Summary
Smc/ScpAB promotes chromosome segregation in prokaryotes, presumably by compacting and resolving nascent sister chromosomes. The underlying mechanisms, however, are poorly understood. Here, we investigate the role of the Smc ATPase activity in the recruitment of Smc/ScpAB to the Bacillus subtilis chromosome. We demonstrate that targeting of Smc/ScpAB to ParB/parS loading sites is strictly dependent on engagement of Smc head domains and relies on an open organization of the Smc coiled coils. We find that dimerization of the Smc hinge domain stabilizes closed Smc rods and hinders head engagement as well as chromosomal targeting. Conversely, the ScpAB sub-complex promotes head engagement and Smc rod opening and thereby facilitates recruitment of Smc to parS sites. Upon ATP hydrolysis, Smc/ScpAB is released from loading sites and relocates within the chromosome—presumably through translocation along DNA double helices. Our findings define an intermediate state in the process of chromosome organization by Smc.
Graphical Abstract
Highlights
-
•
ATP-dependent head engagement is required for chromosomal targeting of Smc/ScpAB
-
•
ATP-dependent head engagement drives coiled-coil opening of Smc in vivo
-
•
Targeting of Smc/ScpAB to parS/ParB requires the head-proximal coiled coil of Smc
-
•
Hinge dimerization antagonizes head engagement, coiled-coil opening, and targeting
Smc/ScpAB is an important chromosome-organizing machine in bacteria. Minnen et al. show that targeting of Smc/ScpAB to chromosomal parS/ParB sites requires ATP-dependent engagement of Smc heads, which promotes disengagement of the Smc coiled coils and drives the complex into a targeting-competent open conformation.
Introduction
Proper segregation of the genetic material during cell division relies on the organization of replicated DNA molecules into compact and individualized sister chromosomes. In eukaryotes, condensation and resolution of chromatin into morphologically distinct chromatids occurs early in mitosis and depends on the interplay of nucleosomes, DNA topoisomerase II and structural maintenance of chromosomes (SMC) protein complexes such as condensin and cohesin (Houlard et al., 2015, Shintomi et al., 2015). In bacteria, segments of the circular chromosome are sequentially partitioned to opposite halves of the cell in line with their duplication by the two replication forks. Resolution of bacterial chromosomes is thus an ordered process, which initiates near the replication origin and concludes with the separation of the replication terminus region. Prokaryotic SMC complexes, called Smc/ScpAB and MukBEF, are enriched in the vicinity of the replication origin on the bacterial chromosome (Badrinarayanan et al., 2012, Danilova et al., 2007, Gruber and Errington, 2009, Minnen et al., 2011, Sullivan et al., 2009, Wilhelm et al., 2015). In rapidly growing cells of Bacillus subtilis, inactivation of Smc/ScpAB is lethal due to a severe block in replication origin separation and nucleoid partitioning (Britton et al., 1998, Gruber et al., 2014, Mascarenhas et al., 2002, Moriya et al., 1998, Soppa et al., 2002, Wang et al., 2014). How Smc/ScpAB enables timely resolution of sister replication origins is largely unclear.
A 50-nm long intramolecular coiled-coil constitutes the central part of SMC proteins, which connects a “hinge” domain with an ATPase “head” domain (Haering et al., 2002, Hirano and Hirano, 2002, Melby et al., 1998). In bacteria, homotypic interaction of two Smc proteins at their hinge supports the alignment of the two Smc coiled coils to produce a highly elongated rod-shaped Smc dimer (Soh et al., 2015). At the hinge-distal end of the Smc rod a single subunit of the kleisin family of proteins (named ScpA in bacteria) binds to the Smc dimer via two separate interfaces (Bürmann et al., 2013, Gruber et al., 2003, Haering et al., 2002, Schleiffer et al., 2003). A helical bundle is formed by ScpA’s N-terminal domain and the “neck” coiled coil of one Smc subunit (Bürmann et al., 2013, Gligoris et al., 2014). ScpA’s C-terminal winged-helix domain attaches to the “cap” of the head in the other Smc subunit (Bürmann et al., 2013, Haering et al., 2004). Asymmetric tripartite rings made up of one ScpA and two Smc proteins are thus formed. Like its eukaryotic descendants, Smc/ScpAB entraps chromosomal DNA molecules within the confines of its SMC/kleisin ring (Cuylen et al., 2011, Gligoris et al., 2014, Wilhelm et al., 2015). DNA entrapment depends on the ScpB subunit, which forms dimers and associates with a central segment of ScpA, as well as on ATP hydrolysis by the Smc complex (Bürmann et al., 2013, Kamada et al., 2013, Wilhelm et al., 2015).
