Abstract
Background:
The aim of this study was to find out the correlation between presence of virulence (gelatinase [gel E], enterococcal surface protein [esp], cytolysin A [cyl A], hyaluronidase [hyl], and aggregation substance [asa1]) and vancomycin-resistant genes (van A and van B) in enterococci, with their phenotypic expression.
Materials and Methods:
A total of 500 isolates (250 each clinical and fecal) were processed. Enterococci were isolated from various clinical samples and from fecal specimens of colonized patients. Various virulence determinants namely asa1, esp, hyl, gel E, and cyl were detected by phenotypic methods. Minimum inhibitory concentration (MIC) of vancomycin was determined by agar dilution method. Multiplex polymerase chain reaction (PCR) was used to detect the presence of virulence and van genes.
Results:
Out of all the samples processed, 12.0% (60/500) isolates carried van A or van B genes as confirmed by MIC test and PCR methods. Genes responsible for virulence were detected by multiplex PCR and at least one of the five was detected in all the clinical vancomycin-resistant enterococci (VRE) and vancomycin-sensitive enterococci (VSE). gel E, esp, and hyl genes were found to be significantly higher in clinical VRE. Of the fecal isolates, presence of gel E, esp, and asa1 was significantly higher in VRE as compared to VSE. The presence of hyl gene in the clinical VRE was found to be statistically significant (P = 0.043) as against the fecal VRE. Correlation between the presence of virulence genes and their expression as detected by phenotypic tests showed that while biofilm production was seen in 61.1% (22/36) of clinical VRE, the corresponding genes, i.e., asa1 and esp were detected in 30.5% (11/36) and 27.8% (10/36) of strains only.
Conclusion:
Enterococcus faecium isolates were found to carry esp gene, a phenomenon that has been described previously only for Enterococcus faecalis, but we were unable to correlate the presence of esp with their capacity to form biofilms.
Keywords: Aggregation substance, cytolysin A, enterococcal surface protein, gelatinase, hyaluronidase
INTRODUCTION
Enterococcus species are now recognized as important causes of urinary tract infections, postsurgical wound infections, bacteremia, endocarditis, meningitis, neonatal sepsis, and infections in transplant patients with Enterococcus faecalis and Enterococcus faecium responsible for the majority of these infections.[1] Nevertheless, the incidence of other species of enterococci from clinical sources shows an alarming increase. This is attributable to their acquisition of various putative virulence determinants and multidrug resistance.[2]
A number of genes encoding for virulence factors including aggregation substance (asa1), enterococcal surface protein (esp), hyaluronidase (hyl), gelatinase (gel E), and cytolysin (cyl) in E. faecalis and E. faecium have been described and their effects have been shown in human and animal studies.[3] asa1, a surface protein adhesin encoded by the gene asa1 has a contribution to virulence together with cyl. It facilitates the aggregation of the donor and recipient bacteria for efficient transfer of transmissible conjugative plasmids.[4] Another enterococcal adhesin is the “esp,” encoded by esp gene that plays a role in biofilm formation and adherence to abiotic surfaces.[5] hyl, which is expressed by the hyl gene, acts on hyaluronic acid and increases bacterial invasion.[6] The gel E gene encodes for an extracellular Zn-metalloendopeptidase that is capable of hydrolyzing gelatin, collagen, casein, hemoglobin, and other biological peptides.[7] The cyl is a cellular toxin, and is capable of lysing a range of prokaryotic and eukaryotic cells.[8]
There is a paucity of information on the virulence genes distributed among enterococcal species.[9,10] The putative virulence genes in enterococcal strains isolated from various clinical sources and colonized patients, and also the possible link between the presence of virulence markers and virulence genes was therefore investigated.
MATERIALS AND METHODS
Study population
The study population included patients of both sexes and all age groups attending the outpatient and inpatient departments of a Tertiary Care Hospital in Eastern Bihar, India. A total of 500 strains of enterococci were collected from samples submitted to the Microbiology laboratory for culture and sensitivity and were used in the present study. This included 250 enterococcal strains collected from clinical samples of the patients, attending the hospital with infections of different types. Another 250 enterococcal strains were isolated from among another 300 patients (otherwise not suffering from any infections), who had been admitted to the hospital and screened for gastrointestinal carriage of vancomycin-resistant enterococci (VRE). Clearance from Institutional Ethics Committee was obtained to carry out this study.
