Abstract
Background:
Multidrug-resistant (MDR) Pseudomonas spp. have been reported to be the important cause of ICU infections. The appearance of ESBL, AmpC and MBL genes and their spread among bacterial pathogens is a matter of great concern. Biofilm production also attributes to antimicrobial resistance due to close cell to cell contact that permits bacteria to more effectively transfer plasmids to one another. This study aimed at determining the incidence of ESBL, AmpC, MBL and biofilm producing Pseudomonas spp. in ICU patients.
Material and Methods:
The clinical specimens were collected aseptically from 150 ICU patients from February 2012 to October 2013. Identification and antimicrobial susceptibility was performed according to Clinical and Laboratory Standards Institute (CLSI) guidelines. ESBLs and AmpC were detected phenotypically and genotypically. MBL was detected by modified Hodge and imipenem-EDTA double-disk synergy test.
Results:
Pseudomonas spp. 35(28%) were the most prevalent pathogen in ICU infections. Multidrug resistance and biofilm production was observed in 80.1% and 60.4% isolates, respectively. Prevalence of ESBL, AmpC and MBL was 22.9%, 42.8% and 14.4%, respectively. The average hospital stay was 25 days and was associated with 20% mortality.
Conclusions:
A regular surveillance is required to detect ESBL, AmpC and MBL producers especially in ICU patients. Carbapenems should be judiciously used to prevent their spread. The effective antibiotics, such as fluoroquinolones and piperacillin-tazobactum should be used after sensitivity testing.
Keywords: ICU, multidrug resistance, pseudomonas
INTRODUCTION
Pseudomonas spp. are one of the most common gram-negative pathogens associated with infections in ICU patients including bacteremia, urinary tract infections, and surgical site infections, but they predominate as agents of lower respiratory tract infections.[1]
Pseudomonas spp. shows a high level of intrinsic resistance to antimicrobial drugs and an ability to become even more drug resistant. These characteristics are caused by selective pressure of mutations in chromosomal genes that lead to production of ESBL and AmpC hyper expression, repression or inactivation of oprD, and over expression of efflux pumps.[2] In addition, Pseudomonas spp. are able to acquire other drug-resistant determinants by horizontal transfer of mobile genetic elements coding for class B carbapenemases (also called metallo-β-lactamases [MBLs]).[3] Because they can be disseminated horizontally through transfer of resistance determinants, MBLs have become a serious concern in hospitals worldwide over the past decade. Such acquired MBLs include the IMP and VIM types SPM-1, GIM-1, SIM-1, AIM-1, KHM-1, NDM-1, and SID-1.[4,5] MBL genes are normally encoded in class 1 integrons along with other resistance determinants, such as the aminoglycoside-modifying enzymes. The integrons are frequently located in plasmids or transposons, the dissemination of which contributes to the global spread of this resistance mechanism.[6,7]
Pseudomonas spp. may also acquire resistance to antibiotics due to permeability barrier of the cell surface in the form of biofilm production. The tendency for bacteria to become surface bound is so ubiquitous in diverse ecosystems that it suggests a strong survival strategy and selective advantage for surface dwellers over their free-ranging counterparts. Virtually any surface, biotic or abiotic (animal, mineral, or vegetable), is suitable for bacterial colonization and biofilm formation. Biofilm is defined as “a structured community of bacterial cells enclosed in a self-produced polymeric matrix adherent to an inert or living surface.” Biofilm-producing organisms are far more resistant to antimicrobial agents than organisms which do not. In some extreme cases, the concentrations of antimicrobials required to achieve bactericidal activity against adherent organisms can be three- to four-fold higher than for those bacteria which do not produce biofilm, depending on the species and drug combination.[8] The versatility and ability of Pseudomonas spp. to combine different resistance mechanisms has led to emergence of strains that are resistant to multiple antimicrobial drugs, which severely limits therapeutic options for treating infections.[9] This emphasizes the need for the detection of isolates that produce these enzymes to avoid therapeutic failures and nosocomial outbreaks.
This study was designed to assess the problem of multidrug-resistant (MDR) Pseudomonas spp. prevalent at various infective foci in ICU patients and to determine the risk factors predisposing to these infections. This study aimed at determining the incidence of ESBL, AmpC, MBL and biofilm producing Pseudomonas spp. in ICU patients.
MATERIALS AND METHODS
The present study was carried out in the Department of Microbiology on ICU patients of J. N. Medical College, Aligarh Muslim University, Aligarh, from February 2012 to October 2013. A total of 150 patients admitted in the ICU were recruited in the study.
Complete history was taken from each patient. One or more clinical samples were obtained from each patient (tracheobronchial aspirate, blood, pus, urine). All specimens were collected aseptically and were promptly sent to the Microbiology laboratory. Standard methods for isolation and identification of aerobic[10,11] bacteria were used.
