Abstract
Background
Engineering lactic acid bacteria (LAB) is of growing importance for food and feed industry as well as for in vivo vaccination or the production of recombinant proteins in food grade organisms. Often, expression of a transgene is only desired at a certain time point or period, e.g. to minimize the metabolic burden for the host cell or to control the expression time span. For this purpose, inducible expression systems are preferred, though cost and availability of the inducing agent must be feasible. We selected the plasmid free strain Lactobacillus plantarum 3NSH for testing and characterization of novel inducible promoters/repressor systems. Their feasibility in recombinant protein production was evaluated. Expression of the reporter protein mCherry was monitored with the BioLector® micro-fermentation system.
Results
Reporter gene mCherry expression was compared under the control of different promoter/repressor systems: PlacA (an endogenous promoter/repressor system derived from L. plantarum 3NSH), PxylA (a promoter/repressor system derived from Bacillus megaterium DSMZ 319) and PlacSynth (synthetic promoter and codon-optimized repressor gene based on the Escherichia colilac operon). We observed that PlacA was inducible solely by lactose, but not by non-metabolizable allolactose analoga. PxylA was inducible by xylose, yet showed basal expression under non-induced conditions. Growth on galactose (as compared to exponential growth phase on glucose) reduced basal mCherry expression at non-induced conditions. PlacSynth was inducible with TMG (methyl β-D-thiogalactopyranoside) and IPTG (isopropyl β-D-1-thiogalactopyranoside), but also showed basal expression without inducer. The promoter PlacSynth was used for establishment of a dual plasmid expression system, based on T7 RNA polymerase driven expression in L. plantarum. Comparative Western blot supported BioLector® micro-fermentation measurements. Conclusively, overall expression levels were moderate (compared to a constitutive promoter).
Conclusions
We evaluated different inducible promoters, as well as an orthologous expression system, for controlled gene expression in L. plantarum. Furthermore, here we provide proof of concept for a T7 RNA polymerase based expression system for L. plantarum. Thereby we expanded the molecular toolbox for an industrial relevant and generally regarded as safe (GRAS) strain.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-016-0448-0) contains supplementary material, which is available to authorized users.
Keywords: L. plantarum 3NSH, BioLector® micro-fermentation system, Orthologous expression system, T7 RNA polymerase, IPTG, Inducible expression
Background
Lactobacillus plantarum is a versatile lactic acid bacterium that is generally regarded as safe (GRAS). It inhabits diverse ecological niches and exhibits probiotic characteristics [42]. L. plantarum is often used as starter or adjunct culture in fermented food and feed production processes like for sausages, cheeses, fermented vegetables, and grass or corn silage [10, 36, 37]. Due to its high oxygen tolerance and robustness in natural fermentation processes, L. plantarum has gained increasing interest also as a host for recombinant protein expression and thus, its use in biotechnological applications is steadily growing [1, 20, 41]. Research involves genomics, transcriptomics, cell engineering and evolutionary strain optimization [9, 37] e.g. for bulk production of chemicals, metabolites and enzymes [23, 28] as well as for in situ delivery of vaccines [8, 11, 12, 32, 50]. Anti-microbial features, such as plantaricin production, are also of growing importance [33].
Specific gene regulatory elements like promoters are a prerequisite for efficient transcription of recombinant genes in any host organism. Accordingly, several constitutive promoters and shuttle vector systems have been established [38, 43, 44, 47]. Often, constitutive expression is preferred, for example for in situ delivery of recombinant proteins in the human body, or when steady-state gene expression is required [38]. Contrarily, constitutive promoters do not allow regulation of gene expression and production levels are directly linked to cellular growth. Continuous transcription throughout the fermentation process poses a limit to the expression of foreign proteins, which are potentially toxic to the host cell or exhibit excessive metabolic burden.
An alternative strategy is to use substrate dependent promoters that can be induced after a certain cell density has been reached. Several inducible promoters for L. plantarum have been described in the literature. The nisin-controlled gene expression (NICE) system is inducible with the bacteriocin nisin from Lactococcus lactis and was established also for L. plantarum [25]. However, the expression is not tightly regulated except if the target expression cassette is integrated into the host’s chromosome [34]. The pSIP system comprises a well-established inducible promoter system and is based on the induction of promoters from Lactobacillus sakei with an inducing peptide [46]. More recently, another inducible promoter based on manganese starvation was described for L. plantarum NC8 [3].
Yet, numerous other substrate induced promoter-repressor systems are present in LAB and other bacteria that eventually may serve to efficiently control transgene expression. Lactobacillus plantarum contains a lac - operon which was expected to be regulated similarly as the well-studied lac-operon of Escherichia coli, where the lac-operon comprises the genes lacZ (β-galactosidase), lacY (lactose permease), lacA (transacetylase) and lacI (repressor). Allolactose is the natural inducer of the lac-operon. In E. coli, thio-galactosides such as IPTG (isopropyl β-D-1-thiogalactopyranoside) and TMG (methyl β-D-thiogalactopyranoside) are the most commonly used inducers in industrial production processes.
We established a synthetic inducible promoter system based on the E. coli derived lac-operon, which we adapted for L. plantarum in the high copy number shuttle vector pCDLbu1 [15, 43]. Based on the inducible synthetic system, we designed and constructed an artificial T7 RNA polymerase regulated dual plasmid expression system and demonstrated its applicability in L. plantarum 3NSH. Additionally, we tested endogenous lac-operon regulatory sequences from L. plantarum 3NSH. This strain is derived from L. plantarum CD033, which was cured of its native plasmid [17]. Plasmid free strains are preferable expression hosts, since native plasmids sometimes interfere with expression vector replication.
Another well-known regulated system is the xylose operon and the xylose promoter/repressor gene from Bacillus megaterium, which is well established for Gram-positive bacteria, and was already used for high yield production of secretory levansucrase in B. megaterium YYBm1 [22] and dextransucrase in B. megaterium MS941 [26]. Moreover, three different recombinant proteins in Brevibacillus choshinensis SP3 under the control of PxylA from B. megaterium have been reported [5]. D-xylose is metabolized by two intracellular enzymes: the D-xylose isomerase (XylA) and the D-xylose kinase (XylB). D-xylose can be transported into the cell by two different mechanisms. One mechanism involves a D-xylose-H+ or –Na+ symporter (xylT) and is regulated by CcpA [40]. Another mechanism is driven by ATP and consists of a high-affinity xylose transporter system involving a periplasmic binding protein. For three species of facultative hetero-fermentative lactobacilli, Lactobacillus pentosus, L. plantarum, and Lactobacillus casei it was shown that EIIMan complex of the phosphoenolpyruvate (PEP): D-mannose phosphotransferase system (PTS) is involved in D-xylose transport via facilitated diffusion [4]. Posno and co-workers [35] reported that L. plantarum does not metabolize D-xylose. For its use as an inducer, this is an advantage, since D-xylose is not degraded and keeps the level of induction constant throughout the process.
In this study, we present the establishment and characterization of different inducible promoter/repressor systems (and their respective inducer) in the high copy number pCDLbu1 shuttle vector for L. plantarum 3NSH. We used mCherry as reporter protein and expression levels were analyzed with the BioLector® micro-fermentation system and confirmed by Western blot immuno-detection. Furthermore, we established an inducible T7 RNA polymerase based system for regulated recombinant gene expression. Summarizing, we present expression plasmids with a set of novel inducible promoters, and expand the toolbox for recombinant protein expression in L. plantarum.
Results and discussion
Different inducible promoter systems were characterized and tested in the plasmid free strain L. plantarum 3NSH [17]. Comparative studies were carried out regarding bacterial growth rates, level of reporter gene expression, effect of inducer and behavioral differences due to varying carbon sources. BioLector® micro-fermentations were established. FlowerPlates (with or without optodes for low pH and dissolved oxygen) for detection of biomass (calculated optical density) and fluorescence were used for L. plantarum fermentation and analysis.
Besides promoter elements and transcription initiation, other factors additionally influence the level of protein expression. Such are terminators, untranslated regions, plasmid copy numbers and the protein itself (amino acids, folding, and toxicity). In our experimental set-up, we chose the ribosomal binding site (RBS) and the spacer between the RBS and start codon to be identical for all promoter constructs (Fig. 1), in order to exclude any translational effects on mCherry expression caused by different RBSs. However, the promoter consensus sequences (including the −35 and −10 region) were specific for each tested promoter.
Fig. 1.
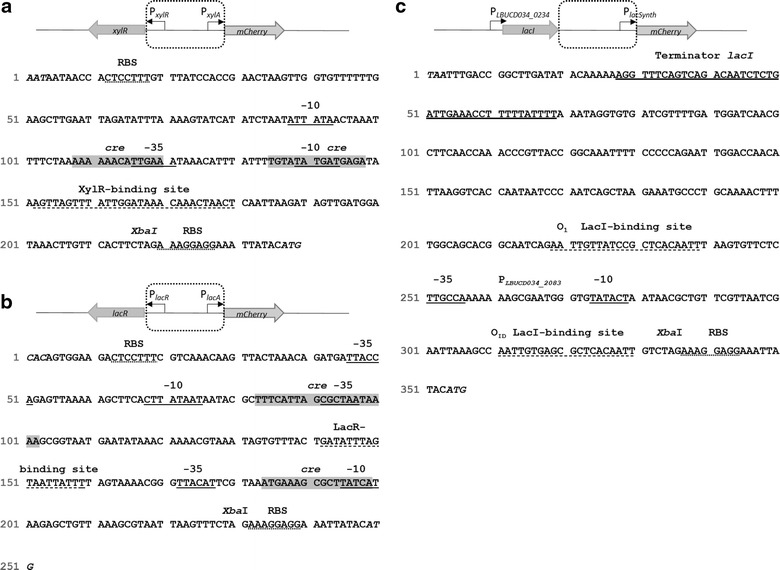
Promoter region sequences of PxylA, PlacA and PlacSynth. Nucleotide sequence from repressor to start codon of mCherry are shown (region within dotted square). The mCherry start codon is indicated in italics, preceded by an identical ribosome binding site (RBS; italics) an XbaI restriction site (bold) and an identical 9 nt spacer sequence was introduced upstream of mCherry start codon. The −35 and −10 promoter region were identified (SoftBerry, BPROM) and are underlined. Primer binding sites for negative controls (for construction of negative controls without promoter) are underlined in dashed line. a PxylA; promoter of xylA gene from B. megaterium DSMZ 319 and promoter of repressor XylR. Operator sequences for XylR binding are underlined; cre sites (catabolite-responsive element) are highlighted. b PlacA; endogenous promoter of LacA from L. plantarum 3NSH and promoter of repressor LacR. LacR-binding site was identified (RegPrecise) and underlined, and putative cre-sites are highlighted. c PlacSynth; promoter P2083 from L. buchneri CD034 with artificially integrated operator binding sites with recommended distance of 93 nt (O1 and OiD from E. coli) are underlined (dotted line), terminator of lacI is underlined (solid line)
The heterologous promoter PxylA is inducible by xylose
The promoter fragment PxylA and the repressor gene cassette XylR were amplified from B. megaterium DSMZ 319 genomic DNA with primers listed in Table 1. Nucleotide sequences of promoters PxylA and promoter PxylR to start codon of mCherry are shown in Fig. 1a, where promoter, RBS, cre (catabolite-responsive element)-sites and XylR binding site are indicated. The xyl-repressor binding motif is indicated according to Stammen and colleagues [45].