Smc/ScpAB localizes in foci within the bacterial cell. These Smc protein clusters are generally positioned in the vicinity of a copy of the replication origin in B. subtilis and Streptococcus pneumoniae (Graumann et al., 1998, Gruber and Errington, 2009, Kleine Borgmann et al., 2013, Minnen et al., 2011, Sullivan et al., 2009). Targeting of Smc/ScpAB to the replication origin region and formation of Smc foci relies on ParB protein and parS sites. ParB binds to short palindromic parS sequences, the six most prominent of which are scattered within a 350 kb region (<10% of the genome) surrounding the replication origin in B. subtilis. In several bacteria, the replicating chromosome displays a distinctive “longitudinal” organization within the cell (Le et al., 2013, Marbouty et al., 2014, Marbouty et al., 2015, Umbarger et al., 2011, Vallet-Gely and Boccard, 2013, Wang et al., 2015): The newly replicated origins are generally found at the outer edges of the elongating chromosome, while other loci on the nascent chromosome are linearly arranged between the replication origin and the more centrally located terminus. Corresponding positions on opposite arms of the chromosome are frequently juxtaposed. The Smc/ScpAB complex as well as ParB protein and parS sites are essential for establishing this longitudinal organization of bacterial chromosomes (Le et al., 2013, Marbouty et al., 2015, Umbarger et al., 2011, Wang et al., 2015). How the loading of Smc/ScpAB by ParB/parS at few genomic positions governs global chromosome organization, however, is unclear (Bürmann and Gruber, 2015).
The SMC head domains share a common fold with nucleotide binding domains (NBD) found in ABC transporters. These domains undergo cycles of engagement and disengagement driven by ATP binding and ATP hydrolysis. In Smc/ScpAB, the Smc ATPase controls DNA binding to the distant hinge domain (Hirano and Hirano, 2006, Soh et al., 2015). Head engagement appears to promote the dissolution of the Smc coiled coil rod and thereby exposes an otherwise occluded binding site for DNA at the Smc hinge (Soh et al., 2015). If, and how, such a potential ATP-driven conformational transition might be relevant for the ParB-dependent recruitment of Smc/ScpAB toward the replication origin region of the bacterial chromosome is unclear. In yeast, ATP hydrolysis by cohesin has been implicated in its chromosomal relocation from sites occupied by the loading complex. However, the underlying molecular mechanisms remain elusive (Hu et al., 2011).
Here, we show that the Smc ATPase cycle controls the dynamic association of Smc/ScpAB with the bacterial chromosome. It determines the initial targeting of Smc/ScpAB to its chromosomal loading sites and subsequent re-distribution into flanking DNA. Smc head engagement is crucial for the recognition of the ParB/parS loading platform. We find that Smc head engagement is remarkably inefficient in Smc/ScpAB, due to the inhibitory action of Smc hinge and Smc rod, which is partially relieved by ScpAB. A head-proximal region of the Smc coiled coil is critical for targeting to ParB/parS. An Smc mutant defective in ATP hydrolysis is highly enriched at parS sites but fails to localize to other parts of the chromosome, including the neighboring replication origin and distant chromosomal arm loci. Smc appears to be released from loading sites to relocate along DNA to other parts of the chromosome in a manner that requires at least one round of ATP hydrolysis. Overall, our results demonstrate that engagement and disengagement of Smc heads define two distinct modes of chromosome association by Smc/ScpAB. Furthermore, they support the intriguing notion that movement of Smc/ScpAB along chromosome DNA is a critical aspect of its activity, which might be related to DNA loop extrusion by Smc.
