Abstract
Nanos (Nos) is one of the evolutionarily conserved proteins known to direct germ-line development. In Drosophila, maternal Nos protein maintains transcriptional quiescence in the germ-line progenitors or pole cells to repress ectopic expression of somatic genes. Here we show that maternal Nos is required to establish and maintain germ-line identity by preventing apoptosis and somatic cell fate. The pole cells lacking maternal Nos were degraded by apoptosis during mid to late embryogenesis. When apoptosis was suppressed by Df(3L)H99, some pole cells lacking Nos adopted somatic cell fates. These pole cells expressed somatic markers ectopically and lost the germ-line marker Vasa. We further found that some Nos-negative pole cells were able to migrate into the gonads, but they failed to develop as functional germ cells during postembryonic stages. We propose a model in which Nos establishes germ-line/soma dichotomy and is also required to maintain germ-line fate.
The mechanism underlying the segregation of germ line from somatic line is a century-old issue in developmental biology. In many animal groups, maternal factors required for germ-line formation are localized in a histologically remarkable region in egg cytoplasm, or germ plasm, and are inherited in the germ-line progenitors (1, 2). In Drosophila, the germ-line progenitors known as pole cells are first formed at the posterior pole of the blastoderm embryos. During later embryogenesis, the pole cells pass through midgut epithelium into hemocoel and migrate within the embryos to reach the embryonic gonads, where they differentiate as functional germ line (1–4). Previous works have demonstrated that none of the pole cells becomes incorporated into any somatic tissues and contributes to somatic development during embryogenesis (5, 6). However, it remains elusive how the developmental fate of pole cells is regulated by germ plasm components.
One of the germ plasm components, Nanos (Nos), is evolutionarily conserved and has a widespread role in germ-line development (7–13). Maternal nos mRNA is enriched in germ plasm, and its protein product is partitioned into pole cells when they are formed and remains detectable in these cells throughout embryogenesis (14). Nos is required in pole cells for their proper migration into the embryonic gonads (15, 16). Within pole cells, Nos is involved in maintaining transcriptional quiescence (17) and is also required for maintenance of a germ-line-specific chromatin status that correlates with transcriptional inactivity (18). In the absence of maternal Nos activity, ectopic expression of somatic genes is detectable in pole cells (17). These results led us to speculate that pole cells lacking Nos may adopt somatic cell fate.
Here, we show that pole cells are able to adopt both germ-line and somatic cell fates and undergo apoptosis. Nos is required to repress the pathway leading to somatic differentiation and apoptosis. Thus, we conclude that Nos is essential for germ-line/soma dichotomy and for germ-line maintenance.
Materials and Methods
Fly Stocks. The nos allele used was nosBN. Df(3L)H99 was as described in ref. 19. PLHΔ23 is described in ref. 20. Using these lines, we generated nosBN Df(3L)H99 PLHΔ23/TM3 (nosBN-H99-PLHΔ23/TM3). In the experiment to examine the fate of pole cells homozygous for Df(3L)H99 and nos-H99 pole cells in the third-instar larvae, we used TM6 ubi-GFP instead of TM3. TM6 ubi-GFP expresses GFP under the control of the ubiquitin promoter in germ and somatic lines (data not shown).
In Situ Hybridization for Midgut Marker Genes. The dechorionated embryos were fixed in 2 ml of a 1:1 mixture of heptane and fixative I [3.7% formaldehyde in PBS (130 mM NaCl/7 mM Na2HPO4/3mMNaH2PO4)]. The fixed embryos were processed for whole-mount in situ hybridization as described in ref. 21 with several modifications. Digoxigenin-labeled RNA probes were synthesized from a 706-bp cDNA fragment from midgut expression 1 (mex1), 3,137-bp cDNA of integrin β neu (bInt-n) (LD09848), 3061-bp cDNA of dGATAe (LD08432), and a 589-bp cDNA fragment from the 3′-terminal region of CG11267. mex1, bInt-n, and dGATAe were reported to be expressed in the embryonic midgut (refs. 22 and 23 and T. Okumura and R. Murakami, personal communication). CG11267 was selected to be expressed in the midgut by our screen. These probes were in situ hybridized with midgut cells after stage 13. However, neither of these genes were expressed in pole cells within the gonads and hemocoel during normal development (data not shown). To label almost all of the midgut epitherial cells intensely, we used mixed probes (mex1/bInt-n and dGATAe/CG11267) for in situ hybridization. Hybridization was performed at 60°C in a solution containing 50% formamide, 5× standard saline citrate (SSC) (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7), 0.1% Tween 20, 0.1 mg/ml yeast tRNA, 10 mM DTT, and 10% dextran sulfate. The embryos were washed six times at 60°C in a solution containing 50% formamide, 5× SSC, and 0.1% Tween 20. Signal was detected with a TSA Biotin System (PerkinElmer). The embryos were then stained for β-galactosidase (β-gal) and Vasa (Vas).