Isolation and identification
Two hundred and fifty enterococci were isolated from various clinical samples (118 isolates from urine, 79 isolates from pus, 34 isolates from blood, and 19 isolates from catheter tip). 300 fecal samples were collected from other patients (as mentioned above) on three separate occasions, i.e., at the time of admission, after 48 h, and after 5 days of admission to screen for VRE. Of the 300 fecal samples, 50 samples showed culture negativity and another 250 samples showed the growth of Enterococcus species. The isolates were identified to species level using standard procedures.[11,12,13]
Hemolytic-assay
Hemolytic activity of enterococci was assessed on two blood agar plates prepared with Muller-Hinton agar (Hi-Media, Mumbai, India) containing 5% defibrinated sheep and human blood, by observation of zone of hemolysis around colonies after incubation for 24 h at 37°C.[14]
Hemagglutination test
Enterococci were grown on brain heart infusion agar supplemented with 10% sheep blood. A loopful of bacteria was mixed on a glass slide with 25 μl of a 3% suspension of sheep, rabbit, human group A, human group O, and human group B erythrocytes. Vibrio cholerae was used as positive control. After 5 min at room temperature, results were recorded as positive or negative.[15]
Physico-chemical properties of the culture filtrates
The effects of physic-chemical agents on hemagglutination test (HA) were investigated by performing HA test after treatment of the bacteria with trypsin, protease K, and pepsin (Hi-Media, Mumbai, India). Bacterial suspensions of test strains were centrifuged and the deposit was added to separate test tubes containing trypsin (1 μg/ml), pepsin (1 μg/ml), and protease K (1 μg/ml) in phosphate-buffered saline (PBS). The test tubes were incubated at 37°C for 60 min. For heat treatment, bacterial suspensions were heated at 50°C for 30 min. HA test was carried out with 20 μl of 3% erythrocyte suspension and 20 μl of enzyme treated and heated culture suspensions on glass slides. The suspensions were mixed, rotated gently for 30 s, and results were recorded as either strong agglutination (+++ −), agglutination (− ++), or no agglutination.[16]
Caseinase production
Casein hydrolysis was detected on Muller-Hinton agar (Hi-Media, Mumbai, India) containing 3% skimmed milk. Plates were streaked with test strains followed by incubation at 37°C for 24 h. The presence of a transparent zone around the colonies indicated caseinase activity. gel E production was detected by stab inoculating the test strain on nutrient agar supplemented with 3% gelatin (Hi-Media, Mumbai, India) kept at 37°C for 24 h followed by refrigeration at 4°C for half an hour. Liquefaction of gelatin was considered as positive.[13,14]
Lipase production
Egg yolk agar (Hi-Media, Mumbai, India) was used for lipase production. The test organism was spot inoculated on the medium and incubated at 37°C for 24-48 h. Positive test result was read as formation of thin iridescent pearly layer overlying the colonies and a confined opalescence in the medium, which was seen when the colonies were scraped off.[12]
Slime layer formation
Brain heart infusion agar (Hi-Media, Mumbai, India) supplemented with 5% sucrose was used to determine the ability of Enterococcus species to produce extracellular polysaccharide on the agar. Test strains grown in Todd-Hewitt broth (Hi-Media, Mumbai, India) were used as the inoculum. The colonies appeared mucoidal, runny, or slimy due to the production of polysaccharide.[13]
Deoxyribonuclease test
Test strains were inoculated on deoxyribonuclease agar (Hi-Media, Mumbai, India). Clearing of the medium around the colonies indicated a positive test.[17]
Biofilm detection assay
The test strains were grown overnight at 37°C in Brain Heart Infusion broth (Hi-Media, Mumbai, India) plus 0.25% glucose. Culture was diluted 1:20 in the same media. 200 μL of this suspension was used to inoculate sterile 96 well polystyrene microtiter plates. After 24 h at 37°C of static incubation, wells were washed with PBS, dried in inverted position, and stained with 1% crystal violet for 15 min. The cells were rinsed once more and solubilized in 200 μl ethanol/acetone (80:20 v/v). The A630 was determined using microtiter plate reader. Biofilm formation was scored as nonbiofilm forming (−), weak - (+), moderate - (++), and strong - (+++) corresponding to the A630 values ≤1, 1-≤2, 2-≤3, and >3, respectively.[18]
Antimicrobial susceptibility and minimum inhibitory concentration tests
Antibiotic susceptibility test was done by Kirby-Bauer disc diffusion method on Mueller-Hinton agar. Minimum inhibitory concentration (MIC) of VRE was determined by agar dilution method using the following concentration of vancomycin 0.5-64 μg/mL. The test was quality controlled using E. faecalis ATCC 51299 and E. faecalis ATCC 29212.[19,20]
Protocol for multiplex polymerase chain reaction
DNA extraction method
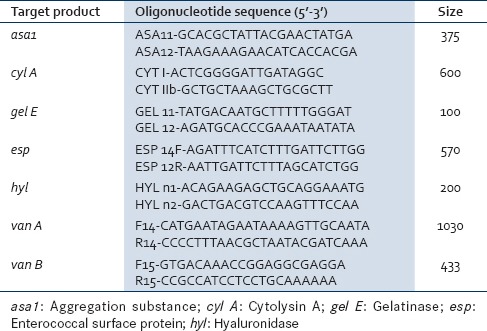
Genomic DNA used as template for polymerase chain reaction (PCR) amplification was prepared using conventional phenol-chloroform DNA extraction method. PCR was performed in PCR system, model number T1 Thermoblock. The oligonucleotide primer pairs used to amplify the virulence genes asa1, cyl A, gel E, esp, and hyl as well as the vancomycin-resistant genes van A, van B, and the expected amplicon sizes are as follows [Table 1]:
Table 1.
Polymerase chain reaction primers and products for the detection of virulent genes and vancomycin-resistant genes
The amplification of virulence genes was carried out as follows: Initial denaturation at 95°C for 15 min followed by denaturation at 94°C for 1 min, annealing at 56°C for 1 min, and extension at 72°C for 1 min. The PCR amplification of the van genes were carried out as follows: Predenaturation at 95°C for 4 min followed by denaturation at 95°C for 30 cycles of 30 s each; 1 min for annealing at 52°C and elongation at 72°C for 1 min. Both positive control and negative control, consisting solely of the PCR reaction mixture without DNA template were included to check the validity of the technique utilized in the study.[21,22]
Analysis of DNA by agarose gel electrophoresis
Twenty-five microliters of respective amplified products were loaded into the wells and electrophoresed at a constant current of 50V for about 45 min using 1.5% agarose gel. A 100 bp DNA ladder marker was included as the standard molecular weight marker. The electrophoresed gel was later subjected to ethidium bromide staining and photographed under UV transillumination [Figures 1–3].