Results
Smc ATPase Activity Controls the Dynamic Distribution of Smc/ScpAB over the Bacterial Chromosome
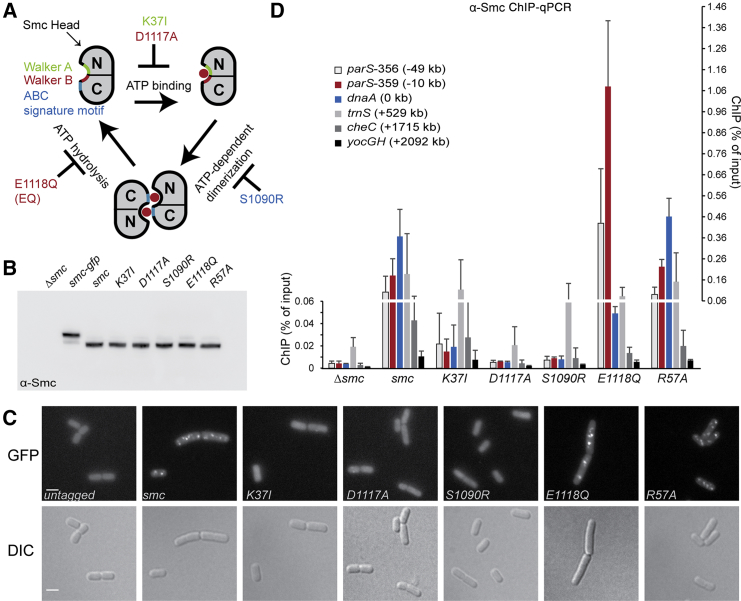
To investigate a potential role of the Smc ATP hydrolysis cycle in the localization of Smc/ScpAB complexes within the bacterial cell and on the bacterial chromosome, we made use of well-characterized, single amino-acid substitutions in the Smc head domain, which specifically block ATP binding (K37I or D1117A) or ATP-dependent head engagement (S1090R), alleviate a proposed stimulatory effect of DNA on ATP hydrolysis (R57A), or strongly reduce Smc ATP hydrolysis (E1118Q; or EQ for short) (Figure 1A) (Hirano et al., 2001, Hirano and Hirano, 2004, Hirano and Hirano, 2006, Lammens et al., 2004, Schwartz and Shapiro, 2011). These mutant proteins, with the exception of Smc(R57A), fail to support growth of B. subtilis on rich medium indicating that the mutations render the Smc protein non-functional (Figure S1A) (Gruber et al., 2014). All non-functional Smc proteins, however, are expressed to normal levels as judged by immunoblotting using an α-Smc antiserum (Figure 1B) and efficiently bind to the kleisin subunit ScpA (Bürmann et al., 2013, Wilhelm et al., 2015). The smc alleles were then tagged at their C terminus with a monomeric version of gfp and integrated into the endogenous locus by allelic replacement. Cells were analyzed by fluorescence imaging on minimal medium, which supports near normal growth of smc mutant strains. Wild-type Smc-GFP protein formed approximately one GFP focus per μm cell length (Figures 1C and S1H) (Graumann et al., 1998, Gruber and Errington, 2009, Sullivan et al., 2009). The cellular localization of the R57A mutant protein was indistinguishable from wild-type Smc. In contrast, K37I, D1117A, and S1090R mutant proteins failed to form any discernible structures in fluorescence images being indicative of a dispersed cellular localization (Mascarenhas et al., 2005). Crucially, the Smc(EQ) protein produced bright GFP foci, which on average occurred slightly less frequently than in wild-type cells (Figures 1C and S1H). These observations demonstrate the involvement of the Smc ATPase activity in the sub-cellular organization of Smc complexes and indicate that Smc is able to localize within the cell when its ATP hydrolysis activity is reduced but not when ATP binding or Smc head engagement is blocked.
Figure 1.
Smc ATPase Activity Determines the Chromosomal Distribution of Smc/ScpAB
(A) Schematic representation of the Smc ATPase cycle.
(B) Immunoblotting of ATPase mutant Smc proteins with α-Smc antiserum. Whole-cell extracts from strains BSG1002, BSG1007–BSG1008, BSG1067, BSG1045–BSG1047, and BSG1083. See also Figure S1B.
(C) Fluorescence images of cells harboring mGFP-tagged Smc alleles: BSG1002, BSG1067–BSG1068, BSG1855–BSG1857, and BSG1881. Scale bar, 2 μm. Differential interference contrast (DIC) (bottom) and GFP fluorescence images (top) are shown. Quantification of foci number per cell is given in Figure S1H.
(D) ChIP-qPCR analysis of cells from strains BSG1002, BSG1007–BSG1008, BSG1045–BSG1047, and BSG1083 using α-Smc antiserum. Error bars were calculated from two independent experiments as SD. Please note that values of ChIP enrichment below and above 0.06% are displayed on different scales given on the left and right side of the graph, respectively. The analysis of chromosomal loci harboring highly transcribed genes (such as the tRNA cluster trnS) generally produce ambiguous results with relatively high levels of ChIP signal in control samples (Δsmc). This seems to be a widely observed phenomenon in ChIP and it remains unclear whether the enrichment is physiologically relevant.
See also Figure S1.
