Abstract
Background:
Urinary tract infections (UTIs) remain a major problem both in hospitalized and outdoor patients. Multidrug-resistant enterococci are emerging as a major nosocomial pathogen with increasing frequency. However, the incidence of community-acquired enterococcal infections and species prevalent in India is not thoroughly investigated.
Objectives:
This study aims to estimate the burden of community-acquired UTIs seen at a tertiary care hospital and to identify the Enterococcus species isolated from these patients. The study also aims to determine the antibiotic susceptibility pattern with reference to high-level aminoglycosides and vancomycin.
Materials and Methods:
Semi-quantitative cultures from a total of 22,810 urine samples obtained from patients seen at various Outpatient Departments were analyzed. From them 115 nonduplicate isolates of enterococci were obtained as significant pure growth (>105 cfu/ml) and speciated. Antibiotic susceptibility was performed by Kirby–Bauer disc diffusion method. Vancomycin resistance screening was performed by the vancomycin screen agar method recommended by Clinical and Laboratory Standards Institute and confirmed by determination of minimum inhibitory concentration by agar dilution method.
Results:
Of 115 enterococcal isolates, 61 were identified as Enterococcus faecalis, 42 as Enterococcus faecium, 3 each as Enterococcus dispar, and Enterococcus pseudoavium. High-level gentamicin resistance (HLGR) was higher in E. faecium (47.6%) than E. faecalis (32.7%) and HLSR also showed the same pattern with 47.6% and 27.9% resistance, respectively. Vancomycin resistant enterococci accounted for 11.3% of the isolates, and out of them 53.8% were E. faecium by agar dilution method.
Conclusion:
High rate of resistance to antibiotics of penicillin group and aminoglycosides was observed in our tertiary care hospital even in community acquired UTIs. Hence, there is an urgent need for more rational and restricted use of antimicrobials.
Keywords: Enterococcus faecalis, Enterococcus faecium, urinary tract infection, vancomycin resistant enterococci
INTRODUCTION
Enterococci are part of the normal resident flora of the gut, oral cavity, and female genital tract. They are well-known as nosocomial opportunistic pathogens.[1] E. faecalis (80–90%) and E. faecium (5–10%) are the two most commonly isolated enterococcal species from clinical samples.[2,3] The most frequent infections caused by these organisms include urinary tract infections (UTIs) followed by intra-abdominal or intrapelvic abscesses and blood stream infections.[4] Community-acquired infections due to enterococci are on the rise due to intensive use of broad spectrum antibiotics. Moreover, since vancomycin-resistant enterococci (VRE) from animal sources such as poultry and human foods of animal origin play an important role in human colonization and infection, a significant level of VRE colonization may be found among persons not associated with the health care setting.[5] High-level aminoglycoside resistance (HLAR), β-lactamase production, and glycopeptides resistance including VRE have been reported among enterococcal species.[6,7] Isolation of E. faecium and other species from clinical samples is on the rise and poses a significant health concern, as they show intrinsic resistance to many commonly used antibiotics and may lead to a treatment failure.[8]
Although a few studies from India have been published which report prevalence of enterococci in hospital settings as well as nosocomial infections with their antimicrobial sensitivity patterns, there is a paucity of information on the status of community-acquired enterococcal infections. This study was undertaken to know the prevalence and antimicrobial susceptibility patterns of enterococci in community-acquired UTI.
MATERIALS AND METHODS
A total of 22,810 urine samples screened for UTI from the patients attending various Outpatient Departments (OPDs) of the All India Institute of Medical Sciences, New Delhi over a period of 14 months from April 2013 to May 2014 were included in the study. Mid-stream urine samples were cultured by a semi-quantitative method on cysteine lactose electrolyte deficient medium (Hi-Media Laboratories, Mumbai, India). All the samples were screened for Enterococcus and nonduplicate isolates obtained in significant colony count (>105 CFU/ml) were included in the study. Community-acquired VRE included VRE cultured from patient <48 h after hospital admission, or as an outpatient. Patients who were previously hospitalized or residing in a nursing home for >24 h during the last 31 days, previous history of positive VRE culture or infection, received vancomycin in recent past, or those patients who had outpatient surgery were excluded. Coexisting medical or other factors possibly associated with healthcare exposure (diabetes mellitus, prior antimicrobial drug therapy, renal failure, and immunosuppression) were assessed for all VRE cases.