Figure 1.
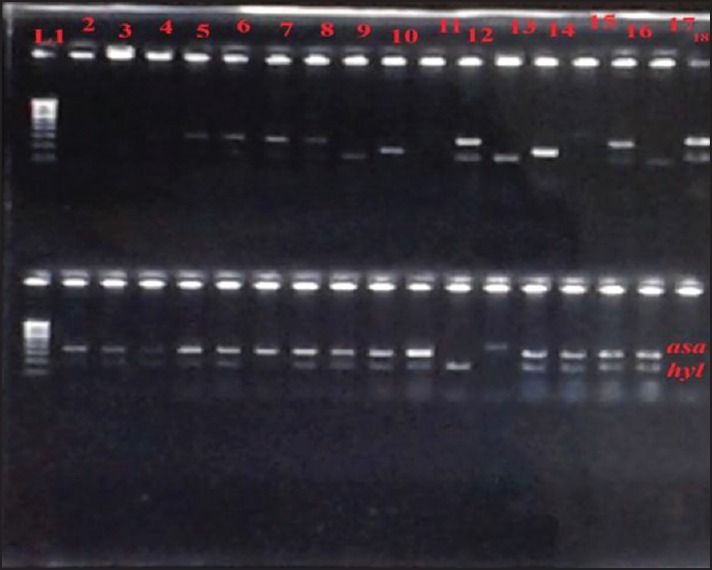
Polymerase chain reaction products of asa1 gene 375bp and hyl gene 200bp. L1 stands for Lane 1 and corresponds to molecular markers from 100bp to 1000bp. asa1: Aggregation substance 1; hyl: Hyaluronidase
Figure 3.
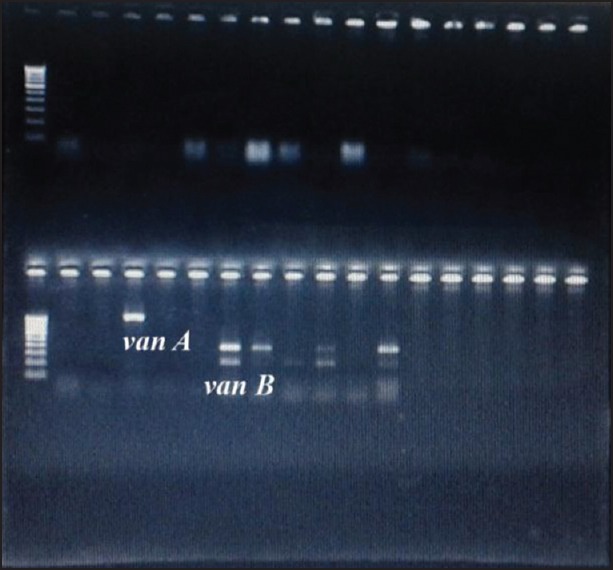
Bands corresponding to 1030 bp (van A) and 433 bp (van B)
Figure 2.
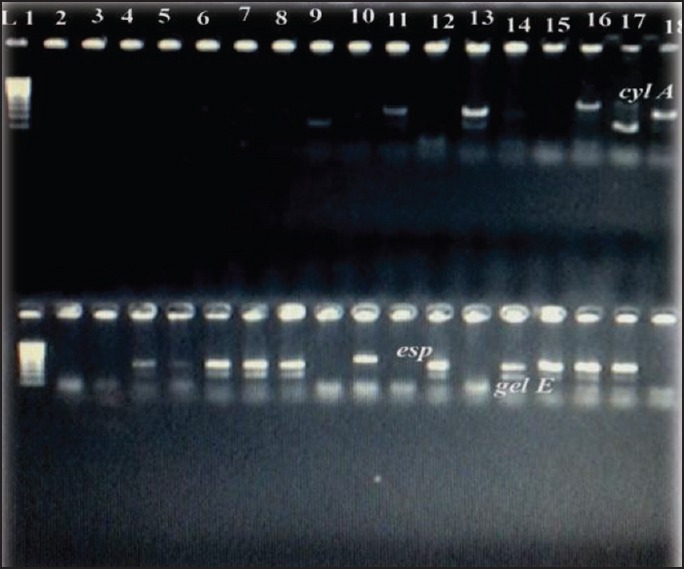
Polymerase chain reaction products of cyl A gene 600bp; esp gene 570bp; gel E 100bp. cyl A: Cytolysin A; esp: Enterococcal surface protein; gel E: Gelatinase
Statistical analysis
Statistical analysis was performed by Chi-square test. P ≤ 0.05 was considered to be significant and P ≤ 0.001 was considered to be highly significant. All statistical analyses were carried out using online statistical software at http://www.physics.csbsju.edu/stats/contingency_NROW_NCOLUMN_form.html. Statistical analysis has been done to find out the prevalence of different virulence markers and virulence genes (gel E, esp, cyl A, hyl, asa1) in VRE, and vancomycin-sensitive enterococci (VSE) were statistically significant/insignificant.