Next, we used untagged alleles of all ATPase mutants to examine their chromosomal distribution in B. subtilis by chromatin immunoprecipitation (ChIP) with an antiserum raised against the Smc protein. qPCR with primer pairs specific for selected regions of the chromosome was performed to measure the co-purification of chromosomal DNA with Smc. As predicted from their inability to form GFP foci in the cell, ATP binding and head engagement mutants resulted in little DNA co-purification similar to the Δsmc control—being consistent with the notion that ATP binding and engagement mutants fail to localize to the chromosome (Figure 1D). Wild-type Smc produced highly significant ChIP enrichment of origin proximal DNA (parS-356, parS-359, and dnaA) as observed before with tagged alleles of smc (Gruber and Errington, 2009). Intriguingly, the Smc(EQ) protein showed on the one hand markedly stronger localization to parS DNA than wild-type Smc and on the other hand quite low enrichment at the juxtaposed dnaA locus. Clearly, Smc(EQ) protein is able to efficiently target specific regions of the chromosome. However, wild-type distribution of Smc on the bacterial chromosome—including its prominent localization to the replication origin—requires hydrolysis of ATP by Smc. Smc(R57A) showed a ChIP-qPCR pattern indistinguishable from wild-type Smc indicating that cellular ATP hydrolysis might be (if at all) only mildly affected by this amino-acid substitution in B. subtilis.
Smc/ScpAB Specifically Recognizes ParB/parS Nucleoprotein Complexes in Its Pre-hydrolysis State
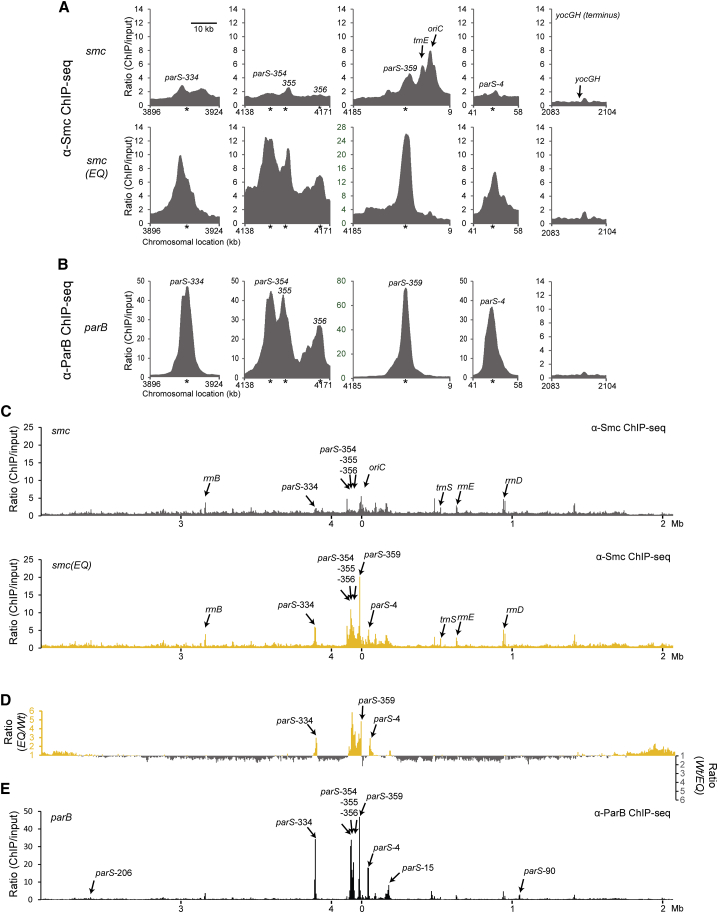
To get a global view of the chromosomal distribution of Smc and Smc(EQ) proteins, we then analyzed ChIP input and eluate fractions by next-generation sequencing. Individual sequence reads were mapped to 1 kb sliding windows spaced at 100-bp intervals (Figure 2A) or to 5 kb bins (Figure 2C). In order to normalize for the copy number differences between origin-proximal and -distal loci caused by ongoing DNA replication, the ratio of the normalized number of reads in input and eluate fractions was calculated for each window. The resulting ChIP sequencing (ChIP-seq) profile for the Smc(EQ) sample showed striking peaks that are overlapping with several parS sites on the chromosome (Figure 2A). The profile of the wild-type sample is markedly different from the Smc(EQ) profile (Figure 2A) (Gruber and Errington, 2009). Its peaks at parS sites are less pronounced. Instead, other more prominent peaks are present at and near the replication origin (oriC) and generally more signal was detected all along the chromosome arms. Largely similar ChIP-seq results were obtained with an antiserum raised against the ScpB subunit (Figures S2 and 7A; discussed below), indicating that the observed enrichments are not caused by antibody artifacts and suggesting that substantial fractions of Smc and ScpB co-localize on the chromosome, presumably by forming Smc/ScpAB holo-complexes (Kleine Borgmann et al., 2013). ChIP-qPCR experiments using several primer pairs for chromosomal arm sites (Figure S2D) confirmed that Smc (but not Smc(EQ)) was significantly enriched at several positions on the two chromosome arms with levels of enrichment inversely correlating with distance from the replication origin. Together, these results confirm the specific localization of the Smc(EQ) protein to parS sites and strongly suggest that Smc head engagement is essential for Smc/ScpAB recruitment to the chromosome. However, a full cycle of ATP hydrolysis appears to be involved in the localization of Smc/ScpAB to other chromosomal sites—such as the replication origin and the chromosome arms (Figure 2A).