Table 1.
List of primers used in this study
| Name | 5´–3´ Sequence |
|---|---|
| PlacSynth_SacI_EcoRI_F | GATGACGAGCTC GAATTCTGGTCTTTATTCTTCAA |
| M13_R_NheI | CGACGAGCTAGCAGCCAGGAAACAGCTATGACC |
| mCherry_RBS_XbaI | GCTGCTTCTAGAAAGGAGGAAATTATACATGTTATCAAAGGGTGAAGAAG |
| mCherry_R_BamHI | CGTCGTGGATCCTTATCACTTGTATAATTCATCCATACC |
| Tldh_amp_R_PstI | CTGCTGCTGCAGAAAAAGATTAAAAAAGCCGCTGC |
| mCherry_seq_R | TGGACGACCTTCACCTTCAC |
| mCherry_seq_F | AACGTATGTACCCAGAAGATG |
| CAT_seq2_back | TACATCATTCTGTTTGTGATGG |
| B_mega_XylOP_out_F | AACATATAAACAGCCAGTTGCC |
| B_mega_XylOP_R(SpeI, ScaI, BamHI) | GTAGTAGGATCC AGT ACTAGTTTCCCCCTTTGATTTAAGTG |
| mCherry_w/o_RBS_XbaI | CGTCGTTCTAGAATGTTATCAAAGGGTGAAGAAGATAAC |
| p256_miniori_for | CATCATAAGCTTCCCGCACGCATAGCGGTGC |
| B_mega_XylOP_F_MfeI, KpnI | GTAGTACAATTG GGTACCAAGGTGAGGGTGGAGACAG |
| Bmega_XylR_newRBS_XbaI_Phos_R | GTATAATTTCCTCCTTTCTAGAAGTGAACAAGTTTATCCAT |
| mCherry_Phos_F | ATGTTATCAAAGGGTGAAGAAG |
| B_mega_XylOP_seq_F | CAATTCCGATATTAATACTGATG |
| B_mega_XylOP_seq_R | CTAGTCGGAATAGGAATTTGTG |
| LacI_Lplant_F_SacI | AGCAGCGAGCTCCCTAATAGAACTGCGGTGGTC |
| LacI_Lplant_R_XbaI | AGCAGCTCTAGAAACTTAATTACGCTTTAACAGC |
| lacR_Gal_seq_R | AATTGAAGTGATGCGGGTCTG |
| lacR_Gal_seq_F | AATTGCGCCAGCTAACACCC |
| T7_RNAP_Lp_RBS | CAGCAGTCTAGATCCTAAAGGAGG |
| T7_RNAP_Lp_Term_R_SalI | CAGCAGGTCGACTTGATATACAAAAAAGG |
| M13_2_F | TTGTAAAACGACGGCCAGTG |
| T7-Promoter_SacI | GCTGCTGAGCTCAGATCGATCTCGATCCCGCG |
| T7-Terminator_SalI | GCTGCTGTCGACTCCGGATATAGTTCCTCCTTTC |
| ery_back_KasI | CATCATGGCGCCTCCGATTGCAGTATAAATTTAACG |
| oripE194_seq_back | AATCAAATCGGTATAAATCTGAC |
| Ery_F_NheI | CATCATGCTAGCTCCGATTGCAGTATAAATTTAACG |
| Pempty_SacI_R | TAGTAGTCTA GA GCTCGAATTCACTGGCCGTCG |
| mCherry_RBS_SacI_F | GCTGCTGAGCTCAAGGAGGAAATTATACATGTTATCAAAGGGTGAAGAAG |
Restriction sites are underlined and highlighted in bold or italics
Description of the operon and its regulation was presented by Schmiedel and colleagues [40]. Preliminary tests were performed with native RBS from XylA from B. megaterium DSMZ 319 in pCDLbu1_PxylA(nativeRBS)_mCherry. The comparison of RBS and spacer sequence of PxylA (native RBS) and PxylA is shown in Additional file 1: Figure S1. We compared mCherry expression with native RBS to the uniform and artificial SOPT#9 spacer RBS and sequence (5´-TCTAGAAAGGAGGAAATTATACATG-3´, from XbaI to start codon), which was established for L. plantarum CD033 [47]. SOPT#9 was used for all constructs and allowed comparison of mCherry expression apart from translational influences. Interestingly, we found that SOPT#9 lead to slightly higher expression levels compared to the native xylA RBS and spacer sequence (data not shown) and was well suited for recombinant protein expression in L. plantarum 3NSH. Parental rolling circle replicating plasmid pCDLbu1 is shown in Fig. 2a. The final shuttle vector pCDLbu1_PxylA_mCherry for PxylA regulated mCherry expression is depicted in Fig. 2b.
Fig. 2.
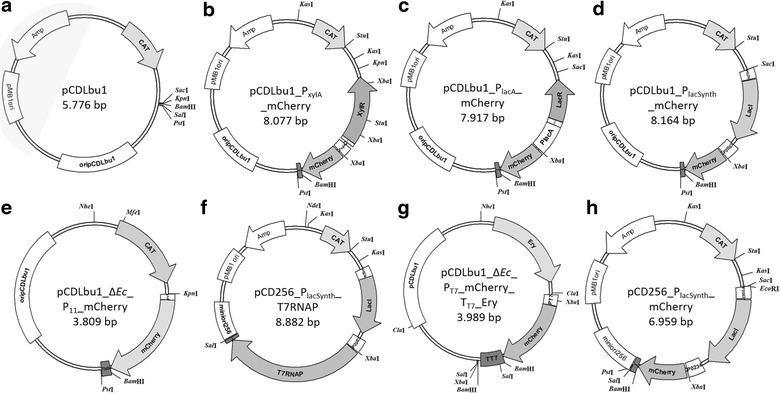
Maps of plasmids used in this study. Annotations and relevant restriction sites are indicated. a pCDLbu1 initial vector backbone (highlighted region are E. coli specific sequences); b pCDblu1_PxylA_mCherry; c pCDLbu1_PlacA_mCherry; d pCDLbu1_PlacSynth_mCherry; e pCDLbu1ΔEc_P11_mCherry (constitutive P11 promoter; internal reference plasmid described by Tauer and colleagues [47]); The term ‘ΔEc’ indicates removal of E. coli specific sequences which are highlighted in plasmid A. f pCD256_PlacSynth_mCherry; g pCD256_PlacSynth_T7RNAP; h pCDLbu1ΔEc_PT7_mCherry _TT7_Ery. OripCDLbu1 and miniori256: origins of replication (ori) for L. plantarum 3NSH; pMB1ori: ori for replication in E. coli; CAT chloramphenicol acetyltransferase gene; Amp ampicillin resistance gene; Ery ermI gene encoding resistance to erythromycin; P promoter; T terminator of transcription. Subscripted characters are specifications. Important restriction sites are indicated
Cells were grown on selective media with either glucose (Fig. 3a, c) or galactose (Fig. 3b, d) as main carbon source, induced with xylose (or absence of inducer) after 2 h and analyzed. Figure 3a, b show relative fluorescence units (RFUs) of mCherry expression (with or without induction) under control of PxylA for 23 h. In related literature the used amount of xylose added as inducer varies from 0.5 % (w/v) in Bacillus megaterium to 0.2 and 2 % in B. subtilis [2, 22, 26]. Figure 3a shows that the addition of 0.2 or 2 % xylose in MRS medium with glucose as main carbon source showed no effect on mCherry expression as compared to non-induced conditions. Figure 3b indicates that growth on galactose and induction with 0.2 or 2 % xylose led to enhanced expression of mCherry expression during exponential growth phase. Moreover, basal expression in medium containing 2 % galactose as main carbon source was repressed during the exponential phase (Fig. 3b), as compared to growth on glucose (Fig. 3a). Lactobacillus plantarum 3NSH is incapable of metabolizing xylose (data not shown), but xylose is efficiently transported into the cell. The use of this promoter/repressor-system in lactobacilli was tested here for the first time.
Fig. 3.