RESULTS
A total of 500 enterococci (250 each clinical and fecal) were processed, out of which 37 (7.4%) isolates were vancomycin-resistant and 23 (5.6%) showed reduced susceptibility to vancomycin by phenotypic agar dilution method. Thus, a total of 60 strains (36 clinical and 24 fecal) which were VRE/VIE by MIC tests were also confirmed to carry van A or van B genes by PCR methods.
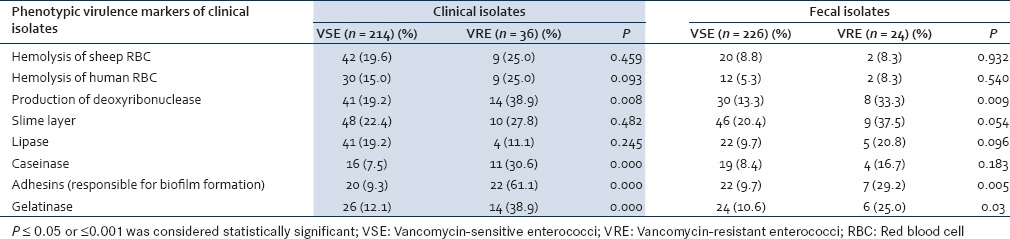
Various virulence factors of enterococci were detected by phenotypic methods for both clinical and fecal isolates. The differences in the presence of virulence factors in clinical VSE and VRE were found to be statistically insignificant for most of the virulence factors except for the production of caseinase, adhesins, and gel E which was significantly higher in VRE than VSE (P = 0.000). For the fecal isolates, production of slime layer and gel E was significantly higher in VRE than VSE (P = 0.05; P = 0.03) [Table 2].
Table 2.
Comparative evaluation of the virulence factors in clinical and fecal isolates of vancomycinsensitive enterococci and vancomycin-resistant enterococci
The presence of genes encoding for potential virulence factors was studied by multiplex PCR in both the clinical and fecal isolates. The predominant virulence gene in clinical VRE was gel E with 16 (44.4%) clinical VRE isolates having the gel E gene as compared to 35 (16.4%) VSE isolates. This finding was found to be statistically significant (P = 0.000). The prevalence of esp (P = 0.001) and hyl (P = 0.047) genes were significantly higher among VRE isolates (27.8% and 36.1%) than among VSE isolates (8.9% and 21.0%). The presence of other virulence genes such as cyl A and asa1 were not found to be statistically significant [Table 3].
Table 3.
Presence of virulence genes in vancomycin-sensitive enterococci and vancomycin-resistant enterococci (clinical and fecal)
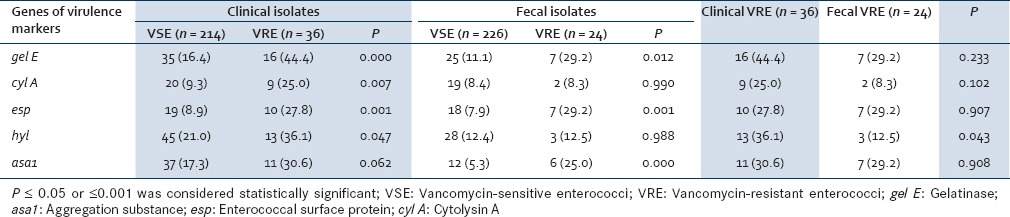
Like the clinical isolates, gel E, as well as esp, was the predominant gene detected in the fecal VRE which was seen in 29.2% of isolates each followed by asa1, seen in 25.0% of strains. The presence of gel E, esp, and asa1 was found to be significantly higher in the VRE than VSE (P = 0.012, 0.001, and 0.000, respectively). The presence of other virulence genes was statistically insignificant [Table 3].
The presence of virulence genes in clinical and fecal VRE was compared. All the virulence genes were encountered more frequently in clinical VRE than in fecal VRE except for esp which was found in 29.2% (7/24) of fecal VRE as compared to 27.8% (10/36) of clinical VRE. However, only the presence of hyl gene was found to be significantly higher in clinical VRE than fecal VRE (P = 0.043) [Table 3].
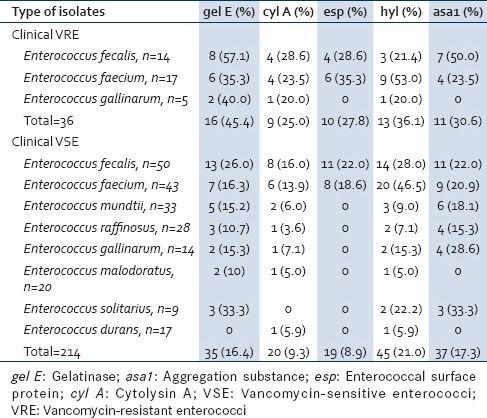
Table 4 shows the presence of various virulence genes in VRE species. The predominant genes in E. faecalis were gel E 57.1%; followed by asa1 50.0%, cyl A and esp being 28.6% each, and hyl 21.4%. The predominant gene in E. faecium was hyl gene (53.0%) followed by gel E, esp 35.3% each, cyl A, and asa1 23.5% each. Enterococcus gallinarum showed the presence of gel E 40.0% followed by cyl A and hyl 20.0% each.
Table 4.