Figure 2.
Hydrolysis Mutant but Not Wild-Type Smc Co-localizes with ParB/parS
(A) Close-up view of ChIP-seq profiles for wild-type Smc (BSG1002) (top panel) and Smc(EQ) (BSG1008) (bottom panel) generated using antiserum raised against the Bs Smc protein. Sequence reads were mapped to 1 kb windows spaced at 100-bp intervals and normalized for input DNA as follows. The number of reads for the ChIP sample in a given window was divided by the number of reads in the input sample for the same window (after normalizing the total number of reads). Raw input and ChIP data are shown in Figure S2. Axes labeled in green color highlights different scaling. Asterisks indicate the positions of parS sites.
(B) Close-up view of the ChIP-seq profile of ParB protein (from BSG1470 cells) generated using antiserum raised against purified BsParB-His6 protein. Data analysis and presentation as in (A).
(C) Whole-genome views of data presented in (A). Sequence reads were mapped to 5-kb windows spaced at 5-kb intervals across the genome and normalized for input DNA.
(D) To highlight differences between the distribution of wild-type Smc and Smc(EQ) the normalized ratios for Smc(EQ) in a given window was divided by the equivalent ratio for Smc(wt). Numbers above one are shown in yellow colors (axis on the left side). For numbers below one, the inverse ratio was calculated and displayed in gray colors (axis on the right side).
(E) Whole-genome view of the ParB ChIP-seq data presented in (B). Data analysis as in (C).
See also Figure S2.
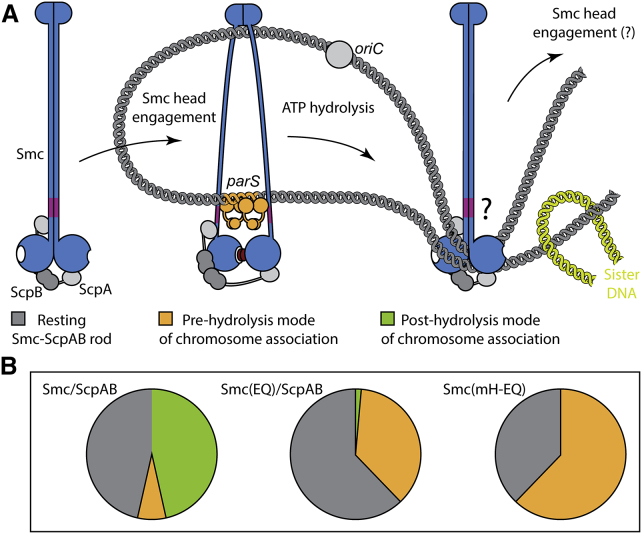
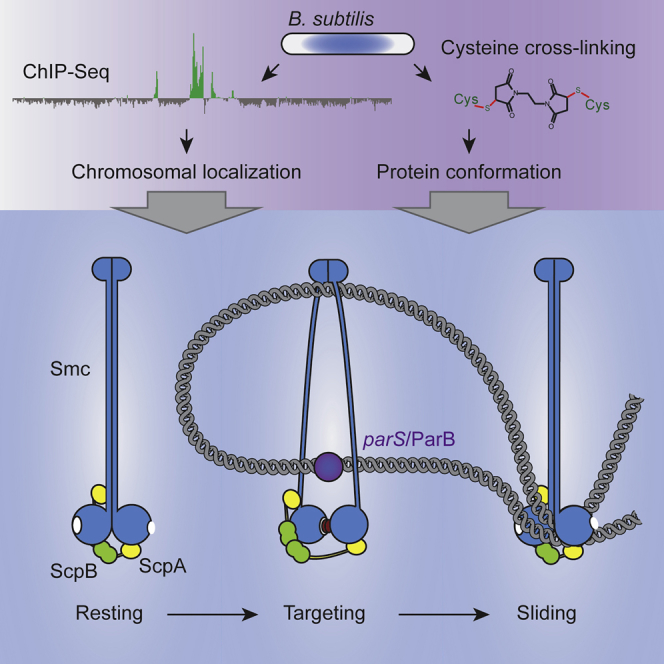
Figure 7.