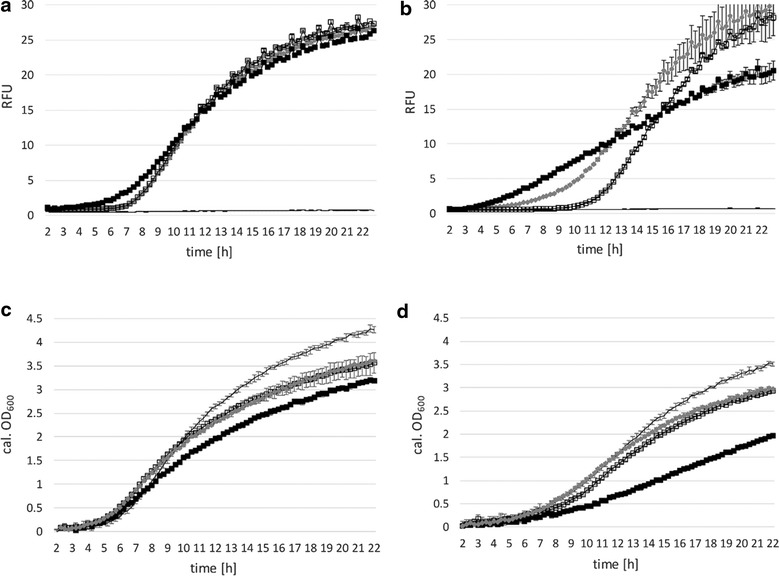
Promoter characteristics of PxylA and growth behavior. a, c MRS medium with glucose and 5 μg ml−1 CM; b, d MRS medium with galactose and 5 μg ml−1 CM, mean values of four replicates are given and standard deviations are indicated. Filled square induced 2 % xylose. Filled diamond induced 0.2 % xylose. Square non-induced. Solid line negative control. a,b specific expression levels of mCherry under control of PxylA after induction with 2, 0.2 % xylose (or no induction) after 2 h in BioLector® micro-fermentation. Change of mCherry expression (RFUs, relative fluorescent units) over time (hours) of PxylA mediated expression in comparison to the negative control is shown. c and d corresponding calculated OD600 values
Additionally, we tested a negative control (expression plasmid without the promoter/repressor fragment), which did not show any mCherry expression (Fig. 3a, b, solid line). Thus, basal expression was caused by weak repression of PxylA through inefficient XylR repressor binding and not by any putative additional regulatory sequences present on the plasmid.
In B. megaterium, the presence of glucose was shown to cause repression of PxylA by CcpA (catabolite control protein A) binding cre-sites within the promoter region and the xylA gene [13]. The xylA promoter in our context (Fig. 1a) contains two cre-sites, which were termed cre*-35.5 and (cre)-8.5 (a cre-like site) by Gösseringer and coworkers [13] who also showed that in B. megaterium the cre + 130.5 (within xylA sequence) and cre*-35.5 are simultaneously bound by CcpA, which results in looping of intervening DNA and tight repression of xylA transcription. Interestingly, we did not observe catabolite repression of mCherry expression by the presence of glucose with our construct (Fig. 3a). A reason for this could be the lack of cre + 130.5 within the mCherry gene, hence, multimer formation and efficient catabolite repression is prevented. Another explanation for the relatively strong basal expression level could be that glucose inhibits DNA binding by XylR and acts as a low-efficiency inducer for XylR as reported by Dahl and co-workers [6]: similar structure of xylose and glucose enable both sugars to utilize the same binding site on repressor XylR.
Plasmid pCDLbu1_PxylA_mCherry containing cells only showed minor growth differences on selective medium with either glucose or galactose (Fig. 3c, d). Growth on galactose slightly increased mCherry expression and decreased basal expression levels, resulting in an improved regulation of the system during exponential phase (Fig. 3b). We hypothesize that galactose interferes less with XylR mediated repression in L. plantarum than glucose and, hence, leads to improved repression of mCherry expression.
Lactobacillus plantarum 3NSH does not metabolize xylose, but effective transportation of xylose was demonstrated through inducibility of expression. The L. plantarum WCFS1 complete genome sequence [21] suggests genes involved in transport (lp_0331, lp_0975), but no xylA or xylB. Chaillou et al. [4] report that EIIMan complex of the phosphoenolpyruvate (PEP): D-mannose PTS is involved in D-xylose transport via facilitated diffusion. For industrial processes, it is considered an advantage, when the inducing substance is not degraded and a constant concentration during cultivation can be maintained. In terms of plant based biomass degradation, where xylose is highly abundant, this expression regime could provide a self-inducing promoter system for the production of e.g. endoglucanases and xylanases, thereby increasing the rate and efficacy of substrate metabolism in ensiling processes.
The endogenous promoter PlacA is inducible by lactose
The promoter of lacA (β-galactosidase) and the promoter of the Lac repressor (lacR) were amplified from L. plantarum 3NSH genomic DNA with primers shown in Table 1. Figure 1b shows the nucleotide sequences of endogenous promoters PlacA and promoter PlacR in divergent orientation. LacR binding site, cre-site and RBS are indicated. The final shuttle vector pCDLbu1_PlacA_mCherry is shown in Fig. 2c.
For promoter characterization, mCherry expression under induced and non-induced conditions was monitored. Lactose as well as the non-metabolizable lactose analogues isopropyl-β-D-thiogalactopyranoside (IPTG) and thiomethyl-β-D-galactoside (TMG) were tested for induction of PlacA. IPTG and TMG failed to induce LacR controlled gene expression (data not shown). This is in contrast to previous findings, where TMG was successfully used for the induction of ß-galactosidase expression in L. plantarum ATCC® 8014™ [19]. Different sugars were tested for induction of PlacA (including lactose, xylose, fructose, glucose, maltose, arabinose and galactose), but PlacA was only induced with lactose.
Lactobacillus plantarum 3NSH harboring plasmid pCDLbu1_PlacA_mCherry were grown on selective media either containing 2 % glucose or 2 % galactose as carbon source and were induced with 0.5 or 2 % lactose after 2 h. Induction of mCherry expression with lactose was weak, but slight increase of RFUs was observed upon the addition of 0.5 or 2 % lactose on glucose (Fig. 4a), but was not observed on galactose (Fig. 4b). Contradicting the observation by Hasan and Durr [14], we did not detect full repression in the presence of glucose.
Fig. 4.
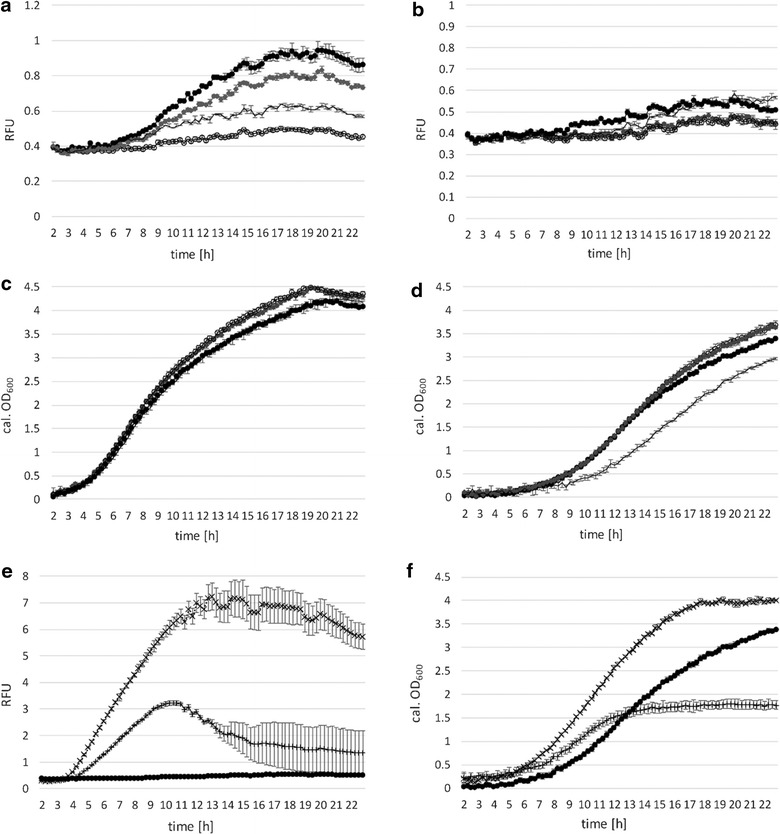
Promoter characteristics of PlacA and growth behavior. a, c MRS medium with glucose and 5 μg ml−1 CM; b, d MRS medium with galactose and 5 μg ml−1 CM; e, f MRS medium with 5 μg ml−1 CM. Mean values of four replicates are given and standard deviations are indicated. Filled circle induced, 2 % lactose. Filled diamond induced, 0.5 % lactose. Circle non-induced. a, b specific expression levels of mCherry under control of PlacA after induction with 0.5 and 2 % lactose (or no induction) after 2 h in BioLector® micro-fermentation. Change of mCherry expression (RFUs, relative fluorescent units) over time (hours) of PlacA mediated expression in comparison to the negative control is shown (solid line). c, d corresponding calculated OD600 values. e, f RFU and growth in selective MRS with lactose as main carbon source; x: induced, 2 % lactose; +: induced, 0.5 % lactose; filled circle induced, 2 % lactose and 2 % galactose as additional carbon source
Catabolite inhibition through diminished entry of lactose into the cell could explain why calculated OD600 does not increase with additional carbon source (Fig. 4c, d). Negative control (without the promoter/repressor fragment; Fig. 4a, b, solid line) grew weaker on galactose without obvious reason.
Compared to the negative control and compared to growth on glucose, minor growth impairment of the plasmid containing cells on 2 % galactose (Fig. 4d) or 2 % lactose (Fig. 4f) as carbon source was observed. Figure 4e, f show mCherry expression and growth on selective MRS medium with 0.5 and 2 % lactose as the sole carbon source and inducer. In contrast to data presented in Fig. 4a, mCherry expression increases, showing the catabolite repressive effect of glucose and galactose on PlacA or on cell entry of lactose. An increase from 0.5 to 2 % lactose increases expression (Fig. 4e) and growth (Fig. 4f). However, obtained calculated OD600 values on galactose and induction with lactose (Fig. 4c, f, filled circle) were comparable and did not increase, albeit the twofold amount of carbon source was available.
The chromosomally encoded lac locus (lp_3468, lp_3469 and lp_3470) as well as existence of a second lac locus (lp_3483, lp_3484), as indicated for L. plantarum WCSF1 genomic sequence [21], might interfere with usage of lactose as inducer, since L. plantarum 3NSH can utilize lactose as carbon source. The expression levels of mCherry under control of endogenous PlacA were rather low on glucose or galactose, but when lactose was used as sole carbon source and inducer, expression levels improved significantly.