Distribution of virulence genes in clinical vancomycin-resistant enterococci and vancomycin-sensitive enterococci
Out of the 14 VR E. faecalis, all the strains produced one or two virulence genes. Twelve (85.7%) strains of E. faecalis produced two virulence genes: 35.7% (5/14) strains showed the concomitant presence of gel E and asa; another 21.4% (3/14) strains expressed gel A and hyl genes; and 28.6% (4/14) strains presented cyl A and esp genes. Of the 17 VR E. faecium, 12 (70.6%) strains were found to produce two genes. The various combination of two virulence genes seen in E. faecium were 35.3% (6/17) expressed gel A with hyl; 17.6% (3/17), each, expressed cyl A with esp and asa1 with esp.
Of the clinical VSE, the predominant genes in E. faecalis were hyl (28.0%); followed by gel E (26.0%), esp and asa1 (22.0%) each, and cyl A (16.0%). hyl (46.5%) was the main gene in E. faecium followed by asa1 (20.9%), esp (18.6%), gel E (16.3%), and cyl A (13.9%). Apart from the esp gene, the remaining four virulence genes were detected in all the nonfaecalis and nonfaecium strains. Enterococcus durans was the least virulent among VSEs as it showed the expression of only cyl A and hyl (5.9%) each [Table 4].
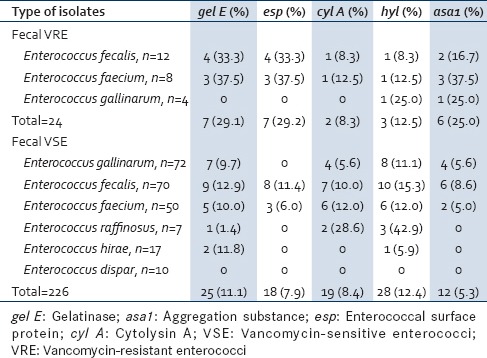
Table 5 shows the distribution of virulence genes in fecal VRE. gel E (33.3%) was the predominant virulence gene in E. faecalis followed by esp (33.3%), asa1 (16.7%), cyl A, and hyl (8.3%) each. The main genes in E. faecium were gel E, esp, and asa1 (37.5%) each. E. gallinarum showed the presence of only hyl and asa1 (25.0%) each. The simultaneous presence of two virulence genes was seen between gel E and asa1 in 16.2% (2/12) of E. faecalis, while 37.5% (3/8) E. faecium carried asa1 and esp gene combination. Of the fecal VSE, E. faecalis and E. faecium showed the presence of all virulence genes. Enterococcus dispar was least virulent species as it did not express any virulence genes. The esp gene was totally absent in all nonfaecalis and nonfaecium strains [Table 5].
Table 5.
Distribution of virulence genes in fecal vancomycin-resistant enterococci and vancomycinsensitive enterococci
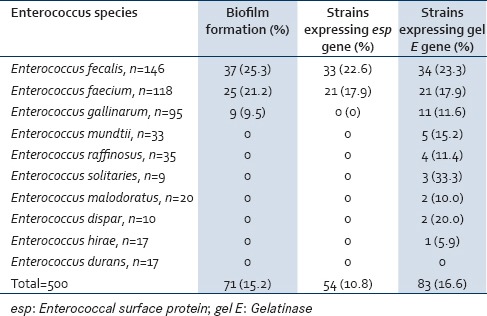
Of the 500 Enterococcus species isolated from clinical and fecal sources, 71 (14.2%) strains were positive for biofilm formation whereas 83 (16.6%) strains were found to show the presence of gel E gene and only 54 (10.8%) strains expressed the esp gene. Of E. faecalis strains, 25.3% (37/146) produced biofilm, 23.3% (34/146) showed the presence of gel E, and 22.6% (33/146) only expressed esp. E. faecium 21.2% (25/118) produced biofilm among which 17.9% (21/118 each) expressed both gel E and esp genes. 9.5% (9/95) E. gallinarum strains that produced biofilm also expressed gel E along with some additional strains accounting 11.6% (11/95). However, these strains lacked the esp gene. An additional 24 strains that included Enterococcus mundtii 15.2% (5/33), followed by Enterococcus raffinosus 11.4% (4/35), Enterococcus solitaries 33.3% (3/9), Enterococcus malodoratus 10.0% (2/20), E. dispar 20.0% (2/10), and Enterococcus hirae 5.9% (1/17) also expressed the gel E gene [Table 6].
Table 6.
Correlation between biofilm formation and the presence of esp and gel E genes in all enterococcus species (clinical and fecal)
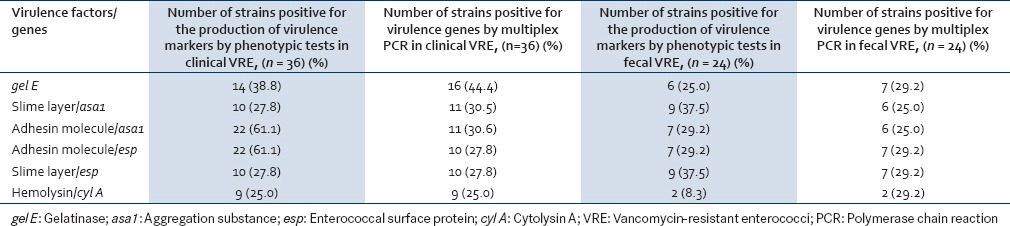
Correlation between the virulence markers and virulence genes as detected by phenotypic and genotypic tests, for clinical and fecal VRE is shown in Table 7. It was seen that those strains that were positive for the production of gel E and hemolysin also showed the presence of the corresponding genes by multiplex PCR for both clinical and fecal strains. However, the number of strains positive for gel E genes by PCR was more than the number of strains showing positive reaction by phenotypic tests [Table 7]. As far as the production of biofilm (adhesin molecule) and slime layer was concerned, the number of strains positive by phenotypic tests was more than the number of strains showing the presence of the corresponding genes by PCR namely asa1 and esp.