Immunostaining. Immunofluorescent staining of embryos was carried out as described in ref. 24. For Vas staining, the dechorionated embryos were fixed in 2 ml of a 1:1 mixture of heptane and fixative II (4% paraformaldehyde in PBS). The fixed embryos were treated with a rat anti-Vas antibody (1:2,000 dilution) and then with a Texas red-conjugated anti-rat IgG (1:50, Cappel).
For triple staining, the in situ hybridized embryos were stained for β-gal and Vas. We used a rabbit anti-β-gal antibody (1:400, Chemicon) and a rat anti-Vas antibody (1:2,000). As secondary antibodies, an Alexa 647-conjugated anti-rat IgG (1:1,000, Molecular Probes) and an Alexa 488-conjugated anti-rabbit IgG (1:400, Molecular Probes) were used. Stained embryos were then mounted in Vectashield (Vector Laboratories) and observed under a confocal microscope (Leica Microsystems, Tokyo).
For immunostaining larval ovaries and testes, the dissected gonads were fixed in fixative II for 20 min and treated three times with methanol. Fixed gonads were rinsed in PBS containing 1% Triton X-100 for 15 min and treated with rabbit anti-β-gal, rat anti-Vas, and mouse anti-GFP (1:400, Wako Pure Chemicals, Osaka) antibodies. As secondary antibodies, an Alexa 647-conjugated anti-rat IgG (1:500, Molecular Probes), an Alexa 568-conjugated anti-rabbit IgG (1:1,000, Molecular Probes), and an Alexa 488-conjugated anti-mouse IgG (1:500, Molecular Probes) were used. The stained samples were then mounted in Vectashield and observed under a confocal microscope (Leica Microsystems).
Double Staining with Terminal Deoxynucleotidyltransferase-Mediated dUTP Nick End Labeling (TUNEL) and Immunostaining Methods. TUNEL was performed essentially according to the method reported in ref. 19 with several modifications. Embryos were fixed with fixative I for 30 min and devitellinized. Those embryos were washed in PBS containing 0.3% Triton X-100 and in TUNEL buffer [TUNEL dilution buffer (Roche Applied Science) containing 0.3% Triton X-100 and 1.5 mM CoCl2]. The embryos were incubated for 3 h at 37°C in TUNEL buffer containing 1× TUNEL enzyme (Roche Applied Science) and 10 μM dUTP [1:2 mix of Bio-16-dUTP and dUTP (Roche Molecular Biochemicals)] and washed in PBS containing 0.3% Triton X-100. The embryos were treated with FITC-avidin DN (1:200 dilution, Vector) for 1 h. The embryos were then stained for Vas protein by the method described above.
Pole Cell Transplantation and β-Gal Staining. Pole cell transplantation and β-gal staining were performed as described in ref. 15.
Results and Discussion
We wished to examine the developmental fate of pole cells lacking Nos (nos pole cells) as embryogenesis proceeds. However, beginning at stage 9/10 most pole cells were lost in the embryos derived from nos homozygous mothers (nos embryos) (24) (Fig. 1A). Because nos pole cells sometimes showed irregular shapes characteristic of apoptotic cells (Fig. 1D) and were TUNEL-positive (25) (Fig. 1 B and F), we concluded that these pole cells were eliminated by apoptosis. To further test this conclusion and potentially to be able to analyze the developmental fate of nos pole cells, we used Df(3L)H99 (H99), a small deletion that uncovers the genomic region including reaper (rpr) (19), head involution defective (hid) (26), and grim (27), which are all involved in apoptosis. As expected, H99 deficiency suppressed apoptosis of nos pole cells. In embryos lacking maternal Nos and zygotic H99 activity (nos-H99 embryo), none of the pole cells showed TUNEL signal (Table 1), showing that apoptosis of nos pole cells requires activities of genes within the H99 deficiency.
Fig. 1.