Susceptibility testing of aerobic and anaerobic isolates was performed using the disk diffusion method as described by the Clinical and Laboratory Standards Institute (CLSI).[12] Antimicrobial disk used were imipenem (10 μg), cefpodoxime (10 μg), cefotaxime (30 μg), cefepime (30 μg), cefixime (5 μg), cefoperazone (75 μg), cefoperazone/sulbactam (75/10 μg), ticarcillin (75 μg), piperacillin (100 μg), piperacillin/tazobactum (100/10 μg), ceftazidime (30 μg), ceftazidime/clavulanic acid (30/10 μg), cefotaxime/clavulanic acid (30/10 μg), ceftriaxone (30 μg), amikacin (30 μg), gentamicin (10 μg), tobramycin (10 μg), ofloxacin (5 μg), levofloxacin (5 μg), polymyxin B (300 units) and colistin (10 μg). All discs were obtained from Hi-Media Labs, Mumbai, India.
Phenotypic methods for ESβL detection
Pseudomonas isolates were first screened for the production of ESBL by the disk diffusion method (screening test) using cefotaxime, ceftriaxone, cefepime, and ceftazidime[12] and later on confirmed by the cephalosporin/clavulanate combination disk (disk potentiation test) method using ceftazidime, ceftazidime + clavulanic acid, cefotaxime, cefotaxime + clavulanic acid, cefoperazone, cefoperazone + sulbactam, and piperacillin, piperacillin + tazobactum. A difference of 5 mm between the zone diameters of either of the cephalosporin disks and their respective cephalosporin/clavulanate disk is taken to be phenotypic confirmation of ESBL production.[13]
Double-disk synergy test (DDST)[13,14] was also used for phenotypic confirmation of ESBL producers in which the test strains were adjusted to the 0.5 McFarland standard and swabbed on a Muller-Hinton agar plate. A susceptibility disk containing piperacillin-tazobactam was placed in the center of the plate. Disks containing one of the oxyimino-β-lactam antibiotics (cefotaxime, ceftazidime and ceftriaxone) were placed 30 mm (center to center) from piperacillin-tazobactam disk. A clear extension of the edge of the oxyimino-β-lactam inhibition zone toward the disk containing clavulanate was interpreted as synergy indicating a positive result. Escherichia coli ATCC 25922 (non-ESBL producer) was used as a control strain.
Phenotypic methods for AmpC detection
Cefoxitin disks were used to screen AmpC producers by the disk diffusion method.[15] Those isolates which were resistant to cefoxitin were considered as potential AmpC producers.
Phenotypic methods for MBL detection
The isolates were tested for sensitivity to imipenem (10 μg) using the Kirby-Bauer method as recommended by CLSI.[12] Isolates from various samples showing zone of inhibition less than 16 mm or heaped up zone were screened for MBL production by the following methods.
Modified hodge test For MBL detection[16]
The indicator organism, E. coli ATCC 25922, at a turbidity of 0.5 McFarland standard, was used to swab inoculate the surface of a Mueller-Hinton agar plate (Becton Dickinson, Cockeysville, Md.), and the test strain was heavily streaked from the center to the plate periphery. After the plate was allowed to stand for 15 min at room temperature, a 10 μg IPM disk was placed at the center, and the plate was incubated overnight. The presence of distorted inhibition zone (cloverleaf) was interpretated as a positive result for carbapenem hydrolysis.
The imipenem-EDTA double-disk synergy test[16]
The test strains were adjusted to the McFarland 0.5 standard and used to inoculate Muller-Hinton agar plates. Disk containing imipenem 10 μg was placed on the plate and a blank filter paper soaked with 10 μl of 0.5 M EDTA solution was placed at a distance of 10 mm (edge to edge). After overnight incubation, the presence of synergistic inhibition zone was interpreted as positive.
Genotypic methods for the detection of ESBL and AmpC production
Preparation of DNA template: Template DNA was prepared from freshly cultured bacterial isolates by suspending bacterial colonies in 50 μL of molecular-grade water and then heating at 95ΊC for 5 min and immediately chilling at 4ΊC.