The synthetic promoter PlacSynth is inducible by IPTG
The lacA promoter and the lacI promoter/repressor are widely used for many different E. coli based expression systems and many mutant versions are available [48]. Therefore, we synthesized a DNA template consisting of the promoter P2083 of L. buchneri CD034 gene LBUCD034_2083 [16], containing two operator binding sites of the E. coli LacI repressor. We inserted a codon optimized version of the E. coli LacI repressor gene (Additional file 2: Figure S2) under control of the constitutive promoter P0234 of the L. buchneri CD034 gene LBUCD034_0234 [16]. Operator binding sites (O1 and Oid) for Lac repressor binding were selected according to Oehler and colleagues [30] and integrated into P2083. In E. coli, a third operator binding site (O2) is encoded within the coding sequence of lacA [31]. This downstream cis-acting regulative sequence is involved in DNA bending and interaction with LacI multimers. But because integration of O2 sequence into the mCherry coding sequence was not realizable, O2 was not included in our constructs.
A synthetic regulative element for mCherry expression (P0234_lacI_P2083_mCherry) was constructed and promoter sequence and regulative elements are shown in Fig. 1c. LacI binding sites, operator binding sites (O1 and Oid), RBS and P2083 are indicated. The consecutive construct is termed PlacSynth and cloned into pCDLbu1 (Table 2; Fig. 2a). The resulting expression vector pCDLbu1_PlacSynth_mCherry is depicted in Fig. 2d.
Table 2.
Plasmids and strains used in this study
| Plasmid | Reference | Size (bp) | Relevant characteristics | |
|---|---|---|---|---|
| pET-30a | Novagen | 5400 | T7 promoter, T7 terminator | |
| pE194 | [18] | 3728 | Erythromycin resistance gene (ermE) | |
| pCD256 | [43] | 4790 | Low copy plasmid in L. plantarum | |
| pCDLbu1 | [15] | 5776 | High copy plasmid in L. plantarum | |
| pCDLbu1_PT7_mCherry_TT7_Ery | This study | 6425 | T7 RNA polymerase specific promoter | |
| pCDLbu1ΔEc_PT7_mCherry_TT7_Ery | This study | 3989 | T7 RNA polymerase specific promoter, without sequences for replication and selection in E. coli | |
| pCDLbu1ΔEc_P11_mCherry | [47] | 3809 | ||
| pCD256_PlacSynth_mCherry | This study | 6959 | Low copy plasmid; promoter PlacSynth; gene of interest mCherry | IPTG (1 mM) |
| pCD256_PlacSynth_T7RNAP | This study | 8882 | Low copy plasmid; promoter PlacSynth; gene of interest T7 RNA polymerase | IPTG (1 mM) |
| pCDLbu1_PlacSynth_mCherry | This study | 8164 | High copy plasmid; promoter PlacSynth; gene of interest mCherry | IPTG (1 mM), TMG (17 mM) |
| pCDLbu1_PlacA_mCherry | This study | 7917 | High copy plasmid; promoter PlacA; gene of interest mCherry | Lactose (0.5 -2 % w/v) |
| pCDLbu1_PxylA_mCherry | This study | 8077 | High copy plasmid; promoter PxylA; gene of interest mCherry | Xylose (0.2 -2 % w/v) |
| pCDLbu1_PxylA(nativeRBS)_mCherry | This study | 8077 | High copy plasmid; promoter PxylA; gene of interest mCherry; native RBS and spacer sequence from B. megaterium DSMZ 319 xylA | Xylose (2 % w/v) |
| Strains | ||||
| B. megaterium DSMZ 319 | DSMZ | |||
| E. coli Neb10β | NEB | |||
| L. plantarum CD033 | [43] | |||
| L. plantarum 3NSH | [17] | Plasmid cured L. plantarum CD033 |
According to E. colilac-operon regulation, we tested mCherry expression subsequent to induction with IPTG. Increasing concentrations in the range of 0.1 to 5 mM (0.1, 0.5, 1.0, 2.0 and 5.0 mM) were tested and showed that already 0.1 and 0.5 mM induce PlacSynth sufficiently in L. plantarum 3NSH. Lower IPTG concentrations (like 0.1 and 0.5 mM) are in the range of common E. coli implementations. Moreover, 1 mM IPTG led to saturated induction of mCherry in L. plantarum 3NSH (Fig. 5a, b) and no further increase of expression was observable at augmented concentrations from 1 to 5 mM (data not shown). Additionally, TMG was tested for PlacSynth induction. We observed similar mCherry expression with induction of 17 mM TMG compared to 1 mM IPTG (data not shown). For L. plantarum NC2 it was shown that ß-galactosides are transported via ATP driven proton motive force [19]. Induction of recombinant gene expression in a fermentation setting (BioLector® measurement) with IPTG (and TMG) is shown here for the first time in L. plantarum.
Fig. 5.
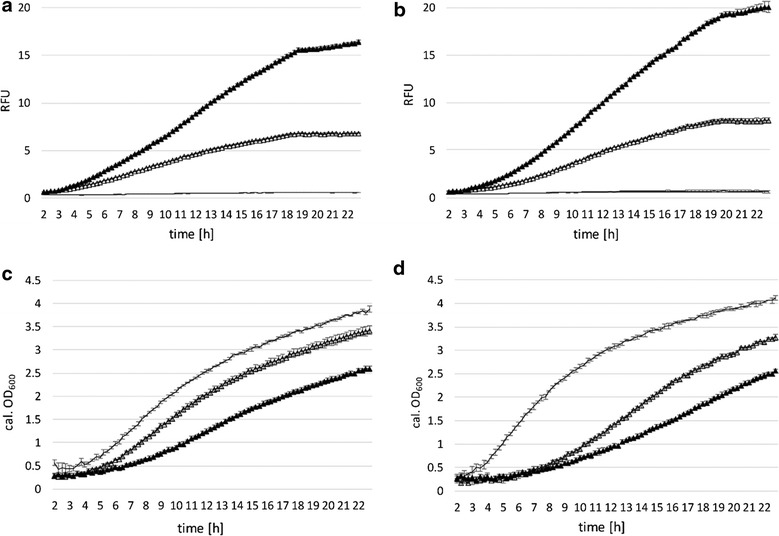
Promoter characteristics of PlacSynth and growth behavior. a, c MRS medium with glucose and 5 μg ml−1 CM; b, d MRS medium with galactose and 5 μg ml−1 CM, mean values of four replicates are given and standard deviations are indicated. Filled triangle induced; triangle non-induced. a, b specific expression levels of mCherry under control of PlacSynth after induction with 1 mM IPTG (or no induction) after 2 h in BioLector® micro-fermentation. Change of mCherry expression (RFUs, relative fluorescent units) over time (hours) of PlacSynth mediated expression in comparison to the negative control is shown (solid line). c, d calculated OD600 values. Solid line negative control
Comparative growth and induction on selective media with glucose or galactose are shown in Fig. 5c and d. Induced cultures show growth impairment (compared to the non-induced cultures) on both carbon sources, though growth on glucose as carbon source is preferred, thus leading to higher expression values and biomass (Fig. 5a, c). Overall, regarding expression levels as well as repression under non-induced conditions in both tested media variations, PlacSynth performed better than PxylA (Fig. 3) and PlacA (Fig. 4). Negative control (without the promoter/repressor fragment) showed no mCherry expression (Fig. 5; solid line). Therefore, measured expression levels correlate to induction of PlacSynth through thiogalactosides, such as IPTG (and TMG), and basal expression might be caused by weak repression of P2083.
Consequently, we suggest limited stoichiometric availability of the repressor LacI, resulting in incomplete repression of PlacSynth by the repressor. LacI binds to the operator sites by forming tetramers, which might not be possible if LacI availability is not sufficient [31, 49]. A stronger promoter for LacI expression (instead of P2083) might increase repressor levels and improve transcription control. Additionally, the operator O2 downstream of the start codon, which is originally present within the coding sequence of β-galactosidase [31], is absent within the mCherry sequence. Therefore, bending of DNA via binding of tetrameric Lac-repressor to two adjacent operators for sufficient repression is not possible. Albeit, it was reported for E. coli that the presence or absence of operator O2 does not have an impact on lac operon expression anyhow [27].
Apparently, on selective medium with glucose as carbon source, the ratio of induced expression to basal expression under non-induced conditions of PlacSynth was highest compared to PxylA or PlacA (Figs. 3, 4). However, the PlacSynth mediated expression after induction is still moderate and thus appropriate for the regulation of T7 RNA polymerase based expression of mCherry. Therefore, this synthetic promoter/repressor fragment was used for establishment of the inducible T7 system in L. plantarum 3NSH.
T7 RNA polymerase driven mCherry expression in L. plantarum 3NSH
In order to establish an orthologous expression system in L. plantarum, we combined the synthetic repressor/promoter system PlacSynth (Fig. 1c). The adapted E. coli phage T7 RNA polymerase was applied to establish two compatible plasmids: one contained a codon optimized version of the T7 RNA polymerase (Additional file 3: Figure S3) under the control of PlacSynth (Fig. 2f) and the second plasmid contained mCherry under control of the T7 RNA polymerase promoter PT7 (Fig. 2g).
For inducing T7 RNA polymerase expression, we used 1 mM IPTG, equally to PlacSynth induction (Fig. 5). Results of the expression experiment are shown in Fig. 6a. Induction with 1 mM IPTG led to augmented expression of the reporter protein compared to non-induced conditions. Some basal expression of the reporter gene was detected under non-induced conditions similarly to results with PlacSynth (Fig. 5a). Therefore, the plasmid containing mCherry under control of the T7 RNA polymerase promoter (pCDLbu1ΔEc_PT7_mCherry_TT7_Ery) was tested in absence of the second plasmid, which provides the T7 RNA polymerase (pCD256_PlacSynth_T7RNAP). Thereby, we observed no mCherry expression neither with nor without IPTG (Fig. 6a, solid line).
Fig. 6.