Table 7.
Correlation between virulence markers and virulence genes in clinical and fecal vancomycin-resistant enterococci
DISCUSSION
Very few reports regarding the distribution of virulence genes in various species of enterococci from clinical samples are available.[23] This study was designed to identify various virulence genes in VRE and VSE as well as to evaluate the correlation between virulence markers and virulence genes both phenotypically and genotypically.
Only gel E, esp, and hyl genes were found to be significantly higher in clinical VRE than VSE. As for fecal isolates, presence of gel E, esp, and asa1 was significantly higher in VRE than VSE. Other studies show that prevalence of esp (P = 0.001) and hyl (P = 0.04) genes were significantly higher among VRE isolates (44.4% and 27.7%) than among VSE isolates (16.4% and 8.8%).[24]
Authors reported esp of 80.0% (32/40) to be the predominant virulence factor followed by gel E of 50.0% (20/40) among VRE. Considering vancomycin resistance as variable, the authors did not find any significant difference in the presence of activity of virulence factors between resistant and susceptible enterococci.[25]
It has been reported that the esp gene has been restricted to vancomycin-resistant strains.[26] In contrast, our result showed the presence of esp gene in both VSE and VRE strains. The presence of esp in isolates susceptible and resistant to different antibiotics indicate that this trait probably emerged prior to the acquisition of resistance not only to vancomycin but also to other antibiotics used in hospital settings.
Of the clinical VRE, all the five virulence genes were detected in E. faecalis and E. faecium. However, virulence genes namely asa1 and esp were absent in E. gallinarum. Until recently, a majority of the infection derived isolates were E. faecalis strains and was regarded as the most pathogenic species. It is known that Enterococcus possess highly efficient gene transfer mechanism. The virulent genes are associated with highly transmissible plasmids which might have led to the dissemination of virulent genes in less virulent E. faecium and E. gallinarum.
Of the VSE, all the five different virulence genes were detected in vancomycin-sensitive E. faecalis and E. faecium. The predominant genes in E. mundtii, E. raffinosus, E. solitaries, and E. gallinarum were asa1 being 18.1%, 15.3%, 33.3%, and 28.6% (6/33, 4/28, 3/9, and 4/14), respectively. The higher prevalence of hyl gene in E. faecalis and E. faecium and asa1 gene in nonfaecalis and nonfaecium strains from our setup depicts that this virulence marker may have permeated more deeply into the species by horizontal transfer and would have acquired it comparatively earlier, thereby enhancing the ability of the organism to cause disease beyond that intrinsic to the species. We observed a considerable number of E. faecium strains expressed hyl and asa1 genes as compared to esp, gel E, and cyl A genes. At present, we cannot say with certainty whether, and to what extent, E. faecium actually makes hyl, and under what conditions this protein may be synthesized or exported. Northern hybridization experiments indicate that the hyl open reading frame is transcribed under nonselective growth conditions in vitro. Therefore, we have compelling reasons to believe that the protein is synthesized at least under some environmental conditions. asa too has an important effect on biofilm formation because this substance promotes the adherence of microorganisms to a surface.[27]
In another study, the distribution of virulence genes in various species was viz: E. faecalis 53.0% (35/66), Enterococcus casseliflavus 50.0% (2/4), E. faecium 40.6% (13/32), and E. mundtii 25.0% (1/4). Enterococcus durans showed the total absence of all virulene genes. E. casseliflavus and E. mundtii showed the presence of only two genes - asa1 and esp 25.0% (1/4).[28] The permeation of these virulence genetic characteristics into different species differs according to the setup, patient demographics, and other extrinsic factors. Thus, the nonfaecalis and nonfaecium strains isolated from our study were more virulent as compared to the strains seen in the above study which may be due to the presence of asa1 in nonfaecalis and nonfaecium strains that promote cell-to-cell contact and help in the transfer of other virulence genes through plasmids.
Of the clinical VRE, 85.7% (12/14) E. faecalis produced two virulence genes: 35.7% (5/14) strains showed the concomitant presence of gel E and asa1. Another 21.4% (3/14) strains expressed gel E and hyl genes and 28.6% (4/14) strains presented cyl A and esp genes. Of the 17 VR E. faecium, 12 (70.5%) strains were found to produce two genes. The various combination of two virulence genes seen in E. faecium were: 35.3% (6/17) gel E with hyl; 17.6% (3/17), each expressing cyl A with esp, and asa1 with esp. Contrasting reports were seen in another study, where the most common combination of genes was between asa1 and gel E from infections of hospitalized patients. This suggests that these traits entered the species earlier than did other toxins such as hemolysin, bacteriocin, and gel E. asa1 is an integral component of the pheromone-responsive plasmid exchange system. Therefore, nosocomial strains of Enterococcus species may be those best equipped to participate in genetic exchange and may be selected by the presence of antibiotic resistance determinants on such plasmids.[25]
Of the fecal VRE, the predominant genes in E. faecalis were gel E (33.3%). The main genes in E. faecium were gel E, esp, and asa1 (37.5% each). E. gallinarum showed the presence of hyl and asa1 (25.0% each) only. Vancomycin-sensitive E. faecalis and E. faecium isolated from feces showed the presence of all five virulence genes. The esp gene was totally absent in nonfaecalis and nonfaecium strains. Contrasting reports were seen in another study from Portugal where virulence genes were detected only in E. faecalis (gel E 75.3%, asa1 30.1%, and esp 4.1%), but rarely found in E. faecium and other unusual species.[29]
When correlation between presence of virulence genes and their expression as detected by phenotypic tests was done, it was found that while biofilm production was seen in 61.1% (22/36) of clinical VRE, but the corresponding genes, i.e., asa1 and esp were detected in 30.5% (11/36) and 27.8% (10/36) of strains only. This suggests the presence of other genes (not detected by multiplex PCR in the present study) responsible for the formation of biofilm in vitro.