Removal of Nos activity causes apoptosis of pole cells during embryogenesis. Embryos from nosBN/TM3 females (C and E) (control embryos) and from nosBN/nosBN females (D and F) (nos embryos) were stained with an anti-Vas antibody (A, C, and D) and double-stained with an anti-Vas antibody and TUNEL labeling (B, E, and F). (A and B) The number of pole cells per embryo (A) and the percentage of pole cells with TUNEL signal (B) in control (red) and nos (blue) embryos are plotted against the developmental stage. Embryos (3–30) of each genotype were examined at each stage. (C–F) Stage-13 embryos. Vas signal is shown in magenta, and TUNEL signal is shown in green. Arrowheads show pole cells with altered morphology associated with apoptosis. (Scale bar = 10 μm.)
Table 1. H99 deficiency suppresses apoptosis of nos pole cells.
| Embryos examined* | No. of pole cells observed | No. of pole cells with TUNEL signal (%) | Significance† |
|---|---|---|---|
| nos | 380 | 75 (19.7) | |
| nos-H99 | 524 | 0 (0) | P < 0.0001 |
Stage 13 embryos derived from nosBN/nosBN females mated with WT males (nos) and nosBN Df(3L)H99/nosBN females mated with Df(3L)H99/TM3 males (nos-H99) were double-stained with an anti-Vas antibody and TUNEL labeling. Because the lack of H99 results in repression of apoptosis in somatic tissues (19), the embryos without TUNEL signal in the somatic region, such as gnathal segments and the dorsal folds, were judged to be homozygous for Df(3L)H99.
Significance was calculated by using Fisher's exact probability test.
To examine the developmental fate of nos-H99 pole cells, we transplanted them into host embryos and examined their developmental fate, because nos-H99 embryos were lethal. As shown in Table 2 and Fig. 2 D–I, nos-H99 pole cells were integrated within somatic tissues, such as midgut epithelium, tracheal epithelium, and gastric ceca, whereas neither control nor nos pole cells ever were. These “integrated” nos-H99 pole cells were morphologically distinct from pole cells that were merely lost in midgut lumen and hemocoel. Pole cells integrated within somatic tissues were morphologically indistinguishable from their neighboring host cells (Fig. 2 D–I). For example, pole cells within midgut epithelium were columnar in shape (Fig. 2 D and G). In contrast, the “lost” nos-H99, nos, and control pole cells were round in shape and were neither associated with nor embedded within somatic tissues (Fig. 2B). Moreover, we found that the integrated pole cells expressed somatic markers ectopically. When nos-H99 pole cells were integrated into the midgut epithelium, all of them expressed the midgut marker genes CG11267 and dGATAe (n = 13 pole cells examined) and bInt-n and mex1 (n = 9) (Fig. 3 B and D). Conversely, the germ-line marker Vas was lost or significantly reduced in these pole cells (Fig. 3C). In contrast, all of the lost pole cells were Vas-positive but failed to express midgut markers (mex1/bInt-n) (n = 11) (Fig. 3 F–J). These results clearly show that nos pole cells can differentiate as somatic cells when their apoptosis is suppressed.
Table 2. Developmental fate of transplanted pole cells.
 |
Fig. 2.
nos-H99 pole cells have the ability to adopt somatic cell fate and to enter the embryonic gonads. Pole cells from nos (B), nos-H99 (C–F), and control (A) embryos were transplanted into hosts, and the hosts were developed until stages 15–17. The transplanted pole cells were identified as blue cells after heat treatment and staining for β-gal (arrowheads). The transplanted control (A) and nos-H99 (C) pole cells were observed within the gonads, whereas nos pole cells remained outside the gonads (B). Some nos-H99 pole cells were integrated within somatic tissues (D–F). (D) nos-H99 pole cell within midgut epithelium is shown. Note that nos-H99 pole cell and the neighboring midgut epithelial cells were columnar in shape at stage 17. nos-H99 pole cells within gastric ceca (E) and tracheal (F) epithelium are also shown. The lower cell indicated by an arrowhead in E was out of focus. (G–I) Drawings tracing the integrated nos-H99 pole cells (blue) and the surrounding somatic cells in D–F, respectively. (Scale bar = 10 μm.)