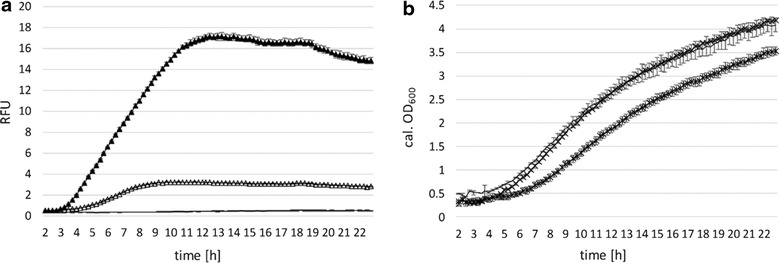
Promoter characteristics and analysis of mCherry expression under control of PT7, regulated by T7 RNA polymerase under control of PlacSynth, and growth behavior. a Change of mCherry expression (RFUs, relative fluorescent units) over time (hours) regulated by T7 RNA polymerase dual plasmid system in BioLector® micro-fermentation is shown. T7 RNAP is under control of PlacSynth and induced with 1 mM IPTG (or non-induced) after 2 h. MRS medium with glucose either 5 μg ml−1 CM or 5 µg ml−1 Ery (or both for the dual plasmid system). –x-: induced; mean values of four replicates are given and standard deviations are indicated. –: non-induced; mean values of three replicates are given and standard deviations are indicated. b Calculated OD600 values. Solid line negative control (n = 3; pCDLbu1ΔEc_PT7_mCherry_TT7_Ery)
Although, constitutive expression using the P11 promoter was significantly higher, inducible expression based on the T7 RNA polymerase system serves as a valuable tool for regulated gene expression at moderate levels. However, growth was not affected by PlacSynth regulating a dual plasmid expression system (Fig. 6b) compared to strains with plasmid pCDLbu1_PlacSynth_mCherry (Fig. 5c). This effect could also be contributed to the different plasmid backbones (pCDLbu1 and pCD256, Fig. 2d, f).
For constructing the T7 polymerase encoding plasmid, the low copy plasmid pCD256 was used (Table 2). The second plasmid (mCherry under control of PT7) was established from pCDLbu1 (Table 2) without E. coli specific sequences. Thereby we generated a smaller plasmid and less genetic load. Intentionally we introduced two different origins of replication within a cell, which is known to be preferred due to plasmid incompatibility [29]. Chromosomal integration of expression cassettes has been shown previously in L. plantarum [24, 36] and would be a feasible strategy for generating a stable T7 RNA polymerase expressing host strain. Such a L. plantarum strain would be the basis for a new T7 based expression system in a food grade host, providing specific regulation and easy exchange of any target gene that is under control of the T7 promoter PT7.
Comparative analysis and semi-quantitative Western blot
The constitutive L. plantarum promoter P11 (expression vector pCDLbu1ΔEc_P11_mCherry) served as a benchmark in a comparative analysis [38, 47]. Plasmid pCDLbu1ΔEc_P11_mCherry is shown in Fig. 2e. Measurements of expression levels with plasmid pCDLbu1ΔEc_P11_mCherry were included for intrinsic comparison, because it was previously shown to yield strongest expression of mCherry amongst several tested variants in L. plantarum CD033 [47], the parental strain of L. plantarum 3NSH.
Expression levels of mCherry under control of P11 were compared to PxylA, PlacA, PlacSynth, and the PlacSynth regulated T7 RNA polymerase, in Fig. 7a. Growth curves of producing strains and wild type are shown in Fig. 7b. MRS selective medium was used with galactose as carbon source and induction with xylose for pCDLbu1_PxylA_mCherry, and with glucose plus induction with 2 % lactose for pCDLbu1_PlacA_mCherry. Growth on glucose and induction with 1 mM IPTG was used for pCDLbu1_PlacSynth_mCherry, and subsequently for the T7 dual plasmid system. P11 driven expression (pCDLbu1ΔEc_P11_mCherry) is more effective (Fig. 7a). Expression levels of PlacA were quite low for direct comparison with promoter P11, but results with PlacA were also included in Fig. 7a and b.
Fig. 7.
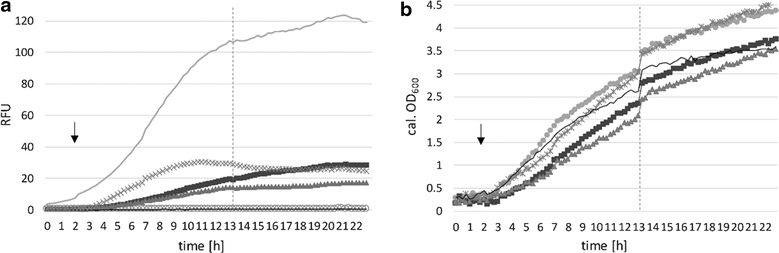
Expression and growth comparison of inducible promoters to constitutive promoter. BioLector® micro-fermentation measurement for 23 h in selective MRS medium with glucose with low pH FlowerPlate at 30 °C (individual values). Dotted vertical line indicates sampling point (200 µl) for semi-quantitative Western blot, 13 h after start, followed by ongoing measurement, therefore OD600 curves are shifted after sampling. Arrow indicates induction time point (or absence of inducer). Solid line mCherry under control of constitutive P11 promoter (pCDLbu1ΔEc_P11_mCherry), –x-: T7 RNA polymerase based dual plasmid system induced with 1 mM IPTG, square PxylA induced with 2 % xylose, triangle PlacSynth induced with 1 mM IPTG, circle PlacA induced with 2 % lactose. a specific expression levels of mCherry under control of inducible promoters with respective inducers. b corresponding calculated OD600 values
A semi-quantitative Western blot of the inducible promoter systems at induced and non-induced conditions on glucose (Fig. 8a, c) or galactose (Fig. 8b) was performed. Sample point is indicated as vertical dotted line in Fig. 7 after 13 h of growth. About 5 µg biomass per slot were applied, the commercially obtained positive control (mCherry-His6; 28.8 kDa) was applied at concentrations of 25 and 50 ng per slot. P11 samples were applied undiluted and 1:5 diluted due to stronger expression compared to the inducible systems (Fig. 7a). The Western blot shows better inducibility of the PlacSynth system on glucose medium (Fig. 8a) whereas PxylA induction is more distinct on galactose medium (Fig. 8b) and the T7 system is inducible on glucose (Fig. 8c). We also observed basal transcription under non-induced conditions for all compared promoter/repressor systems. This is in accordance with the BioLector® measurements.
Fig. 8.
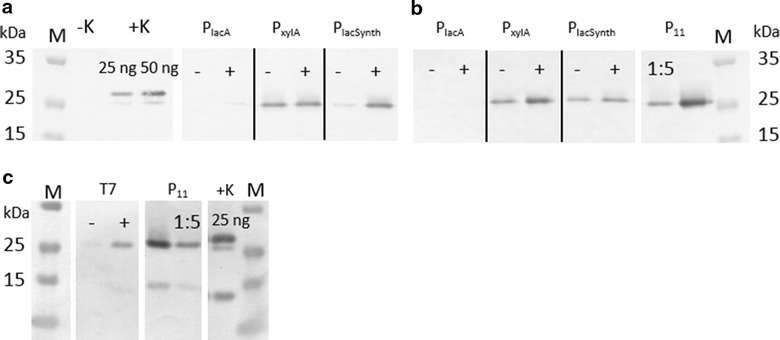
Semi-quantitative Western blot analysis. Evaluation of intracellular mCherry (26.7 kDa) expression after 13 h of BioLecture® micro-fermentation measurement with low pH FlowerPlate at 30 °C. Five µg biomass per slot were applied. a MRS medium with glucose and 5 μg ml−1 CM; b MRS medium with galactose and 5 μg ml−1 CM; c T7 RNAP dual plasmid system, MRS medium with glucose and 5 μg ml−1 CM and 5 μg ml−1 Ery. − non-induced, + induced with the respective inducer. P11 sample (mCherry under control of constitutive P11 promoter) is additionally diluted 1:5 to adjust to expression levels of inducible promoters (in accordance BioLecture® results). M pre-stained protein ladder; +K positive control mCherry(His6) 28.8 kDa; −K L. plantarum 3NSH wild type
Suitable and controllable expression levels were achieved by PlacSynth induction with IPTG on glucose and galactose (Fig. 5a, b). Apparently, the ratio of induced expression to basal expression under non-induced conditions of PlacSynth on selective medium with glucose as carbon source was best (compared to PlacA or PxylA). Moreover, the PlacSynth mediated expression after induction was still moderate and thus, more appropriate for the regulation of T7 RNA polymerase based expression of mCherry. By using a stronger promoter for lacI, expression, repression of PlacSynth might be improved. If necessary, this strategy could also be applied for xylR promoter and lacR promoter. Best regulation of induced and non-induced conditions while yielding similar expression levels was achieved by T7 RNA polymerase dual plasmid based system induced with 1 mM IPTG (Fig. 7a, 8c). The adapted T7 RNA polymerase was successfully established here for L. plantarum.
Conclusions
In this study, we tested and compared three different promoter-repressor systems for induced recombinant protein expression (red fluorescent protein mCherry) in plasmid free L. plantarum 3NSH. Reporter gene and regulatory elements were cloned into the high copy number plasmid pCDLbu1. The endogenous LacA promoter (PlacA) derived from L. plantarum 3NSH showed only weak reporter gene expression upon induction with 2 % lactose, which was found to be the exclusive inducer so far. Glucose and galactose acted as repressors of PlacA. With lactose as single carbon source better expression levels were obtained. The XylA promoter (PxylA) derived from B. megaterium DSMZ 319 was tested in combination with the expression of the repressor XylR. Upon induction with 0.2–2 % xylose, we measured increased mCherry expression during exponential phase, and repression under non-induced conditions with galactose as the carbon source. A synthetic promoter (PlacSynth), based on the E. coli derived lac operon resulted in moderate expression levels after induction with IPTG and TMG. PlacSynth was used efficiently for the establishment of a dual plasmid system for well-regulated T7 RNA polymerase expression, and transcription of mCherry under control of the T7 RNA polymerase promoter.
Some feasible suggestions for inducible recombinant protein expression in L. plantarum 3NSH are presented in this study. Expression levels of recombinant protein are however, much lower as compared to expression levels driven by the constitutive P11 promoter. Plasmid pCDLbu1ΔEc_P11_mCherry served as a benchmark and has been described previously [47]. Additionally, differences of expression in exponential phase, initiated by varying promoters, decreased during prolonged fermentation. In stationary phase (after 22 h) mCherry levels of tested inducible promoters are aligned. Yet, depending on the recombinant protein (e.g. amino acid composition and post-translational modifications) or experiment outlook (e.g. short time setting or production of cell toxic products) promoters, which are inducible by conventional sugars or well-established inducers are of particular interest. Although general knowledge of recombinant protein expression (e.g. therapeutics or metabolites) in lactobacilli steadily increases, efficiency and expression levels are not yet comparable to E. coli based systems. Recombinant gene expression usually exerts additional metabolic burden for the host. This often results in unstable genetic constructs, inhibition of cell growth and/or plasmid loss. Therefore, inducible expression systems where transcription of the target gene can be tightly controlled are preferable. The presented expression systems might behave different in other Lactobacillus strains, the adaption of new promoter/repressor systems, and in particular a T7 RNA polymerase based expression systems for L. plantarum, is anticipated to contribute to a flexible genetic tool box for cell engineering and recombinant protein expression in lactic acid bacteria.