The strains were identified and speciated according to standard laboratory procedures as per the scheme of Facklam and Collins.[9] Antimicrobial susceptibility was determined by Kirby–Bauer disc diffusion method. Various antibiotics tested were ampicillin (10 μg), amoxyclav (30 μg), norfloxacin (10 μg), tetracycline (30 μg), ciprofloxacin (5 μg), nitrofurantoin (300 µg), vancomycin (30 μg), teicoplanin (30 μg), fosfomycin (200 μg), and linezolid (15 μg) (Hi-Media Laboratories, Mumbai, India).
HLAR was determined by disc diffusion method using high-level gentamicin (120 µg) and streptomycin (300 μg) discs (Hi-Media Laboratories Pvt., Ltd., Mumbai, India). Vancomycin screen agar test was performed using brain heart infusion agar with 6 μg/ml vancomycin (Hi-Media Laboratories, Mumbai, India) to look for resistance to vancomycin. One or more colonies or a film of growth on this screen agar was considered to be resistant to vancomycin. Control strains included were Staphylococcus aureus ATCC 25923, E. faecalis ATCC 51299, and E. faecalis ATCC 29212.
Minimum inhibitory concentration testing
Determination of minimum inhibitory concentration (MIC) of vancomycin for enterococcal isolates which grew on vancomycin agar screen was done by agar dilution method.[10] Muller-Hinton agar was supplemented with different concentrations of vancomycin. Ten microliter of bacterial culture was spot inoculated after adjusting the turbidity with McFarland 0.5 standard. The plates were incubated at 37°C for 24 h and examined for growth. The minimum concentration of vancomycin which inhibited bacterial growth was considered MIC. Enterococci which had MIC >32 μg/ml were considered resistant; 8–16 μg/ml as intermediately resistant; and MIC of 4 μg/ml as susceptible to vancomycin as per Clinical and Laboratory Standards Institute guidelines.[10] Data were analyzed, and descriptive percentages were mentioned using SPSS statistical software (version 20) [IBM Corp. NY, USA].
RESULTS
Of the 22,810 urine samples screened for UTI, 115 (0.5%) Enterococcus isolates were obtained during the study period. The ages of the study population ranged between 1 and 75 years with a mean age of 36.8 ±20.0 years. Of 115 patients, 62 (53.9%) were males, and 53 (46.1%) were females. Most cases were from Urology OPD (24.3%) followed by Medicine OPD (18.3%) in our hospital. The most common enterococcal isolate was E. faecalis (61/115 [53%]), followed by E. faecium (42/115 [36.5%]), E. pseudoavium (3/115 [2.6%]), E. dispar (3/115 [2.6%]), E. casseliflavus (2/115 [1.7%]), E. avium (2/115 [1.7%]), and E. gallinarum (2/115 [1.7%]).
Antimicrobial resistance profile of enterococcal isolates showed that resistance was most frequently observed with tetracycline (77.4%), ciprofloxacin (74.8%), and norfloxacin (66.1%). E. faecium strains as compared to E. faecalis display a higher degree of drug resistance to multiple other antibiotics including ampicillin, amoxyclav, ciprofloxacin, gentamicin, nitrofurantoin, norfloxacin, streptomycin, and teicoplanin. All the enterococcal strains were sensitive to fosfomycin and linezolid by standard disc diffusion method.
Sixteen enterococcal isolates (13.9%) were vancomycin resistant by Kirby–Bauer disc diffusion method (E. faecalis-8, E. faecium- 6, E. gallinarum-2) [Table 1]. Risk factors and demographic characteristics for community-acquired VRE patients are summarized in Table 2. By vancomycin agar screen method, fifteen isolates showed growth, giving an overall VRE positivity of 13%. Therefore, the concordance between the two methods was more than 93.7%. Thirteen isolates showed MIC of vancomycin ≥128 μg/ml and were resistant to vancomycin by agar dilution method. Two enterococcal isolates were intermediately resistant to vancomycin with MIC of 8 μg/ml and 16 µg/ml, respectively. Therefore, 13 isolates were identified as VRE and two isolates showed intermediate resistance.
Table 1.
Antibiotic resistance (%) among the enterococcal isolates by Kirby-Bauer disc diffusion method
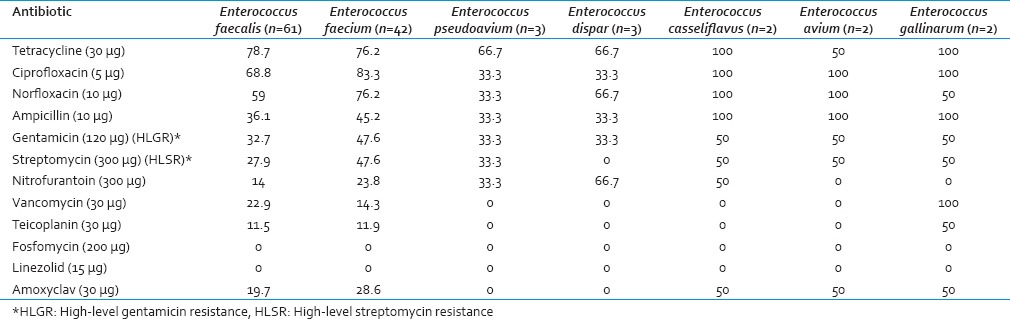
Table 2.