Detection of bla genes by polymerase chain reaction: Molecular detection of blaCTX-M, blaTEM, blaSHV, blaAmpC was performed by using polymerase chain reaction (PCR) according to methods described previously with minor modifications.[17,18] The primers and cycling conditions for detection of bla genes were the same as those described by Shahid et al.[18] and Feria et al.[19]
Detection of biofilm in Pseudomonas isolates
In vitro biofilm forming ability of Pseudomonas isolates was tested by the tube method, as described by Hassan et al.[20] and Mathur et al.,[21] with slight modification. 0.5 ml (1.5 × 108 organism/ml) of 48 hour culture saline washed suspension was inoculated into a polysterene tube containing 4.5 ml of trypticase soy broth (TSB) with 1% glucose.[21] Tubes were incubated at 37ΊC for 48 hours without agitation. After 48 hours, the culture broth in the tube was aspirated, and the tubes were washed twice with distilled water. The walls of tube were stained with 0.1% crystal violet after media and cells were discarded. Excess stain was removed and tubes were washed with water. Tubes were than dried in inverted position and observed for biofilm formation.
RESULTS
From 150 patients admitted to ICU, total number of isolates was 160. 121 (75.6%) were gram-negative bacilli, 22 (13.8%) were gram-positive cocci and 10.6% were fungal isolates. Pseudomonas spp. represented 35 (28%) of isolates as shown in Figure 1.
Figure 1.
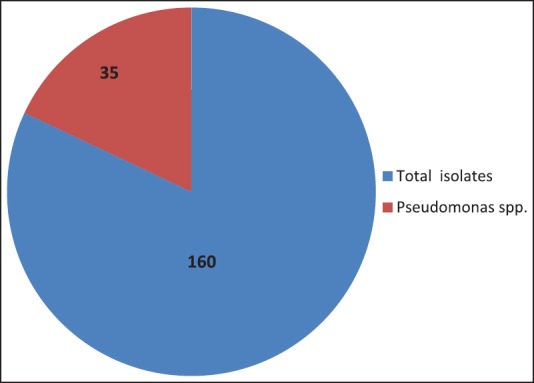
The various strains of micro-organisms isolated from the patients admitted in ICU
Table 1 shows that Pseudomonas spp. were commonly isolated from endotracheal aspirate (40%), followed by wound (24.1%) then urine (26.3%) and blood (10%) with no statistically significant difference.
Table 1.
Distribution of Pseudomonas spp. according to the type of specimen

The most important risk factors significantly associated with infections caused by Pseudomonas spp. in ICU were endotracheal intubation and mechanical ventilation (P = <0.0001). Comorbid conditions significantly associated with ICU infections were COPD (P = 0.001) and hypertension (P = 0.004). Duration of ICU stay of 7-15 days (P = 0.03) and > 15 days (P = 0.0006) were also statistically significant factor associated with ICU infections [Table 2].
Table 2.
Number and percentage of total and Pseudomonas spp.-positive cases in relation to risk factors
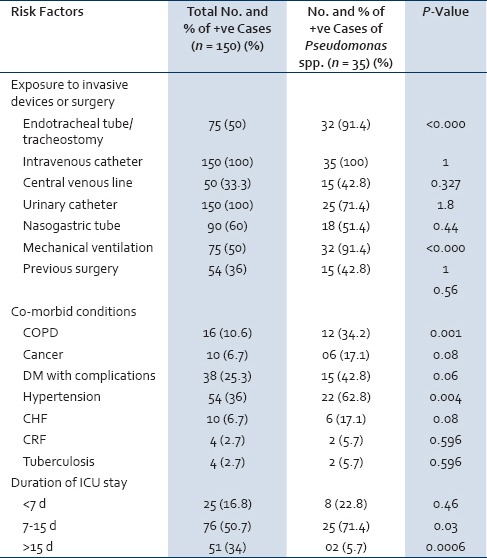
Figure 2 shows antibiotic susceptibility pattern of Pseudomonas spp. detected by the disk diffusion method. Pseudomonas spp. exhibited high degree of antibiotic resistance against ofloxacin (80.0%), ticarcillin (80.0%), piperacillin (74.2%), gentamycin (71.4%), ceftazidime (68.5%) and cefpodoxime (65.7%). Five isolates (14.3%) of Pseudomonas spp. were resistant to imipenem. There was no resistance for polymyxin B and colistin.
Figure 2.
Antibiotic resistance pattern of Pseudomonas spp. detected by the disk diffusion method
All the Pseudomonas spp. isolates were further screened for the production of ESBL, AmpC and MBL activity first by phenotypic methods. The positivity for ESBL, AmpC and MBL by phenotypic methods is shown in Tables 3–5, respectively.
Table 3.
Number and percentage of ESâL producing Pseudomonas spp. detected by phenotypic methods

Table 5.
Number and percentage of MBL producing Pseudomonas spp. detected by phenotypic methods

Table 4.
Number and percentage of AmpC producing Pseudomonas spp. detected by phenotypic methods

Those isolates which were confirmed phenotypically as ESBL and AmpC producers were subjected to PCR for detection of blaCTX-M, blaSHV, blaTEM, blaAmpC genes. The frequency of blaCTX-M, blaSHV, blaTEM, blaAmpC genes is depicted in Table 6. BlaCTX-M was the most prevalent gene. [Figures 3 and 4] shows positive results of PCR for blaCTX-M and blaAmpC gene respectively.