Methods
Enzymes and gene synthesis
All restriction and modifying enzymes, as well as Q5 DNA polymerase, were purchased from New England Biolabs (NEB). Primers (Table 1) were obtained from Integrated DNA Technologies (IDT) and phosphorylated primers were synthesized by Sigma-Aldrich.
The ribosome binding site (RBS) was identical for all constructs. Identical Shine-Dalgarno sequence (SDS) and spacer region (bold) was selected for every construct according to SOPT#9 (5´-AAGGAGGAAATTATACATG-3´), tested for efficient mCherry (start codon underlined) expression in L. plantarum CD033 [47].
Reporter gene mCherry and the synthetic LacR repressor/promoter fragment (PlacSynth) were codon optimized for L. plantarum WCSF1 using http://www.jcat.de/ and synthesized by GeneArt® (life technologies). Promoter PlacSynth, T7 RNA polymerase and transcriptional terminator from L. buchneri CD034 D-lactate hydrogenase gene (AFS00145.1) [16] were also codon optimized as described above and synthesized by GeneArt®. Nucleotide sequence of the codon optimized synthetic promoter/LacI repressor system is shown in Additional file 2 and T7 RNA polymerase is shown in Additional file 3.
Strains and cultivation conditions
Plasmids were constructed and propagated in E. coli Neb10β and clones were selected on LB agar plates with 100 µg ml−1 Ampicillin at 37 °C. Sequence positive plasmids were amplified for transformation into plasmid cured L. plantarum 3NSH [17]. Clones were selected on either MRS agar plates with either 5 µg ml−1 chloramphenicol (CM), 5 µg ml−1 erythromycin (Ery) or both combined at 30 °C.
In liquid medium, E. coli strains were cultivated under agitation at 37 °C in LB-medium. L. plantarum 3NSH was cultivated at 30 °C under oxygen limitation without agitation in MRS medium [7], supplemented with either 2 % (w/v) d-glucose or 2 % (w/v) D-galactose. B. megaterium DSMZ 319 was purchased from the “Deutsche Sammlung von Mikroorganismen und Zellkulturen” (Braunschweig, Germany) and was cultivated aerobically at 30 °C in nutrient medium. Antibiotics were added as required equally to solid media preparations.
Plasmid extraction was performed with plasmid purification kit for high-copy E. coli plasmids (NucleoSpin® Plasmid, Macherey–Nagel). After sequence verification, plasmids were used to transform L. plantarum 3NSH [44]. Plasmid isolation from L. plantarum 3NSH was performed according to Sambrook and Russel [39] with addition of 10 mg ml−1 lysozyme (Merck, 105281) and RNase (R6513, Sigma) to the resuspension buffer and incubation for 30 min at 37 °C before cell lysis.
Construction of the PxylA/xylR-plasmid
The xylR repressor/promoter fragment (PxylA) was amplified from genomic DNA of B. megaterium DSMZ 319. Therefore, an overnight culture was used for DNA extraction, with pre-treatment described for Gram-positive bacteria (DNeasy Blood and Tissue Kit, Quiagen). Primers B_mega_XylOP_out_F and B_mega_XylOP_R(SpeI, ScaI, BamHI) were used for amplification of the xylR repressor and xylA promoter genes with native RBS of xylA. Primers mCherry_w/o_RBS_XbaI and p256_miniori_for were used for amplification of reporter gene mCherry and Terminator Tldh from L. casei BL23 (L-lactate dehydrogenase gene, LCABL-06930) from pCD256_PlacSynth_mCherry (Fig. 2h). Constructs were ligated at SpeI and XbaI complementary overhangs and amplified via a PCR using B_mega_XylOP_F_MfeI, KpnI and Tldh_amp_PstI_R, digested with KpnI and PstI and cloned into the pCDLbu1 plasmid (Fig. 2a) with an origin of replication for E. coli and L. plantarum [43].
For generating expression constructs with identical RBS and spacer sequence, we exchanged the native RBS of xylA (Additional file 1) with the RBS sequence SOPT#9 [47]. Nucleotide sequence of final PxylA is shown in Fig. 1a. Therefore, we performed a continuous PCR around the ligated plasmid with phosphorylated primers B_mega_XylOP_newRBS_XbaI_Phos_R and mCherry_Phos_F. After ligation, plasmid pCDLbu1_PxylA_mCherry (Fig. 2b) was transformed into L. plantarum 3NSH. For screening and sequencing purposes, primers B_mega_XylOP_seq_F and B_mega_XylOP_seq_R were used.
Construction of the PlacA/lacR-plasmid
The plasmid pCDLbu1_PxylA_mCherry (Fig. 2b) was SacI and XbaI digested and fused with the LacR repressor/promoter fragment (PlacA). This fragment was amplified from genomic DNA of L. plantarum 3NSH (DNeasy Blood and Tissue Kit, Quiagen) with the primers LacI_Lplant_F_SacI and LacI_Lplant_R_XbaI and sequenced (sequence of PlacR and PlacA is shown in Fig. 1b). BLASTn analysis showed 99 % coverage (three mismatches) with the transcription regulator lacR gene of L. plantarum WCSF1. After ligation and transformation into E. coli Neb10β, sequence positive plasmid pCDLbu1_PlacA_mCherry (Fig. 2c) was recovered and transformed into L. plantarum 3NSH. For screening and sequencing purposes, primers lacR_Gal_seq_R and lacR_Gal_seq_F were used.
Construction of the PlacSynth/lacI-plasmid
Consecutive arrangement of synthetic PlacSynth/LacI regulon (P0234_lacI_P2083) is shown in Fig. 1c. The promoter from L. buchneri CD034 gene LBUCD034_0234 [16] was selected for transcription of lacI, encoding the E. coli derived codon optimized LacI repressor (Additional file 2). The promoter from L. buchneri CD034 gene LBUCD034_2083 [16] was selected for regulation of the reporter gene mCherry. Operator binding sites [30] were artificially inserted into P2083 sequence. Operator sequence O1 und Oid were adapted from E. coli [31]. Both constitutive L. buchneri CD034 promoters were identified within our group in previous promoter library experiments (data not shown).
The synthetic promoter was amplified from the synthetic GeneArt® construct with primers PlacSynth_F_SacI_EcoRI and M13_R_NheI, digested with EcoRI and BamHI and ligated into EcoRI and BamHI digested pCD256 [43], receiving pCD256_PlacSynth. This plasmid was amplified and proliferated in E. coli. Reporter gene mCherry was amplified from pCDLbu1ΔEc_P11_mCherry [47] with primers mCherry_RBS_XbaI and mCherry_R_BamHI, digested and ligated with pCD256_PlacSynth (cut XbaI and BamHI) plasmid. After transformation of E. coli and positive colony screening, pCD256_PlacSynth_mCherry (Fig. 2h) was recovered and the insert was amplified via PCR with primers PlacSynth_F_SacI_EcoRI and Tldh_amp_R_PstI, followed by digestion with EcoRI and PstI and ligation into digested pCDLbu1 (Fig. 2a) vector. The plasmid pCDLbu1_PlacSynth_mCherry (Fig. 2d) was amplified in E. coli and subsequently introduced into L. plantarum 3NSH. For screening and sequencing purposes, primers mCherry_seq_R, mCherry_seq_F and Cat_seq 2_back were used.
Construction of the T7 RNA polymerase based dual plasmid expression system
DNA was amplified with the primers T7_RNAP_Lp_RBS and T7_RNAP_Lp_Term_R_SalI from a synthetic template. The fragment was XbaI and SalI digested and ligated into the XbaI and SalI digested pCD256_PlacSynth_mCherry plasmid (Fig. 2h), thus receiving the plasmid pCD256_PlacSynth_T7RNAP (Fig. 2f).
The second plasmid (pCDLbu1ΔEc_PT7_mCherry_TT7_Ery, Fig. 2g) was cloned stepwise. The reporter gene mCherry was amplified with primer M13_2_F and mCherry_R_BamHI from plasmid pCDLbu1_PlacSynth_mCherry (Fig. 2d), digested with XbaI and BamHI and ligated into digested pET-30a plasmid (Table 2). Primers T7-Promoter_SacI and T7-Terminator_SalI were used to amplify the PT7_mCherry_TT7 fragment from the established pET30a-mCherry plasmid. The erythromycin resistance gene (ermE) was amplified with primers ery_KasI_back and oripE194_seq_back from pE194 [18]. The ermE fragment was digested with ClaI to fuse it with the ClaI digested PT7_mCherry_TT7 fragment, followed by an enrichment PCR with primers ery_KasI_back and T7-Terminator_SalI. The resulting fragment was digested with KasI and BspEI and ligated into the KasI and XmaI digested plasmid pCDLbu1 [15], resulting in the plasmid pCDLbu1_PT7_mCherry_TT7_Ery. After amplification of the plasmid in E. coli JM109, E. coli specific sequences (pMB1 origin of replication and ampicillin resistance gene) were removed by PCR with primers Ery_F_NheI and M13_R_NheI. The PCR product of plasmid pCDLbu1ΔEc_PT7_mCherry_TT7_Ery (Fig. 2g) was digested with NheI, circularized by ligation and directly used to transform L. plantarum 3NSH [44].
Subsequent to sequence verification of a colony harboring plasmid pCDLbu1ΔEc_PT7_mCherry_TT7_Ery was used for establishing competent cells and transformed with plasmid pCD256_PlacSynth_T7RNAP, resulting in a strain carrying two different expression vectors. After transformation, cells were selected on MRS plates with 5 µg ml−1 CM and 5 µg ml−1 Ery. Colonies were screened for both expression plasmids verified by sequencing.