Risk factors and demographic characteristics for community-acquired vancomycin-resistant enterococci patients

Of the 13 isolates of Enterococcus found to be resistant to vancomycin by MIC testing, 12 were found to be resistant to teicoplanin by disc diffusion method also. These 12 isolates (92.3% of VRE), therefore, showed the Van A type of phenotype (resistance to both vancomycin and teicoplanin). On speciation, 6 of these were identified as E. faecalis, 4 E. faecium, and 2 as E. gallinarum. One isolate of E. faecium showed the Van B type of phenotype (resistant to vancomycin, sensitive to teicoplanin).
High-level gentamicin resistance (HLGR) was more in E. faecium (47.6%) than E. faecalis (32.7%) and HLSR was also same with 47.6% and 27.9% resistance, respectively [Table 1]. HLGR was detected in 39.1% and HLSR in 35.6% of the total isolates. Forty-one isolates (35.6%) showed both HLGR and HLSR which included 14.7% (n = 17) of the E. faecalis and 17.4% (n = 20) of the E. faecium isolates. Out of thirteen vancomycin-resistant isolates, HLGR and HLSR were found in 92.3% and 84.6% isolates, respectively.
DISCUSSION
In this study, E. faecalis (53%) was found to be the most predominant isolate in community acquired enterococcal UTIs. Most clinical infections were due to either E. faecalis or E. faecium. Most of the studies done on enterococci support the similar findings which could be attributed to the predominance of E. faecalis in the endogenous flora of the body.[11]
The sources and reservoirs that play a role in the resistance to antibiotics of enterococci that are community acquired are not known. Reports from developed countries suggest that VRE colonization can frequently occur in the community, suggesting a reservoir in the community.[12,13] VRE have also been found in sewage, from stools of healthy farm animals and animal products. In many countries, glycopeptides containing animal feeds are believed to be responsible for VRE intestinal colonization in farm animals and pets which ultimately results in colonization of humans via the food chain.[14] Therefore, transmission of VRE from these sources may result in an increased human reservoir of VRE colonization. Control of these cases, therefore, requires community-based initiative.
E. faecium strains as compared to E. faecalis display a higher degree of drug resistance to multiple other antibiotics. This emphasizes the need to speciate enterococcal isolates from clinical samples.
The past records of the patients did not indicate prior exposure to vancomycin in any of these patients. In our study, 11.3% were found to be VRE, of which 53.8% were E. faecalis. Interestingly our observations were in in accordance with two Indian studies, one from Gujarat and one from South India, in which E. faecalis was found to be a predominant isolate among VRE.[15,16] However, in most of the other studies E. faecium was found to be the most common isolate among VRE.[17] Emergence of VRE is of concern due to the limited therapeutic options.[17,18,19] This study shows that 92.3% of VRE isolates had Van A phenotype. This finding is in accordance with a previous study by Mathur et al., who also reported Van A phenotype in 80% of their VRE isolates.[19]
In present study, HLAR was seen in 39.1% and 35.6% of the enterococcal isolates for gentamicin (HLGR) and streptomycin (HLSR), respectively. Among E. faecalis and E. faecium isolates resistance rate was more against streptomycin than gentamicin (HLSR), as reported by others which is similar to previous studies.[19,20] This observation is important because the HLAR in enterococci isolates can well nullify the efficacy of combination therapy. The high prevalence of HLSR strains even if it is due to intrinsic low-level resistance for streptomycin, restricts the clinical use of aminoglycosides for enterococci.
CONCLUSION
The vancomycin resistance rate among the Enterococcus isolates was 11.3% in our study which was high as compared to other reports from North India. The most common phenotype of glycopeptide resistance seen in our study was the Van A phenotype (resistance to both vancomycin and teicoplanin). Other phenotypes seen were Van B (resistant to vancomycin, sensitive to teicoplanin).