Table 6.
bla (CTX-M, SHV, TEM) and blaAmpC gene distribution among Pseudomonas isolates from ICU patients

Figure 3.
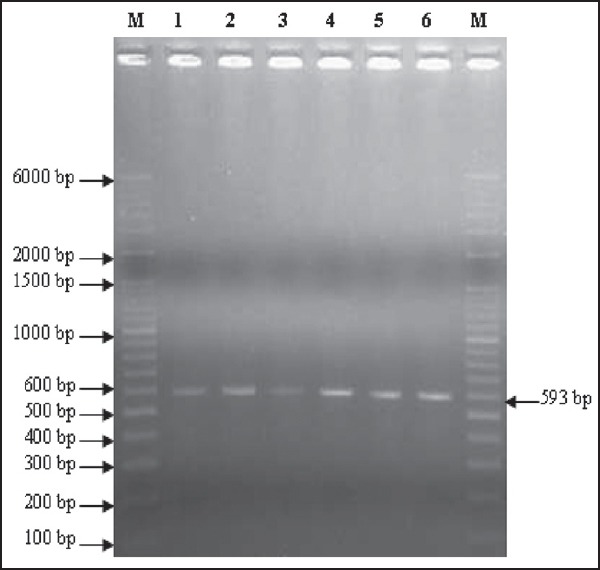
2% agarose gel electrophoresis showing results of pcr for the detection of BLActx-m gene. lanes m are showing ladders (o’ range rulertm 100 bp + 500 bp). Lanes 2-6 are showing test strains positive for BLActx-m gene (593 bp). Lane 1- positive control
Figure 4.
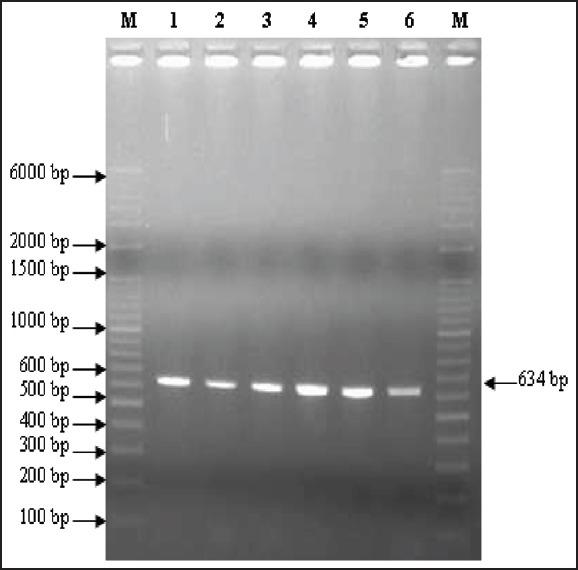
2% agarose gel electrophoresis showing results of pcr for the detection of BLAampc gene. Lanes m are showing ladders (o’ range rulertm 100 bp + 500 bp). Lanes 2-6 are showing test strains positive for BLAampc gene (634 bp). Lane 1 1- positive control
Figure 5 depicts the biofilm production in Pseudomonas isolates from ICU patients. Among the 35 isolates, 21 (60.4 %) were biofilm producers. Figure 6 shows antibiotic resistance profile of biofilm-positive (BFP) and biofilm-negative (BFN) Pseudomonas isolates.
Figure 5.
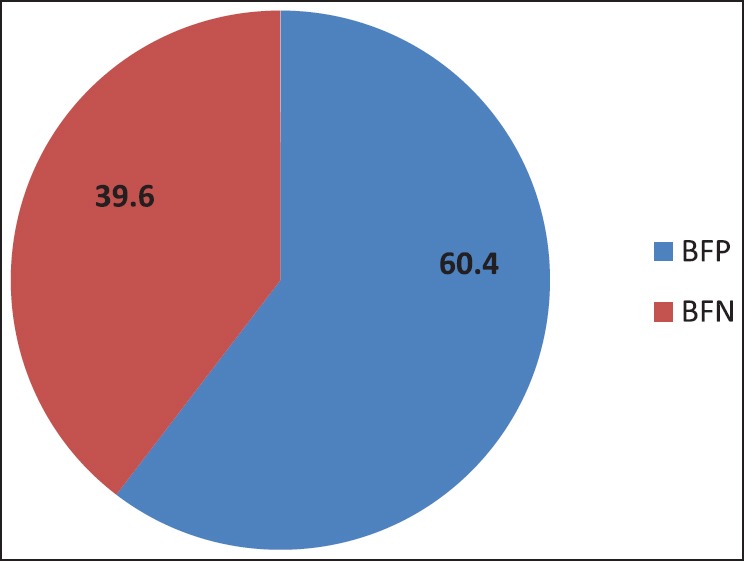
Biofilm production in Pseudomonas isolates from ICU patients
Figure 6.