Fig. 3.
nos-H99 pole cells within midgut epithelium expressed midgut marker genes but lost germ-line marker Vas. We examined expression of midgut marker genes and Vas protein in nos-H99 pole cells within midgut epithelium (A–E), hemocoel (lost pole cells) (F–J), and gonad (K–O) of host embryos. The transplanted pole cells were identified as the cells expressing β-gal (marked by arrowheads). The embryos at stages 13–15 were triple-stained with probes for the midgut marker genes (bInt-n/mex1; red) (B, G, and L) and antibodies against β-gal (green) (A, F, and K) and Vas protein (blue) (C, H, and M). nos-H99 pole cell integrated within midgut epithelium (mg), like the neighboring host cells, was cuboidal at stage 14 (E). The integrated pole cell expressed midgut marker genes (B) but not Vas protein (C). In contrast, nos-H99 pole cells, which were found within hemocoel in the vicinity of midgut (J), maintained Vas expression (H), but never expressed midgut marker genes (G). Within the gonad, nos-H99 pole cells and the host pole cells were both negative for midgut markers (L). Arrows in K and N indicate nonspecific staining of tracheal lumen. Merged images (D, I, and N) and photographs taken with a compound microscope equipped with Nomarski optics (E, J, and O) are shown. Dotted lines indicate midgut epithelia in E and J and gonad (go) in O. (Scale bar =10 μm.)
We have shown that transplanted nos pole cells never migrate into the host gonads (15). To our surprise, their ability to enter the gonads was restored by suppressing apoptosis. In ≈25% of the host embryos, nos-H99 pole cells were incorporated within the gonads (Figs. 2C and 3 K–O and Table 2). This percentage is less than that observed when control pole cells were transplanted into the hosts, presumably because only half of the nos-H99 embryos used as donors are homozygous for H99 (Table 2). Thus, the ability of nos pole cells to migrate into the gonads is fully restored by suppressing apoptosis in our transplantation experiments.
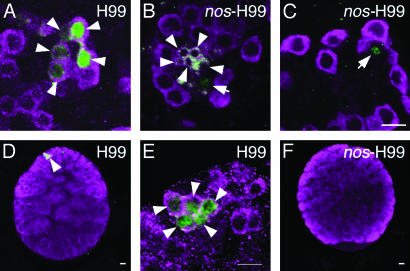
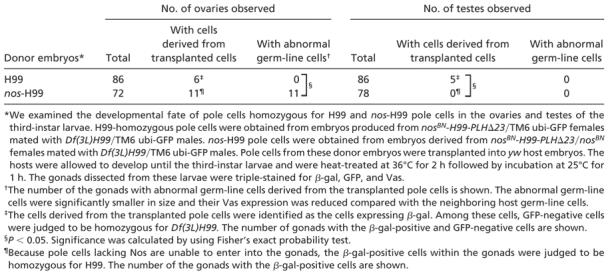
nos-H99 pole cells incorporated within the embryonic gonads were indistinguishable from the host pole cells in their morphology and Vas staining (Figs. 2C and 3 K–O). However, these pole cells did not contribute to egg and sperm production. In the ovaries, the cells derived from transplanted nos-H99 pole cells were detectable until at least the end of the third instar, but their size was much smaller than that of normal germ-line cells, and their Vas expression was significantly reduced (Fig. 4 B and C and Table 3). In the testes of the third-instar larvae, the cells derived from transplanted nos-H99 pole cells were no longer detectable (Fig. 4F and Table 3). These results indicate that nos-H99 pole cells are unable to complete gametogenesis. As a control, we transplanted pole cells from H99-homozygous embryos into hosts. These pole cells were able to migrate into the embryonic gonads (data not shown) and were found normally within the ovaries and testes of the third-instar larvae (Fig. 4 A, D, and E).
Fig. 4.
The developmental fate of nos-H99 pole cells incorporated into the gonads. We examined the developmental fate of pole cells homozygous for H99 (A, D, and E) and nos-H99 (B, C, and F) pole cells in ovaries (A–C) and testes (D–F) in third-instar larvae. H99-homozygous pole cells were obtained from embryos produced from nosBN-H99-PLHΔ23/TM6 ubi-GFP females mated with Df(3L)H99/TM6 ubi-GFP males (for details, see Materials and Methods). The germ-line cells derived from the transplanted pole cells were identified as the cells expressing β-gal (green, marked by arrowheads in A, D, and E). Among these cells, GFP-negative cells were judged to be homozygous for H99 (data not shown). nos-H99 pole cells were obtained from embryos derived from nosBN-H99-PLHΔ23/nosBN females mated with Df(3L)H99/TM6 ubi-GFP males. Because pole cells lacking maternal Nos alone are unable to enter the gonads, the β-gal-positive cells within the gonads were judged to be homozygous for H99 (green, marked by arrows and arrowheads in B and C). Germ-line cells from nos-H99 pole cells were much smaller than normal germ-line cells (B, arrowheads), and their Vas expression (magenta) was significantly impaired (B and C, arrows). β-gal signal was frequently seen in the nuclei of the transplanted cells (A–E), but some transplanted cells showed β-gal signal in their cytoplasm (B, arrowheads). (E) A higher-magnification image of β-gal-positive germ-line cells shown in D. nos-H99 pole cells were no longer discernible in a testis of third-instar larvae (F). (Scale bars = 10 μm.)