Some authors reported that 74.0% (37/50) Enterococcus species were biofilm producers. However, esp genes were detected in 76.0% (38/50) and gel E gene in 60.0% (30/50) isolates. As with esp, gel E gene participation in biofilm formation is controversial. Authors say that presence of gel E enzyme can affect the virulence and the process of biofilm formation in Enterococcus species.[25]
Our study showed that the number of strains that were positive for the production of hemolysin also showed the presence of corresponding gene by multiplex PCR both for clinical and fecal VRE. However, the number of strains positive for this gel E gene by PCR was more than the number of strains showing positive reaction by phenotypic tests. In our hands, results obtained by phenotypic tests always revealed a lower percentage of strains that produced gel E compared to genotypic characterization. This may be due to the presence of silent genes that are expressed only under in vivo conditions or due to the presence of undetected gene mutations.
In another study, it was observed that the asa1 and esp genes were equally identified in phenotypic and genotypic assays. cyl A gene (hemolysin) was detected in 82 strains genotypically as compared to only 54 in phenotypic assay. Similarly for gel E, gel E gene was identified in 83 strains in comparison to 77 phenotypically gel E positive strains.[30]
CONCLUSION
Multiplex PCR protocol used in the study for simultaneous detection of five different virulence genes and van genes proved to be a reliable and rapid alternative to phenotypic testing and uniplex PCR. The number of strains positive for gel E gene by PCR was more than the number of strains showing positive reaction by phenotypic tests, which might be due to the presence of silent genes. Thus, it seems necessary to perform both phenotypic and genotypic assays for better characterization of the strains. Biofilm formation could not be linked to any specific gene. In fact, this phenomenon is multifactorial and depends on a number of genes working together along with extrinsic factors. So far, several other genes or gene sets have been reported as auxiliaries in biofilm formation in enterococcus, which highlights the complexity and the multifactorial nature of this trait. We found the presence of esp in the vancomycin-resistant isolates as well as in the sensitive isolates. The prevalence of various virulence genes in nonfaecalis and nonfaecium strains proves widespread dissemination of virulence genes through horizontal gene transfer mechanism among the less virulent species.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
I would like to thank Dr. Gopal Nath, Professor and HOD, Department of Microbiology, IMS (BHU), for his guidance and technical support for conducting molecular tests.
REFERENCES
- 1.Jett BD, Huycke MM, Gilmore MS. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–78. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prakash PV, Rao RS, Parija CP. Emergence of unusual species of enterococci causing infection, South India. BMC Infect Dis. 2004;5:14–9. doi: 10.1186/1471-2334-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giraffa G. Enterococci from foods. FEMS Microbiol Rev. 2002;26:163–71. doi: 10.1111/j.1574-6976.2002.tb00608.x. [DOI] [PubMed] [Google Scholar]
- 4.Pillar CM, Gilmore MS. Enterococcal virulence—Pathogenicity island of E. Faecalis. Front Biosci. 2004;9:2335–46. doi: 10.2741/1400. [DOI] [PubMed] [Google Scholar]
- 5.Toledo-Arana A, Valle J, Solano C, Arrizubieta MJ, Cucarella C, Lamata M, et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. Appl Environ Microbiol. 2001;67:4538–45. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giridhara Upadhyaya PM, Ravikumar KL, Umapathy BL. Review of virulence factors of Enterococcus: An emerging nosocomial pathogen. Indian J Med Microbiol. 2009;27:301–5. doi: 10.4103/0255-0857.55437. [DOI] [PubMed] [Google Scholar]
- 7.Makinen PL, Clewell DB, An F, Makinen KK. Purification and substrate specificity of a strongly hydrophobic extracellular metalloendopeptidase (gelatinase) from Streptococcus faecalis (strain OG1-10) J Biol Chem. 1989;264:3325–34. [PubMed] [Google Scholar]
- 8.Booth MC, Boogie CP, Sahl HG, Seizen RJ, Hatter KL, Gilmore MS. Structural analysis and proteolytic activation of Enterococcus faecalis cytolysin, a novel lantibiotic. J Mol Microbiol Biotechnol. 1996;21:1175–84. doi: 10.1046/j.1365-2958.1996.831449.x. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes SC, Dhanashree B. Drug resistance & virulence determinants in clinical isolates of Enterococcus species. Indian J Med Res. 2013;137:981–5. [PMC free article] [PubMed] [Google Scholar]