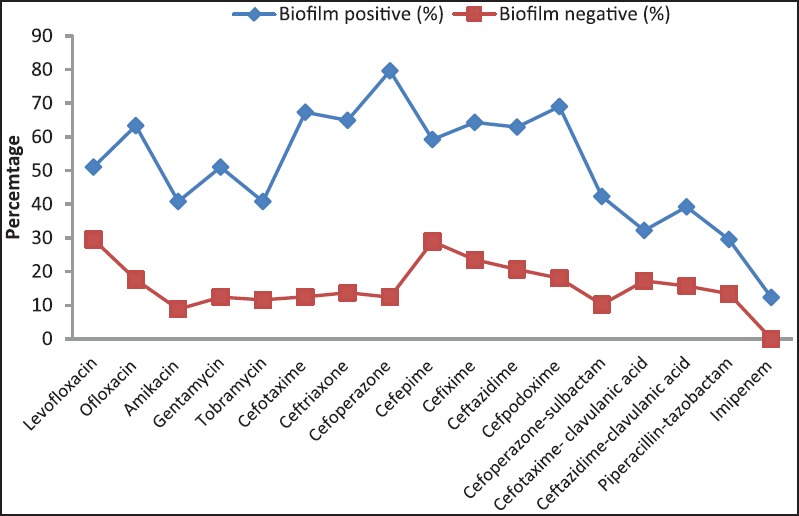
Antibiotic resistance profile of BFP and BFN Pseudomonas isolates
DISCUSSION
Pseudomonas spp. are one of the most common gram-negative pathogens associated with infections in ICU patients including bacteremia, urinary tract infections, and surgical site infections, but they predominate as agents of lower respiratory tract infections.[1]
In this study, Pseudomonas spp. represented 28% of isolates, similar results were reported by Hadadi et al.,[22] on other hand Chim et al.[23] and Izquierdo-Cubas et al.[24] found that Pseudomonas spp. represented 15.6% of the total isolates.
In this study lower respiratory tract infections (LRTIs) were the most common infection in ICU patients (34.3%). Our results agreed with Hadadi et al.,[22] who found that LRTI in ICU represented 29.5%. and also with that of Yetkin et al.[25] and Izquierdo-Cubas et al.[24] On the other hand, this result was not comparable with that of Al-Ghamdi et al.,[26] who reported that LRTIs represented 8.9% of cases.
In our study Pseudomonas spp. were frequently isolated from LRTIs (40%). This agreed with Abd El-Fattah[27] and El Masry.[28]
In our study, age distribution revealed that patients >50 years had higher risk for acquiring infections than younger patients. This result was in agreement with Stéphan et al.,[29] who reported that elderly patients had a higher incidence of infection in ICU compared with younger patients because the aging process leads to variable decline of physiologic functions and differential changes in other organ systems.
Our study showed that the most important risk factors significantly associated with infections caused by Pseudomonas spp. in ICU were endotracheal intubation and mechanical ventilation (P = <0.0001). Comorbid conditions significantly associated with ICU infections were COPD (P = 0.001) and hypertension (P = 0.004). Duration of ICU stay of 7-15 days (P=0.03) and >15 days (P = 0.0006) were also statistically significant factors associated with ICU infections. These results agreed with Giamarellou et al.[30] and Baran et al.[31]
Management of infections due to Pseudomonas spp. represents a difficult therapeutic challenge due to the increasing resistance levels of these organisms to most classes of antimicrobial agents. Our study revealed that on an average 76.1% and 70.2% of the isolated Pseudomonas spp. were resistant to fluoroquinolones and aminoglycosides, respectively. The problem of bacterial resistance to commonly used antibiotics is worldwide. Antibiotic resistance is a greater problem in developing countries especially due to easy availability of antibiotics over the counter.[32]
Resistances of Pseudomonas spp. to ceftazidime, cefotaxime and cefpodoxime were 68.5%, 67.1% and 65.7%, respectively. Comparable results were reported by McGowan[33] and Hadadia et al.[22]
ESβL-producing gram-negative bacilli have emerged as a serious problem in hospitalized patients. They pose a serious threat to the current β-lactam therapy as well as other antimicrobial agents. Screening by the disk diffusion method in this study revealed that 14.3% of Pseudomonas isolates were ESβL producers. These results were comparable to that of Aggarwal et al.,[34] however lower than results reported by Jiang et al.[35]
Confirmation with DDST and disk potentiation tests revealed that 8.5% and 11.4% Pseudomonas isolates in this study were ESβL producers in agreement with Jiang et al.[35] and Supriya et al.[36]
For AmpC production, both disk diffusion test and screening by cefoxitin disk revealed the same results that 51.4% of Pseudomonas spp. were AmpC producers. Similar results were reported by Altun et al.[37]