Table 3. The ability of pole cells to become germ-line cells in the gonads of the third-instar larvae.
 |
Our results imply that pole cells are multipotent, in that they are able to adopt both germ-line and somatic cell fates and undergo apoptosis. Nos is required to repress the pathway leading to somatic differentiation and apoptosis and, thus, to direct germ-line development. Removal of Nos and H99 activities causes pole cells to differentiate into soma. However, some pole cells retain the ability to migrate properly into the gonads. These different behaviors of nos-H99 pole cells could be explained by differences in the cellular environment encountered by the pole cells. Alternatively, there may be a heterogeneity among pole cells, one with and one without the competence to adopt somatic cell fate. This idea is supported by the observation that there are Nos-independent transcriptional repression mechanisms in pole cells. Somatic gene expression is derepressed only in a subset of nos pole cells (17, 18). Furthermore, a nucleosomal histone modification, methylated lysine 4 on histone H3, that correlates well with open chromatin is accumulated only in a subset of nos pole cells (18). We propose that transcriptional derepression is a prerequisite for somatic differentiation of pole cells, although it remains unclear how somatic cell types are specified.
Our data also show that Nos is not essential for pole cell migration because nos-H99 pole cells can be normally incorporated within the gonads. It is reasonable to conclude that Nos represses apoptosis in pole cells, allowing their proper migration into the gonads. However, nos-H99 pole cells do not complete gametogenesis, suggesting that Nos has another function in later germ-line development (16). It has been reported that zygotic Nos is required in the germ-line cells to prevent their precocious entry into oogenesis during larval development (28). However, in the larvae lacking zygotic Nos, the prematurely formed cysts fail to execute oogenesis and eventually degenerate. It is possible that maternal Nos may also be required in pole cells to repress their premature differentiation. Alternatively, the defect of nos-H99 pole cells could result simply from their failure to establish germ-line fate.
In nematode, zebrafish, and mouse embryos, Nos homologues are required for maintenance of the germ-line progenitors (10, 12, 13). These results, and those we have observed in Drosophila, indicate that Nos is involved in an evolutionarily conserved mechanism required for germ-line maintenance. Moreover, in Caenorhabditis elegans and Drosophila, Nos is required to establish germ-line-specific histone modifications that correlate with transcriptionally inactive chromatin (18). We propose that Nos also acts as a part of conserved mechanism repressing somatic gene expression and differentiation to establish germ/soma dichotomy.
Acknowledgments
We thank Dr. P. Lasko for critical reading of the manuscript, Drs. M. Asaoka and M. Mukai for valuable comments, Dr. R. Lehmann for nosBN flies, Drs. A. Nakamura and P. Lasko for an anti-Vas antibody, Dr. R. Murakami for cDNAs of dGATAe and bInt-n, and the Bloomington Drosophila Stock Center for Df(3L)H99 and TM6 ubi-GFP strains. This work was supported by grants from Japan Science and Technology Agency (Core Research for Evolutional Science and Technology Project), the Ministry of Education, Science, and Culture of Japan, and the Japan Society for the Promotion of Science (Research for the Future).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: β-gal, β-galactosidase; Nos, Nanos; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling; Vas, Vasa.
References
- 1.Beams, H. W. & Kessel, R. G. (1974) Int. Rev. Cytol. 39, 413–479. [DOI] [PubMed] [Google Scholar]
- 2.Eddy, E. M. (1975) Int. Rev. Cytol. 43, 229–280. [DOI] [PubMed] [Google Scholar]
- 3.Rongo, C. & Lehmann, R. (1996) Trends. Genet. 12, 102–109. [DOI] [PubMed] [Google Scholar]
- 4.Campos-Ortega, J. A. & Hartenstain, V. (1985) The Embryonic Development of Drosophila melanogaster (Springer, Heidelberg), pp. 9–84.