The acquisition of HLAR and vancomycin resistance has limited the therapeutic options available for clinicians. Therefore, it is suggested that clinical microbiology laboratories should establish effective detection methods for vancomycin resistance. Sources for community-acquired VRE should be investigated on top priority, to contain the spread of VRE to the hospital environment and other populations.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones ME, Draghi DC, Thornsberry C, Karlowsky JA, Sahm DF, Wenzel RP. Emerging resistance among bacterial pathogens in the intensive care unit – A European and North American Surveillance study (2000-2002) Ann Clin Microbiol Antimicrob. 2004;3:14. doi: 10.1186/1476-0711-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jett BD, Huycke MM, Gilmore MS. Virulence of enterococci. Clin Microbiol Rev. 1994;7:462–78. doi: 10.1128/cmr.7.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Low DE, Keller N, Barth A, Jones RN. Clinical prevalence, antimicrobial susceptibility, and geographic resistance patterns of enterococci: Results from the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis. 2001;32(Suppl 2):S133–45. doi: 10.1086/320185. [DOI] [PubMed] [Google Scholar]
- 5.Mathur P, Singh S. Multidrug resistance in bacteria: A serious patient safety challenge for India. J Lab Physicians. 2013;5:5–10. doi: 10.4103/0974-2727.115898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDonald LC, Kuehnert MJ, Tenover FC, Jarvis WR. Vancomycin-resistant enterococci outside the health-care setting: Prevalence, sources, and public health implications. Emerg Infect Dis. 1997;3:311–7. doi: 10.3201/eid0303.970307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heuer OE, Hammerum AM, Collignon P, Wegener HC. Human health hazard from antimicrobial-resistant enterococci in animals and food. Clin Infect Dis. 2006;43:911–6. doi: 10.1086/507534. [DOI] [PubMed] [Google Scholar]
- 8.O'Driscoll T, Crank CW. Vancomycin-resistant enterococcal infections: Epidemiology, clinical manifestations, and optimal management. Infect Drug Resist. 2015;8:217–30. doi: 10.2147/IDR.S54125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Facklam RR, Collins MD. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–4. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. Clinical and Laboratory Standards Institute. 21st International Supplements. CLSI Document M100-S21. Wayne, Pennysylavania, USA: CLSI; 2011. Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 11.Silverman J, Thal LA, Perri MB, Bostic G, Zervos MJ. Epidemiologic evaluation of antimicrobial resistance in community-acquired enterococci. J Clin Microbiol. 1998;36:830–2. doi: 10.1128/jcm.36.3.830-832.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wegener HC. Antibiotics in animal feed and their role in resistance development. Curr Opin Microbiol. 2003;6:439–45. doi: 10.1016/j.mib.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 13.Bates J, Jordens JZ, Griffiths DT. Farm animals as a putative reservoir for vancomycin-resistant enterococcal infection in man. J Antimicrob Chemother. 1994;34:507–14. doi: 10.1093/jac/34.4.507. [DOI] [PubMed] [Google Scholar]
- 14.Bates J. Epidemiology of vancomycin-resistant enterococci in the community and the relevance of farm animals to human infection. J Hosp Infect. 1997;37:89–101. doi: 10.1016/s0195-6701(97)90179-1. [DOI] [PubMed] [Google Scholar]
- 15.Shah L, Mulla S, Patel KG, Rewadiwala S. Prevalence of enterococci with higher resistance level in a tertiary care hospital: A matter of concern. Natl J Med Res. 2012;2:25–7. [Google Scholar]
- 16.Praharaj I, Sujatha S, Parija SC. Phenotypic and genotypic characterization of vancomycin resistant Enterococcus isolates from clinical specimens. Indian J Med Res. 2013;138:549–56. [PMC free article] [PubMed] [Google Scholar]
- 17.Taneja N, Rani P, Emmanuel R, Sharma M. Significance of vancomycin resistant enterococci from urinary specimens at a tertiary care centre in northern India. Indian J Med Res. 2004;119:72–4. [PubMed] [Google Scholar]
- 18.Kapoor L, Randhawa VS, Deb M. Antimicrobial resistance of enterococcal blood isolates at a pediatric care hospital in India. Jpn J Infect Dis. 2005;58:101–3. [PubMed] [Google Scholar]
- 19.Mathur P, Kapil A, Chandra R, Sharma P, Das B. Antimicrobial resistance in Enterococcus faecalis at a tertiary care centre of Northern India. Indian J Med Res. 2003;118:25–8. [PubMed] [Google Scholar]
- 20.Scagnelli M, Pellizer G, de Lalla F, D'Emilio A, Rassu M, Bragagnolo L, et al. Epidemiological analysis of vancomycin-resistant enterococci in a large tertiary-care hospital in Northern Italy. Eur J Clin Microbiol Infect Dis. 2001;20:609–16. doi: 10.1007/s100960100573. [DOI] [PubMed] [Google Scholar]