Model for the Recruitment of Smc/ScpAB to and Release from parS Sites
(A) Model for the targeting to and release from ParB/parS by holo-Smc/ScpAB. Most Smc/ScpAB exists as a rod-shaped structure, which is unable to bind to DNA via its hinge or to ParB/parS via the coiled coils. Dissolution of the Smc rod and engagement of Smc head domains are prerequisites for the targeting of Smc/ScpAB to parS. Upon ATP hydrolysis, the ring-like structure might revert to the rod conformation and is released from parS DNA. Sister DNA segments (in green colors) might be excluded from the Smc rod due to steric restrictions. Repetitive rod-ring-rod transitions might drive DNA loop extrusion.
(B) Pie charts displaying rough estimates for the relative occupancy of the different states illustrated in (A) based on Smc head cross-linking efficiency (Figure 4B). Please note that the fraction of wild-type Smc complexes on and off the chromosome (depicted as green and gray pies in the left chart) is unknown. A tiny fraction of chromosomally loaded Smc(EQ)/ScpAB has been detected (Wilhelm et al., 2015).
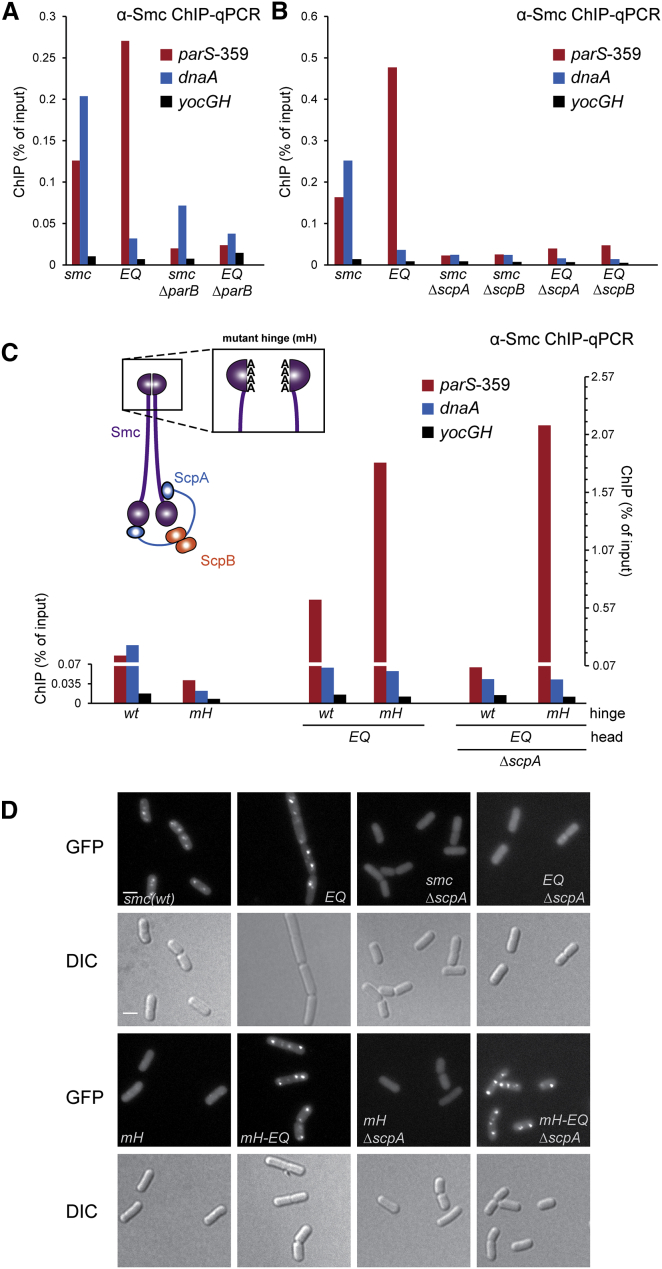
Furthermore, we found that the high levels of enrichment of Smc and Smc(EQ) at parS-359 are fully dependent on the presence of ParB protein (Figure 3A). To compare the distribution of Smc and ParB proteins near parS sites, we next performed ChIP-seq analysis using an antiserum against the ParB protein. ChIP-seq peaks of ParB and Smc(EQ) (but not Smc) proteins at parS sites are very similar in terms of positioning and shape, strongly indicating that the two proteins are closely co-localized on chromosomal DNA (Figures 2A–2C and 2E).
Figure 3.
Dimerization at the Smc Hinge Hinders Chromosomal Association of Smc/ScpAB
(A) ChIP-qPCR was performed on exponentially growing cells of strains BSG1051–BSG1052, BSG1406, and BSG1387 using α-Smc antiserum. Quantification of input and eluate material was done by qPCR using primer pairs specific for the indicated loci.
(B) As in (A) using strains BSG1889–BSG1894.