Those isolates confirmed phenotypically as ESBL and AmpC producers were subjected to PCR for the detection of gene responsible for the production of ESBL and AmpC. The most prevalent gene for ESBL production was blaCTX-M which was detected in 11.4% of Pseudomonas isolates while blaTEM and blaSHV genes were not detected in any of the isolates. These results are in agreement with Picao et al.[38]
PCR detected the blaAmpC gene in 42.8%. These results are comparable to that of Khanal et al.[39]
According to the disk diffusion method, results of our study revealed that 14.3% of Pseudomonas isolates were carbapenem resistant. The modified Hodge test revealed that 8.5% of Pseudomonas isolates were MβL producers. When tested by the imipenem-EDTA DDST, 11.4% of Pseudomonas spp. were MBL producers. However, no resistance was observed for polymyxin B and colistin. This result agreed with Behera et al.[40]
Comparison between modified Hodge test and imipenem-EDTA DDST in our study revealed that the imipenem-EDTA DDST was more sensitive for detecting MBL. The same observation was reported by Jesudason et al.[41] Mereuţă et al.[42] reported that double-disk and combined disk tests are useful, simple and accessible to clinical laboratories but PCR is needed to confirm the presence of the MβL gene in bacteria and to determine type of the enzymes.
Biofilms have an enormous impact on healthcare and are estimated to be associated with 65% of infections in ICU patients. Biofilm growth is associated with an increased level of mutations as well as with quorum-sensing-regulated mechanisms. Antimicrobial resistance is an innate feature of bacterial biofilms.[43] Many studies have shown that biofilm formation is higher in MDR strains. In this study, among 35 isolates, 21 (60.4 %) were biofilm producers. Carlos et al. reported biofilm formation in P. aeruginosa in 83% of clinical strains and that biofilm formation was prevalent among isolates with a MDR phenotype.[44] In our study we found higher antibiotic resistance in biofilm producers as compared to the negative biofilm producers.
CONCLUSION
In this study amikacin, tobramycin, imipenem, polymyxin B and colistin demonstrated maximum sensitivity against Pseudomonas species. Therefore, use of these antibiotics should be restricted to severe infections especially in critically ill ICU patients, in order to avoid rapid emergence of resistant strains.
Carbapenem resistance not only has enormous therapeutic implications, but is also important from the point of view of infection control. Such stains are known for rapid intra institutional spread and therefore, must be notified to infection control teams. Higher antibiotic resistances were seen in strong biofilm producers are due so testing for biofilm formation.
It is important to implement an easy, discriminatory, reproducible and cheap detection of MβL, ESβL and AmpC producers in our hospitals. Double disk synergy method for detection of ESBL and MBLs and estimation of cefoxitin resistance by the disk diffusion method for AmpC detection are not only cost effective and highly sensitive but also easy to perform.
Regular antimicrobial susceptibility surveillance is essential. An effective national and state level area-wise monitoring of the resistance patterns antibiotic policy and draft guidelines should be introduced to preserve the effectiveness of antibiotics and for better patient management.
Footnotes
Source of Support: Nil.
Conflict of Interest: None declared.