- 10.Ira P, Sujatha S, Chandra PS. Virulence factors in clinical and commensal isolates of Enterococcus species. Indian J Pathol Microbiol. 2013;56:24–30. doi: 10.4103/0377-4929.116144. [DOI] [PubMed] [Google Scholar]
- 11.Al Khafaji JK, Samaan SF, Al Saeed MS. Virulence factors of Enterococcus faecalis. Med J Babylon. 2010;7:579–83. [Google Scholar]
- 12.Koneman EW, Allen SD, Janda WM, Schreckenberger PC, Winn WC, editors. Colour Atlas and Text Book of Diagnostic Microbiology. 6th ed. Philadelphia: Lippincott; 2006. Gram positive cocci. Part 2: Streptococci, enterococci and the streptococcus like bacteria; pp. 725–33. [Google Scholar]
- 13.Betty AF, Daniel LS, Weissfeld AS, editors. Bailey & Scott's Diagnostic Microbiology. 12th ed. Missouri: Mosby Elsevier; 2007. Overview of bacterial identifications methods and strategies; pp. 216–41. [Google Scholar]
- 14.Furumura MT, Figueiredo PM, Carbonell GV, Darini AL, Yano T. Virulence associated characteristics of Enterococcus fecalis strains isolated from clinical sources. Braz J Microbiol. 2006;37:230–6. [Google Scholar]
- 15.Gulhan T, Aksakal A, Ekin HI, Savasan S, Boynukara B. Virulence factors of Enterococcus fecium and Enterococcus fecalis strains isolated from humans and pets. Turk J Vet Anim Sci. 2006;30:477–82. [Google Scholar]
- 16.Carvalho Mda G, Teixeira LM. Hemagglutination properties of Enterococcus. Curr Microbiol. 1995;30:265–8. doi: 10.1007/BF00295499. [DOI] [PubMed] [Google Scholar]
- 17.Collee JG, Duguid JP, Fraser AG, Marmion BS, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology. 14th ed. New York: Churchill Livingstone; 2006. Laboratory strategy in the diagnosis of infective syndromes; pp. 113–29. [Google Scholar]
- 18.Prabakaran K, Aberna AR. Evaluation for the association of virulence determinants among E. fecalis with its clinical outcome. Int J Biol Med Res. 2011;2:523–7. [Google Scholar]
- 19.CLSI. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-third Informational Supplement, M100-S23. Vol. 33. Wayne, PA, USA: CLSI; 2013. pp. 90–3. [Google Scholar]
- 20.Sapkota AR, Hulet RM, Zhang G, McDermott P, Kinney EL, Schwab KJ, et al. Lower prevalence of antibiotic-resistant enterococci on U.S. conventional poultry farms that transitioned to organic practices. Environ Health Perspect. 2011;119:1622–8. doi: 10.1289/ehp.1003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. 2004;42:4473–9. doi: 10.1128/JCM.42.10.4473-4479.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akhi TM, Farzaneh F, Oskouei M. Study of enterococcal susceptibility patterns isolated from clinical specimens in Tabriz, Iran. Pak J Med Sci. 2009;25:1–7. [Google Scholar]
- 23.Anupurba S, Banerjee T. Drug resistance in clinical isolates of enterococci with special reference to vancomycin, from North India. J Pure Appl Microbiol. 2012;6:807–12. [Google Scholar]
- 24.Duprè I, Zanetti S, Schito AM, Fadda G, Sechi LA. Incidence of virulence determinants in clinical Enterococcus faecium and Enterococcus faecalis isolates collected in Sardinia (Italy) J Med Microbiol. 2003;52(Pt 6):491–8. doi: 10.1099/jmm.0.05038-0. [DOI] [PubMed] [Google Scholar]
- 25.Comerlato BC, Resende CC, Caierao J, Azevedo AP. Presence of virulence factors in Enterococcus faecalis and Enterococcus faecium susceptible and resistant to vancomycin. Int J Antimicrob Agents. 2013;108:590–5. doi: 10.1590/0074-0276108052013009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kafil SK, Ashgarzadi M. Vancomycin-resistant Enteroccus faecium and Enterococcus faecalis isolated from education hospital of Italy. J Med Microbiol. 2013;9:323–7. [PMC free article] [PubMed] [Google Scholar]
- 27.Trevedi K, Cupakova S, Karpiskova R. Virulence factors and antibiotic resistance in enterococci isolated from food-stuffs. J Vet Med. 2012;56:352–7. [Google Scholar]
- 28.Bittencourt de Marques E, Suzart S. Occurrence of virulence-associated genes in clinical Enterococcus faecalis strains isolated in Londrina, Brazil. J Med Microbiol. 2004;53:1069–73. doi: 10.1099/jmm.0.45654-0. [DOI] [PubMed] [Google Scholar]
- 29.Poeta P, Costa D, Rodrigues J, Torres C. Antimicrobial resistance and the mechanisms implicated in faecal enterococci from healthy humans, poultry and pets in Portugal. Int J Antimicrob Agents. 2006;27:131–7. doi: 10.1016/j.ijantimicag.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 30.Revathy S, Sridharan KS, Elumalai AS, Umasekar Phenotypic detection of high-level aminoglycoside resistance (HLAR) in Enterococcus species in a tertiary care centre. J Biosci Biotechnol Res. 2010;6:751–6. [Google Scholar]