- 5.Underwood, E. M., Caulton, J. H., Allis, C. D. & Mahowald, A. P. (1980) Dev. Biol. 77, 303–314. [DOI] [PubMed] [Google Scholar]
- 6.Technau, G. M. & Campos-Ortega, J. A. (1986) Roux's Arch. Dev. Biol. 195, 489–498. [DOI] [PubMed] [Google Scholar]
- 7.Wang, C. & Lehmann, R. (1991) Cell 66, 637–647. [DOI] [PubMed] [Google Scholar]
- 8.Mosquera, L., Forristall, C., Zhou, Y. & King, M. L. (1993) Development (Cambridge, U.K.) 117, 377–386. [DOI] [PubMed] [Google Scholar]
- 9.Pilon, M. & Weisblat, D. A. (1997) Development (Cambridge, U.K.) 124, 1771–1780. [DOI] [PubMed] [Google Scholar]
- 10.Subramaniam, K. & Seydoux, G. (1999) Development (Cambridge, U.K.) 126, 4861–4871. [DOI] [PubMed] [Google Scholar]
- 11.Mochizuki, K., Sano, H., Kobayashi, S., Nishimiya-Fujisawa, F. C. & Fujisawa, T. (2000) Dev. Genes Evol. 210, 591–602. [DOI] [PubMed] [Google Scholar]
- 12.Köprunner, M., Thisse, C., Thisse, B. & Raz, E. (2001) Genes Dev. 15, 2877–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuda, M., Sasaoka, Y., Kiso, M., Abe, K., Haraguchi, S., Kobayashi, S. & Saga, Y. (2003) Science 301, 1239–1241. [DOI] [PubMed] [Google Scholar]
- 14.Wang, C., Dickinson, L. K. & Lehmann, R. (1994) Dev. Dyn. 199, 103–115. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi, S., Yamada, M., Asaoka, M. & Kitamura, T. (1996) Nature 380, 708–711. [DOI] [PubMed] [Google Scholar]
- 16.Forbes, A. & Lehmann, R. (1998) Development (Cambridge, U.K.) 125, 679–690. [DOI] [PubMed] [Google Scholar]
- 17.Deshpande, G., Calhoun, G., Yanowitz, J. L. & Schedl, P. D. (1999) Cell 99, 271–281. [DOI] [PubMed] [Google Scholar]
- 18.Schaner, C. E., Deshpande, G., Schedl, P. D. & Kelly, W. G. (2003) Dev. Cell 5, 747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White, K., Grether, M. E., Abrams, J. M., Young, L., Farrell, K. & Steller, H. (1994) Science 264, 677–683. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi, S., Kitamura, T., Sasaki, H. & Okada, M. (1993) Development (Cambridge, U.K.) 117, 885–893. [DOI] [PubMed] [Google Scholar]
- 21.Tautz, D. & Pfeifle, C. (1989) Chromosoma 98, 81–85. [DOI] [PubMed] [Google Scholar]
- 22.Robert, A. S., Xie, X., Andres, A. J. & Galewsky, S. (1991) Dev. Biol. 143, 206–211. [DOI] [PubMed] [Google Scholar]
- 23.Yee, G. H. & Hynes, R. O. (1993) Development (Cambridge, U.K.) 118, 845–858. [DOI] [PubMed] [Google Scholar]
- 24.Asaoka-Taguchi, M., Yamada, M., Nakamura, A., Hanyu, K. & Kobayashi, S. (1999) Nat. Cell Biol. 1, 431–437. [DOI] [PubMed] [Google Scholar]
- 25.Gavrieli, Y., Sherman, Y. & Ben-Sasson, S. A. (1992) J. Cell Biol. 119, 493–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grether, M. E., Abrams, J. M., Agapite, J., White, K. & Steller, H. (1995) Genes Dev. 9, 1694–1708. [DOI] [PubMed] [Google Scholar]
- 27.Chen, P., Nordstrom, W., Gish, B. & Abrams, J. M. (1996) Genes Dev. 10, 1773–1782. [DOI] [PubMed] [Google Scholar]
- 28.Wang, Z. & Lin, H. (2004) Science 303, 2016–2019. [DOI] [PubMed] [Google Scholar]