REFERENCES
- 1.Nseir S, Di Pompeo C, Brisson H, Dewavrin F, Tissier S, Diarra M, et al. Intensive care unit-acquired Stenotrophomonas maltophilia: Incidence, risk factors, and outcome. Critical Care. 2006;10:R143. doi: 10.1186/cc5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aloush V, Navon-Venezia S, Seigman-Igra Y, Cabili S, Carmeli Y. Multidrug-resistant Pseudomonas aeruginosa: Risk factors and clinical impact. Antimicrob Agents Chemother. 2006;50:43–8. doi: 10.1128/AAC.50.1.43-48.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Livermore DM. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: Our worst nightmare? Clin Infect Dis. 2002;34:634–40. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 4.Queenan AM, Bush K. Carbapenemases: The versatile β-lactamases. Clin Microbiol Rev. 2007;20:440–58. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornaglia G, Giamarellou H, Rossolini GM. Metallo-β-lactamases: A last frontier for β-lactams? Lancet Infect Dis. 2011;11:381–93. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 6.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: The quiet before the storm? Clin Microbiol Rev. 2005;18:306–25. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poirel L, Naas T, Nicolas D, Collet L, Bellais S, Cavallo JD, et al. Characterization of VIM-2, a carbapenem-hydrolyzing metallo β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob Agents Chemother. 2000;44:891–7. doi: 10.1128/aac.44.4.891-897.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunne WM., Jr Bacterial adhesion: Seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155–66. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Obritsch MD, Fish DN, MacLaren R, Jung R. National surveillance of antimicrobial resistance in Pseudomonas aeruginosa isolates obtained from intensive care unit patients from 1993 to 2002. Antimicrob Agents Chemother. 2004;48:4606–10. doi: 10.1128/AAC.48.12.4606-4610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collee JG, Duguid JP, Fraser AG, Marmion BP, Simmons A. Mackie and McCartney Practical Microbiology. 14th ed. London: Churchill Livingstone; 1996. Laboratory strategy in the diagnosis of infective syndrome; pp. 53–94. [Google Scholar]
- 11.Collee JG, Marr W. Mackie and McCartney Practical Microbiology. 14th ed. London: Churchill Livingstone; 1996. Culture of bacteria; pp. 113–30. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: Seventeenth informational supplement. M100-S17. Wayne, PA: Clinical and Laboratory Standards Institute; 2007. [Google Scholar]
- 13.Peterson DL, Bonomo RA. Extended spectrum beta lactamases: A clinical update. Clin Microbiol Rev. 2005;18:657–86. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended-spectrum beta lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: Hospital prevelance and susceptibility patterns. Rev Infect Dis. 1988;10:867–78. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 15.Shahid M, Malik A, Agarwal M, Singhal S. Phenotypic detection of the extended spectrum and AmpC β-lactamases by a new spot inoculation method and modified three-dimensional extract test: Comparison with conventional three- dimensional extract test. J Antimicrob Chemother. 2004;54:684–7. doi: 10.1093/jac/dkh389. [DOI] [PubMed] [Google Scholar]
- 16.Lee K, Lim JB, Yum JH, Yong D, Chong Y, Kim JM, et al. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob Agents Chemother. 2002;46:1053–8. doi: 10.1128/AAC.46.4.1053-1058.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shahid M, Ensor VM, Hawkey PM. Emergence and dissemination of Enterobacteriaceae with plasmid-mediated CMY-6 and CTX-M-15 beta-lactamases in a community in North-India. World J Microbiol Biotech. 2009;25:1439–46. [Google Scholar]
- 18.Shahid M, Malik A, Adil M, Jahan N, Malik R. Comparison of beta-lactamase genes in clinical and food bacterial isolates in India. J Infect Dev Ctries. 2009;3:593–8. doi: 10.3855/jidc.550. [DOI] [PubMed] [Google Scholar]
- 19.Feria C, Ferreira E, Correia JD, Goncalves J, Canica M. Patterns and mechanisms of resistance to beta-lactams and beta-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J Antimicrob Chemother. 2002;49:77–85. doi: 10.1093/jac/49.1.77. [DOI] [PubMed] [Google Scholar]
- 20.Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15:305–11. [PubMed] [Google Scholar]
- 21.Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: Evaluation of three different screening methods. Indian J Med Microbiol. 2006;24:25–9. doi: 10.4103/0255-0857.19890. [DOI] [PubMed] [Google Scholar]