Construction of negative controls
For the inducible promoter/repressor constructs, a negative control plasmid was established by removing the whole inserted promoter/repressor fragment (plasmid with mCherry coding sequence and terminator; termed empty). The plasmid backbone, which is identical for every construct, was amplified with primers Pempty_SacI_R and mCherry_RBS_SacI_F from pCDLbu1_PlacSynth_mCherry. The thereby established plasmid pCDLbu1_X_mCherry allows testing for mCherry expression, driven by possible read through from upstream regulatory sequences or possible unknown upstream promoter sequences.
As negative control for the T7 RNA polymerase dual-plasmid system, we used a clone harboring only plasmid pCDLbu1_PT7_mCherry_TT7_Ery. Thereby we tested if any other factors except T7 RNA polymerase contributes to mCherry expression.
Induction conditions
Over-night cultures were adjusted to OD600 0.2 in the respective liquid medium. After 2 h of growth at 30 °C in the BioLector® micro-fermentation system, cultures were induced with the respective inducer 1:10 into each well, thus requiring that preparations of each inducer is tenfold concentrated in MRS-medium. Non-induced cells were prepared and tested simultaneously, but without the inducer (MRS medium only).
Tested sugars were used in the D(+)-configuration and weighted as solids (weight) per volume medium (w/v). The promoter PxylA was induced with xylose. Either 0.2, 1 or 2 % xylose were used for induction. Therefore, 100 or 200 g l−1 D-xylose was added to the medium (MRS 5 µg ml−1 CM without glucose), heated in a water bath and sterile filtrated (0.2 µm) and diluted accordingly. Other preliminary tested sugars such as fructose, arabinose, maltose, as well as galactose were prepared likewise.
The synthetic promoter PlacSynth was induced with IPTG (VWR) and TMG (M8146, Sigma). Standard final concentration for IPTG was 1 mM. Therefore, 10 mM IPTG was dissolved in selective MRS medium and 1:10 diluted into respective wells. For testing minimum and maximum induction concentrations, we used dilutions ranging from 0.1 to 5 mM IPTG per well (0.1, 0.5, 1, 2 and 5 mM). 17 mM TMG was also tested for induction of PlacSynth in selective MRS medium, as well as 2 % lactose. For PlacA standard conditions were selective MRS medium with 2 % glucose, 2 % maltose or 2 % galactose or without additional carbon source and induction with 0.5 or 2 % lactose after 2 h.
BioLector® and Tecan reader measurements of intracellular mCherry expression
Pre-measurements were performed in an Infinite® M1000 PRO Tecan microplate reader as described elsewhere [47]. The BioLector® micro-fermentation system (m2p-labs Germany) was also used for online measurement. Overnight cultures (in selective MRS medium with glucose or galactose) were diluted to an OD600 of 0.2 in the respective liquid medium. 720 μl of each sample were pipetted per well of MTP-48-BOH FlowerPlate® (low pH, Lot No. 1408) or MTP-48-B FlowerPlate® (without optodes, Lot No. 1402) in quadruplicates and sealed with sterile tape adhesive sealing (Nunc, 732-2610). Samples were induced after 2 h of growth. Under sterile conditions 80 µl of the particular inducer (tenfold concentrated in MRS medium) was pipetted into the respective well. 80 µl MRS medium were added to non-induced samples and controls, and plates were covered again with sealing tape. Results were analyzed after 23 h using the BioLection 2.3.13 software using a previously described calibration curve for L. plantarum [47]. Calibration parameters were set for 30 °C according to the manufacturer’s recommendations.
SDS-PAGE and Western blot analysis
For Western blot analysis of intracellular mCherry (to compare the expression levels of under induced and non-induced conditions in selective MRS medium with glucose as carbon source) cells were collected at late exponential phase after 13 h of growth (dotted vertical line in Fig. 7). Recombinant purified mCherry with His6-tag (28.8 kDa) was purchased from BioVision (4993-100) and used as a positive control in defined concentrations per slot (25 and/or 50 ng). Per slot we applied samples corresponding to 5 µg biomass each (calculated as described below). The reference strain (pCDLbu1ΔEc_P11_mCherry) was applied undiluted and 1:5 diluted for adaption to mCherry yields obtained by induction of the inducible promoters.
The pellet of 200 µl culture was washed with PBS, centrifuged and pellet was re-suspended in 200 µl PBS. OD600 was measured of each sample. For analyzing equal amount of biomass 0.4/OD600 for each sample was calculated and used for intracellular analysis. A spatula tip of zirconium beads (BMBZ 100-250-17) was added to each sample, followed by alternating 30 s vortex and 30 s on ice; repeated for ten times. To remove cell debris and beads, samples were centrifuged at 4 °C full speed and supernatant was transferred into a fresh tube. A volume of 15 µl of each sample were mixed with 2 × LDS loading buffer and incubated at 99 °C for 10 min. Afterwards, 15 µl per sample and 5 µl protein ladder (Fermentas, SM0671) were loaded onto a NuPAGE® 12 % BisTris gels and electrophoresis was run with MOPS buffer. The gel was blotted onto a PVDF membrane. Anti mCherry antibody (Biovision, 5993-100; 1:10.000) and AP-linked anti-rabbit secondary antibody (Sigma A9919; 1:20.000) were used for detection of mCherry. BCIP/NBT Color Development Substrate (Promega, S3771) was used for staining the blot.
Authors’ contributions
The work presented here was carried out in collaboration between all authors. S Heinl, RG and S Heiss defined the research theme and designed the experiments. S Heiss, AH, CT, MS and EE carried out the laboratory experiments. S Heiss analyzed the data, interpreted the results and prepared this manuscript with input, feedback and advice from S Heinl and RG. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Christian Doppler Research Association, Vienna, Austria.
Competing interests
The authors declare that they have no competing interests.
Additional files
10.1186/s12934-016-0448-0 Comparison of RBS and spacer sequence of PxylA (native RBS) and PxylA.
10.1186/s12934-016-0448-0 Sequence of the codon optimized version of the E. coli lacI repressor gene and PlacSynth.
10.1186/s12934-016-0448-0 Sequence of the codon optimized version of the T7 RNA polymerase gene.
Contributor Information
Silvia Heiss, Email: silvia.heiss@boku.ac.at.
Angelika Hörmann, Email: te14m023@technikum-wien.at.
Christopher Tauer, Email: Ctauer@groupwise.boku.ac.at.
Margot Sonnleitner, Email: margot.sonnleitner@sandoz.com.
Esther Egger, Email: eegger@groupwise.boku.ac.at.
Reingard Grabherr, Email: reingard.grabherr@boku.ac.at.
Stefan Heinl, Phone: 0043-1-47654-6926, Email: stefan.heinl@boku.ac.at.
References
- 1.Arasu MV, Jung MW, Kim dH, Ilavenil S, Jane M, Park HS, Al-Dhabi NA, Jeon BT, Choi KC. Enhancing nutritional quality of silage by fermentation with Lactobacillus plantarum. Indian J Microbiol. 2014;54:396–402. doi: 10.1007/s12088-014-0473-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhavsar AP, Zhao X, Brown ED. Development and characterization of a xylose-dependent system for expression of cloned genes in Bacillus subtilis: conditional complementation of a teichoic acid mutant. Appl Environ Microbiol. 2001;67:403–410. doi: 10.1128/AEM.67.1.403-410.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böhmer N, König S, Fischer L. A novel manganese starvation-inducible expression system for Lactobacillus plantarum. FEMS Microbiol Lett. 2013 doi: 10.1111/1574-6968.12105. [DOI] [PubMed] [Google Scholar]
- 4.Chaillou S, Pouwels PH, Postma PW. Transport of D-xylose in Lactobacillus pentosus, Lactobacillus casei, and Lactobacillus plantarum: evidence for a mechanism of facilitated diffusion via the phosphoenolpyruvate: mannose phosphotransferase system. J Bacteriol. 1999;181:4768–4773. doi: 10.1128/jb.181.16.4768-4773.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Urzo N, Martinelli M, Nenci C, Brettoni C, Telford JL, Maione D. High-level intracellular expression of heterologous proteins in Brevibacillus choshinensis SP3 under the control of a xylose inducible promoter. Microb Cell Fact. 2013;12:12. doi: 10.1186/1475-2859-12-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahl MK, Schmiedel D, Hillen W. Glucose and glucose-6-phosphate interaction with Xyl repressor proteins from Bacillus spp. may contribute to regulation of xylose utilization. J Bacteriol. 1995;177:5467–5472. doi: 10.1128/jb.177.19.5467-5472.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Man JC, Rogosa M, Sharpe ME. A medium for the cultivation of Lactobacilli. J Appl Microbiol. 1960;130–135.