(C) The scheme indicates the disruptive effect of mutations in the Smc hinge domain on dimerization. ChIP-qPCR was performed with α-Smc antiserum on strains BSG1620–BSG1621, BSG1624, BSG1890, and BSG1892–BSG1893.
(D) Fluorescence imaging of Smc-mGFP fusion proteins in cells of strains BSG1067–BSG1068, BSG1378, BSG1413, BSG1662, BSG1677, and BSG1798–BSG1799. Scale bar, 2 μm. Quantification of foci number per cell is given in Figure S4C. Same experiments as in Figure 1C.
See also Figure S3.
In the absence of ScpA or ScpB, the Smc protein is non-functional and Smc-GFP fails to form foci in vivo (Lindow et al., 2002, Mascarenhas et al., 2002). We observed that the chromosomal localization of wild-type Smc measured by ChIP-qPCR is strongly reduced when scpA or scpB is deleted (Figure 3B). Poor localization was also observed in Smc(EQ) cells in the absence of ScpA or ScpB. We thus conclude that ATP-dependent engagement of Smc heads as well as ScpA and ScpB proteins are crucial for efficient localization of Smc to ParB/parS on the chromosome. Apparently, a particular conformation of Smc/ScpAB recognizes ParB/parS nucleoprotein structures.
Dimerization at the Smc Hinge Controls Chromosomal Association of Smc/ScpAB
Next, we investigated the role of the Smc hinge domain in the localization of Smc/ScpAB to the bacterial chromosome. We made use of a previously characterized mutation of four highly conserved glycine residues to alanine at the Bs Smc hinge-hinge interface (designated as “mH” for mutant hinge) to block formation of stable dimers at the Smc hinge domain (Hirano and Hirano, 2002) (Figures S3C and S3D). The ChIP-qPCR enrichment of Smc(mH) was clearly reduced at parS-359 as well as dnaA (Figure 3C). Dimerization of Smc at the hinge thus seems to be important for localization of wild-type Smc/ScpAB to the chromosome. This is consistent with the notion that Smc/ScpAB associates with the chromosome by entrapping the chromosomal DNA double helix within its SMC/kleisin ring (Wilhelm et al., 2015). In stark contrast, however, the enrichment of Smc(mH-EQ) protein at parS-359 (but not at dnaA) was strongly enhanced (∼4-fold) compared to Smc(EQ) (Figure 3C). Thus, hinge dimerization has strikingly antagonistic effects on the association of Smc and Smc(EQ) with the chromosome. Remarkably, ScpA and ScpB are dispensable for targeting of Smc(mH-EQ) to the chromosome (Figures 3C and S3H), while they are essential for wild-type Smc and Smc(EQ) to localize to the chromosome (Figure 3B). A plausible explanation for these striking observations is that the ScpAB sub-complex counteracts an inhibitory function of hinge dimerization on chromosomal targeting of Smc.
To ensure that these surprising findings are not caused by artifacts in the chromatin immunoprecipitation procedure, we have analyzed a set of Smc mutants by live-cell imaging of fluorescently labeled Smc proteins (Figure 3D). As predicted from the ChIP experiments, Smc(EQ)-GFP failed to form chromosomal foci when scpA is deleted, while Smc(mH-EQ)-GFP produced bright foci irrespective of the presence and absence of scpA. Together, these findings corroborate the view that Smc complexes associate with the bacterial chromosome in two fundamentally distinct manners, which are defined by the state of the Smc ATPase.