- 22.Hadadi A, Rasoulinejad M, Maleki Z, Yonesian M, Shirani A, Kourorian Z. Antimicrobial resistance pattern of Gram-negative bacilli of nosocomial origin at 2 university hospitals in Iran. Diag Microbiol Infect Dis. 2008;60:301–5. doi: 10.1016/j.diagmicrobio.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 23.Chim H, Hock Tan B, Song C. Five-year review of infections in a burn intensive care unit: High incidence of Acinetobacter baumannii in a tropical climate. Burns. 2007;33:100–14. doi: 10.1016/j.burns.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Izquierdo-Cubas F, Zambrano A, Frómeta I, Gutiérrez A, Bastanzuri M, Guanche H, et al. National Prevalence of Nosocomial Infections. Cuba 2004. J Hosp Infect. 2008;68:234–40. doi: 10.1016/j.jhin.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 25.Yetkin G, Otlu B, Cicek A, Kuzucu C, Durmaz R. Clinical, microbiologic, and epidemiologic characteristics of Pseudomonas aeruginosa infections in a University Hospital. Am J Infect Control. 2006;34:188–92. doi: 10.1016/j.ajic.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 26.Al-Ghamdi S, Gedebou M, Bilal NE. Nosocomial infections and misuse of antibiotics in a provincial community hospital, Saudi Arabia. J Hosp Infec. 2002;50:115–21. doi: 10.1053/jhin.2001.1149. [DOI] [PubMed] [Google Scholar]
- 27.Abd El-Fattah SM. M.Sc Thesis in Medical Microbiology an Immunology. Faculty of Medicine. Giza, Egypt: Cairo University; 2008. Evaluation of antibiotic resistance among Gram-ve bacilli isolated from critically ill patients: Relation to risk factors and liberal use of antibiotics; pp. 66–7. [Google Scholar]
- 28.El Masry E. M.Sc Thesis in Medical Microbiology and Immunology. Faculty of Medicine. Menoufiya University; 2007. Extended spectrum beta-lactamase-producing Enterobacteriaceae and P. aeruginosa causing nosocomial pneumonia in Menoufiya university hospitals; pp. 97–106. [Google Scholar]
- 29.Stéphan F, Cheffi A, Bonnet F. Nosocomial Infections and Outcome of Critically Ill Elderly Patients after Surgery. Anesthesiology. 2001;94:407–14. doi: 10.1097/00000542-200103000-00008. [DOI] [PubMed] [Google Scholar]
- 30.Giamarellou H, Antoniadou A, Kanellakopoulou K. Acinetobacter baumannii: A universal threat to public health? J Hosp Infect. 2007;67:245–52. doi: 10.1016/j.ijantimicag.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 31.Baran G, Erbay A, Bodur H, Ongürü P, Akinci E, Balaban N, et al. Risk factors for nosocomial imipenem-resistant Acinetobacter baumannii infections. Int J Infect Dis. 2007;12:16–21. doi: 10.1016/j.ijid.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Martinez JL, Baquero F. Interactions among strategies associated with bacterial infection: Pathogenicity, epidemicity and antibiotic resistance. Clin Microbiol Rev. 2002;15:647–79. doi: 10.1128/CMR.15.4.647-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGowan JE., Jr Resistance in nonfermenting gram-negative bacteria: multidrug resistance to the maximum”. Am J Med. 2006;119(Suppl 1):S29–36. doi: 10.1016/j.amjmed.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 34.Aggarwal R, Chaudhary U, Bala K. Detection of extended-spectrum beta-lactamase in Pseudomonas aeruginosa. Indian J Pathol Microbiol. 2008;51:222–4. doi: 10.4103/0377-4929.41693. [DOI] [PubMed] [Google Scholar]
- 35.Jiang X, Zhang Z, Li M, Zhou D, Ruan F, Lu Y. Detection of extendedspectrum beta-lactamases in clinical isolates of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2006;50:2990–5. doi: 10.1128/AAC.01511-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Supriya S, Jalgaonkar V, Ahamad S, Hassani U. Evaluation of extended spectrum beta lactamase in urinary isolates. Indian J Med Res. 2004;120:553–6. [PubMed] [Google Scholar]
- 37.Altun Ş, Tufan ZK, Yağcı S, Önde U, Bulut C. Extended Spectrum Betalactamases, AmpC and Metallo Beta-lactamases in emerging multi-drug resistant Gram-negative bacteria in intensive care units. 2013;2:707. doi:10.4172/scientificreports. [Google Scholar]
- 38.Picão RC, Gales AC. Extended-spectrum beta-lactamase producing (ESBL) Pseudomonas aeruginosa: Nightmare or imagination? Prática Hospitalar. 2007;49:79–84. [Google Scholar]
- 39.Khanal S, Joshi DR, Bhatta DR, Devkota U, Pokhrel BM. β-Lactamase-Producing Multidrug-Resistant Bacterial Pathogens from Tracheal Aspirates of Intensive Care Unit Patients at National Institute of Neurological and Allied Sciences, Nepal. ISRN Microbiol. 2013;2013:847569. doi: 10.1155/2013/847569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behera B, Mathur P, Das A, Kapil A, Sharma V. An evaluation of four different phenotypic techniques for detection of metallo-beta-lactamase producing Pseudomonas aeruginosa. Indian J Med Microbiol. 2008;26:233–7. doi: 10.4103/0255-0857.39587. [DOI] [PubMed] [Google Scholar]
- 41.Jesudason MV, Kandathi AJ, Balaji V. Comparison of the methods for the detection of the carbapenamase and the metallo-beta- lactamases production in the clinical isolates. Indian J Med Res. 2005;121:780–3. [PubMed] [Google Scholar]
- 42.Mereuţă AI, Poiati A, Tuchiluş C, Dorneanu O, Nistor S, Copăcianu B. Screening methods for detection of metallo-beta-lactamase producing gram-ve rods. Rev Med Chir Soc Med Nat Iasi. 2008;109:387–91. [PubMed] [Google Scholar]
- 43.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–32. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez CJ, Jr, Mende K, Beckius ML, Akers KS, Romano DR, Wenke JC, et al. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis. 2013;13:47. doi: 10.1186/1471-2334-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]