- 8.del Rio B, Seegers JF, Gomes-Solecki M. Immune response to Lactobacillus plantarum expressing Borrelia burgdorferi OspA is modulated by the lipid modification of the antigen. PLoS One. 2010 doi: 10.1371/journal.pone.0011199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Douillard FP, de Vos WM. Functional genomics of lactic acid bacteria: from food to health. Microb Cell Fact. 2014 doi: 10.1186/1475-2859-13-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feld LE, Bielak E, Hammer K, Wilcks A. Characterization of a small erythromycin resistance plasmid pLFE1 from the food-isolate Lactobacillus plantarum M345. Plasmid. 2009;61:159–170. doi: 10.1016/j.plasmid.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 11.Fredriksen LC, Kleiveland R, Hult LT, Lea T, Nygaard CS, Eijsink VG, Mathiesen G. Surface display of N-terminally anchored invasin by Lactobacillus plantarum activates NF-κB in monocytes. Appl Environ Microbiol. 2012;78:5864–5871. doi: 10.1128/AEM.01227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geoffroy MC, Guyard C, Quatannens B, Pavan S, Lange M, Mercenier A. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl Environ Microbiol. 2000;66:383–391. doi: 10.1128/AEM.66.1.383-391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gösseringer R, Küster E, Galinier A, Deutscher J, Hillen W. Cooperative and non-cooperative DNA binding modes of catabolite control protein CcpA from Bacillus megaterium result from sensing two different signals. J Mol Biol. 1997;266:665–676. doi: 10.1006/jmbi.1996.0820. [DOI] [PubMed] [Google Scholar]
- 14.Hasan N, Durr IF. Induction of beta-galactosidase in Lactobacillus plantarum. J Bacteriol. 1974;120:66–73. doi: 10.1128/jb.120.1.66-73.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinl S, Spath K, Egger E, Grabherr R. Sequence analysis and characterization of two cryptic plasmids derived from Lactobacillus buchneri CD034. Plasmid. 2011;66:159–168. doi: 10.1016/j.plasmid.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Heinl S, Wibberg D, Eikmeyer F, Szczepanowski R, Blom J, Linke B, Goesmann A, Grabherr R, Schwab H, Pühler A, Schlüter A. Insights into the completely annotated genome of Lactobacillus buchneri CD034, a strain isolated from stable grass silage. J Biotechnol. 2012;161:153–166. doi: 10.1016/j.jbiotec.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Heiss S, Grabherr R, Heinl S. Characterization of the Lactobacillus plantarum plasmid pCD033 and generation of the plasmid free strain L. plantarum 3NSH. Plasmid. 2015;81:9–20. doi: 10.1016/j.plasmid.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Iordănescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch Roum Pathol Exp Microbiol. 1976;35:111–118. [PubMed] [Google Scholar]
- 19.Jeffrey SR, Dobrogosz WJ. Transport of beta-galactosides in Lactobacillus plantarum NC2. Appl Environ Microbiol. 1990;56:2484–2487. doi: 10.1128/aem.56.8.2484-2487.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerovuo J, Tynkkynen S. Expression of Bacillus subtilis phytase in Lactobacillus plantarum 755. Lett Appl Microbiol. 2000;30:325–329. doi: 10.1046/j.1472-765x.2000.00660.x. [DOI] [PubMed] [Google Scholar]
- 21.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MW, Stiekema W, Lankhorst RM, Bron PA, Hoffer SM, Groot MN, Kerkhoven R, de Vries M, Ursing B, de Vos WM, Siezen RJ. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci USA. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korneli C, Biedendieck R, David F, Jahn D, Wittmann C. High yield production of extracellular recombinant levansucrase by Bacillus megaterium. Appl Microbiol Biotechnol. 2013;97:3343–3353. doi: 10.1007/s00253-012-4567-1. [DOI] [PubMed] [Google Scholar]
- 23.Ladero V, Ramos A, Wiersma A, Goffin P, Schanck A, Kleerebezem M, Hugenholtz J, Smid EJ, Hols P. High-level production of the low-calorie sugar sorbitol by Lactobacillus plantarum through metabolic engineering. Appl Environ Microbiol. 2007;73:1864–1872. doi: 10.1128/AEM.02304-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leer RJ, Christiaens H, Verstraete W, Peters L, Posno M, Pouwels PH. Gene disruption in Lactobacillus plantarum strain 80 by site-specific recombination: isolation of a mutant strain deficient in conjugated bile salt hydrolase activity. Mol Gen Genet. 1993;239:269–272. doi: 10.1007/BF00281627. [DOI] [PubMed] [Google Scholar]
- 25.Maischberger T, Mierau I, Peterbauer CK, Hugenholtz J, Haltrich D. High-level expression of Lactobacillus beta-galactosidases in Lactococcus lactis using the food-grade, nisin-controlled expression system NICE. J Agric Food Chem. 2010;58:2279–2287. doi: 10.1021/jf902895g. [DOI] [PubMed] [Google Scholar]
- 26.Malten M, Hollmann R, Deckwer WD, Jahn D. Production and secretion of recombinant Leuconostoc mesenteroides dextransucrase DsrS in Bacillus megaterium. Biotechnol Bioeng. 2005;89:206–218. doi: 10.1002/bit.20341. [DOI] [PubMed] [Google Scholar]
- 27.Marbach A, Bettenbrock K. lac operon induction in Escherichia coli: Systematic comparison of IPTG and TMG induction and influence of the transacetylase LacA. J Biotechnol. 2012;157:82–88. doi: 10.1016/j.jbiotec.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen TT, Nguyen HM, Geiger B, Mathiesen G, Eijsink VG, Peterbauer CK, Haltrich D, Nguyen TH. Heterologous expression of a recombinant lactobacillal β-galactosidase in Lactobacillus plantarum: effect of different parameters on the sakacin P-based expression system. Microb Cell Fact. 2015;14:30. doi: 10.1186/s12934-015-0214-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Novick RP. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oehler S, Amouyal M, Kolkhof P, von Wilcken-Bergmann B, Müller-Hill B. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 1994;13:3348–3355. doi: 10.1002/j.1460-2075.1994.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oehler S, Eismann ER, Krämer H, Müller-Hill B. The three operators of the lac operon cooperate in repression. EMBO J. 1990;9:973–979. doi: 10.1002/j.1460-2075.1990.tb08199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh Y, Varmanen P, Han XY, Bennett G, Xu Z, Lu T, Palva A. Lactobacillus plantarum for oral peptide delivery. Oral Microbiol Immunol. 2007;22:140–144. doi: 10.1111/j.1399-302X.2007.00338.x. [DOI] [PubMed] [Google Scholar]
- 33.Pal G, Srivastava S. Scaling up the production of recombinant antimicrobial plantaricin E from a heterologous host, Escherichia coli. Probiotics Antimicrob Proteins. 2015 doi: 10.1007/s12602-015-9193-7. [DOI] [PubMed] [Google Scholar]
- 34.Pavan S, Hols P, Delcour J, Geoffroy MC, Grangette C, Kleerebezem M, Mercenier A. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl Environ Microbiol. 2000;66:4427–4432. doi: 10.1128/AEM.66.10.4427-4432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Posno M, Heuvelmans PT, van Giezen MJ, Lokman BC, Leer RJ, Pouwels PH. Complementation of the inability of Lactobacillus strains to utilize D-xylose with D-xylose catabolism-encoding genes of Lactobacillus pentosus. Appl Environ Microbiol. 1991;57:2764–2766. doi: 10.1128/aem.57.9.2764-2766.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi F, Capodaglio A, Dellaglio F. Genetic modification of Lactobacillus plantarum by heterologous gene integration in a not functional region of the chromosome. Appl Microbiol Biotechnol. 2008;80:79–86. doi: 10.1007/s00253-008-1527-x. [DOI] [PubMed] [Google Scholar]
- 37.Rossi F, Rudella A, Marzotto M, Dellaglio F. Vector-free cloning of a bacterial endo-1,4-beta-glucanase in Lactobacillus plantarum and its effect on the acidifying activity in silage: use of recombinant cellulolytic Lactobacillus plantarum as silage inoculant. Antonie Van Leeuwenhoek. 2001;80:139–147. doi: 10.1023/A:1012223220427. [DOI] [PubMed] [Google Scholar]
- 38.Rud I, Jensen PR, Naterstad K, Axelsson L. A synthetic promoter library for constitutive gene expression in Lactobacillus plantarum. Microbiology. 2006;152:1011–1019. doi: 10.1099/mic.0.28599-0. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Russel DW. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory; 2001. [Google Scholar]
- 40.Schmiedel D, Kintrup M, Küster E, Hillen W. Regulation of expression, genetic organization and substrate specificity of xylose uptake in Bacillus megaterium. Mol Microbiol. 1997;23:1053–1062. doi: 10.1046/j.1365-2958.1997.2881654.x. [DOI] [PubMed] [Google Scholar]
- 41.Schultz M, Veltkamp C, Dieleman LA, Grenther WB, Wyrick PB, Tonkonogy SL, Sartor RB. Lactobacillus plantarum 299 V in the treatment and prevention of spontaneous colitis in interleukin-10-deficient mice. Inflamm Bowel Dis. 2002;8:71–80. doi: 10.1097/00054725-200203000-00001. [DOI] [PubMed] [Google Scholar]
- 42.Siezen RJ, van Hylckama Vlieg JE. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb Cell Fact. 2011;10(Suppl 1):S3. doi: 10.1186/1475-2859-10-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spath K, Heinl S, Egger E, Grabherr R. Lactobacillus plantarum and Lactobacillus buchneri as expression systems: evaluation of different origins of replication for the design of suitable shuttle vectors. Mol Biotechnol. 2012;52:40–48. doi: 10.1007/s12033-011-9471-x. [DOI] [PubMed] [Google Scholar]
- 44.Spath K, Heinl S, Grabherr R. Direct cloning in Lactobacillus plantarum: electroporation with non-methylated plasmid DNA enhances transformation efficiency and makes shuttle vectors obsolete. Microb Cell Fact. 2012;11:141. doi: 10.1186/1475-2859-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stammen S, Müller BK, Korneli C, Biedendieck R, Gamer M, Franco-Lara E, Jahn D. High-yield intra- and extracellular protein production using Bacillus megaterium. Appl Environ Microbiol. 2010;76:4037–4046. doi: 10.1128/AEM.00431-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sørvig E, Mathiesen G, Naterstad K, Eijsink VG, Axelsson L. High-level, inducible gene expression in Lactobacillus sakei and Lactobacillus plantarum using versatile expression vectors. Microbiology. 2005;151:2439–2449. doi: 10.1099/mic.0.28084-0. [DOI] [PubMed] [Google Scholar]
- 47.Tauer C, Heinl S, Egger E, Heiss S, Grabherr R. Tuning constitutive recombinant gene expression in Lactobacillus plantarum. Microb Cell Fact. 2014;13:150. doi: 10.1186/s12934-014-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson CJ, Zhan H, Swint-Kruse L, Matthews KS. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell Mol Life Sci. 2007;64:3–16. doi: 10.1007/s00018-006-6296-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson TH, Kashket ER. Isolation and properties of thiogalactoside transacetylase-negative mutants of Escherichia coli. Biochim Biophys Acta. 1969;173:501–508. doi: 10.1016/0005-2736(69)90014-5. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Vadlani PV. Lactic acid production from biomass-derived sugars via co-fermentation of Lactobacillus brevis and Lactobacillus plantarum. J Biosci Bioeng. 2015;119:694–699. doi: 10.1016/j.jbiosc.2014.10.027. [DOI] [PubMed] [Google Scholar]


