A, et al. Phosphorylation and Degradation of MdmX is Inhibited by Wip1 Phosphatase in the DNA Damage Response. Cancer research. 2009;69:7960–8. doi: 10.1158/0008-5472.CAN-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–12. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 43.Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, et al. p53DINP1, a p53-Inducible Gene, Regulates p53-Dependent Apoptosis. Molecular Cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- 44.Wu GS, Burns TF, McDonald ER, Jiang W, Meng R, Krantz ID, et al. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet. 1997;17:141–3. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 45.Ashkenazi A, Dixit VM. Death Receptors: Signaling and Modulation. Science. 1998;281:1305–8. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 46.Lin YP, Ma WL, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nature Genet. 2000;26:122–5. doi: 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- 47.Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–6. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- 48.Deng CX, Zhang PM, Harper JW, Elledge SJ, Leder P. Mice lacking p21(CIP1/WAF1) undergo normal development, but are defective in G1 Checkpoint control. Cell. 1995;82:675–84. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 49.Eldeiry WS, Harper JW, Oconnor PM, Velculescu VE, Canman CE, Jackman J, et al. WAF1/CIP1 is induced in p53-mediated G(1) arrest and apoptosis. Cancer Research. 1994;54:1169–74. [PubMed] [Google Scholar]
- 50.Lindqvist A, de Bruijn M, Macurek L, Brás A, Mensinga A, Bruinsma W, et al. Wip1 confers G2 checkpoint recovery competence by counteracting p53-dependent transcriptional repression. The EMBO Journal. 2009;28:3196–206. doi: 10.1038/emboj.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Research. 2015;43:D805–D11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.