Smc Hinge Dimerization Inhibits Smc Head Engagement
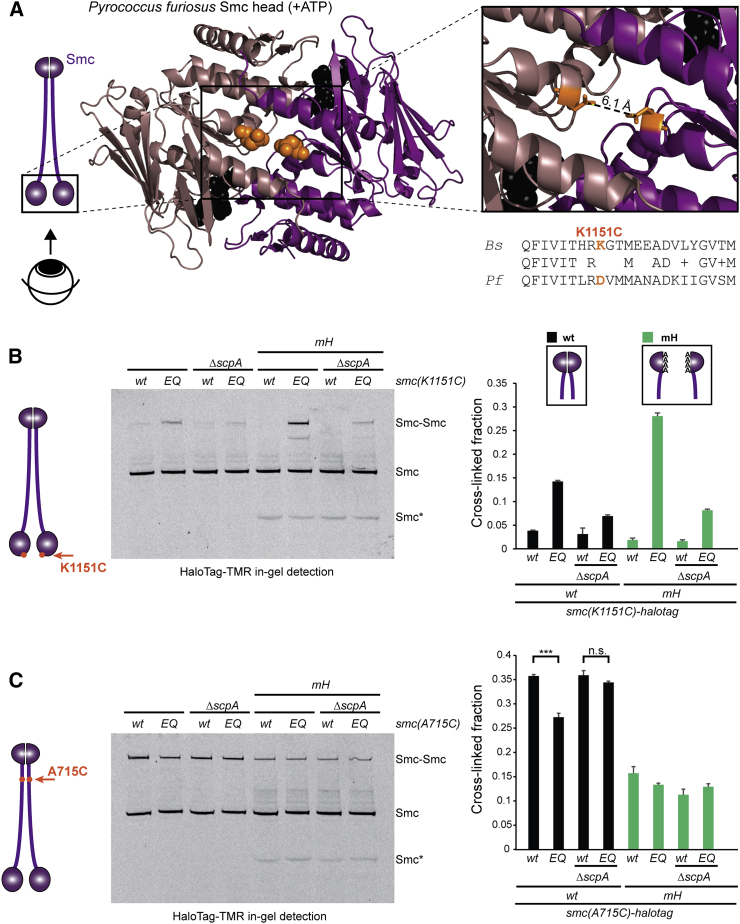
How might the ScpAB sub-complex and dimerization at the Smc hinge control targeting of Smc to chromosomal parS sites in antagonistic ways? Conceivably, ScpAB and the Smc hinge might positively and negatively influence the engagement of Smc head domains, respectively, and thereby regulate the recruitment of Smc/ScpAB to parS. If so, then the levels of head engagement should correlate with the efficiency of chromosomal targeting. To address this, we made use of the efficient chemical cross-linking of closely juxtaposed pairs of cysteines by a thiol-specific bis-maleimide compound (BMOE) in B. subtilis (Bürmann et al., 2013, Soh et al., 2015). Based on the crystal structure of the Pyrococcus furiosus Smc(EQ) head dimer in the presence of ATP (Lammens et al., 2004), we engineered a cysteine residue into the bottom surface of the Bs Smc head (K1151C), so that it is located in close proximity to its pair mate at the 2-fold symmetry axis of the dimer (Figure 4A). In order to be able to precisely quantitate the levels of cross-linking, we fused the cysteine bearing smc gene at its C terminus with a HaloTag thus allowing in-gel fluorescence detection of Smc. In addition, the four endogenous cysteines in Smc were replaced by serine residues to reduce the propensity for any off-target cross-linking (Hirano and Hirano, 2006). The corresponding smc allele supports growth on nutrient rich medium, implying that it is functional (Figure S4A). Cross-linking of K1151C in otherwise wild-type Smc was barely detectable (<4%), while the ATP hydrolysis mutant Smc produced a low but substantial fraction (∼14%) of cross-linked Smc dimers (Figure 4B). Intriguingly, K1151C cross-linking was undetectable in Smc(mH) but very pronounced in Smc(mH-EQ). The latter is cross-linked with an efficiency comparable to those previously observed for other Smc-Smc, Smc-ScpA, and DnaN-DnaN interfaces, which strongly suggests that the K1151C residue is a good reporter for head-head association (Bürmann et al., 2013, Soh et al., 2015, Wilhelm et al., 2015). The low levels of cross-linking observed with wild-type Smc and Smc(EQ) are therefore in most likelihood due to poor efficiency of head engagement. Deletion of scpA or scpB decreased the cross-linking of K1151C in Smc and Smc(EQ) protein even further (Figures 4B and S4B) being consistent with the notion that ScpAB might stimulate the targeting of Smc(EQ) to the chromosome (Figures 3B and S3H)—at least partly—by promoting head engagement (Bürmann et al., 2013, Kamada et al., 2013).
Figure 4.
Hinge Dimerization and Head Engagement Control the Conformation of Smc/ScpAB
(A) Structure of ATP engaged Pf Smc head domains (PDB: 1XEX) in brown and pink colors, respectively, (bottom view). Residue D1131 is indicated in ball representation in orange colors (middle panel). The distance between the carboxyl carbon atom in the side chains of the D1131 symmetry mates is estimated to be ∼6 Å (right panel). A sequence alignment between PfSmc and BsSmc shows that K1151 in BsSmc corresponds to D1131 in PfSmc.
(B) In vivo BMOE crosslinking of Smc(K1151C)-HaloTag in cells of strains BSG1488, BSG1509, BSG1512, BSG1547, BSG1597–BSG1598, BSG1791, and BSG1800. Four endogenous cysteine residues were replaced by serines. Cross-linked Smc-Halotag species were detected by in-gel fluorescence of the HaloTag-TMR substrate (left panel). Smc∗ indicates a degradation product of Smc(mH). The graph (right panel) shows mean values and SDs from three replicates.
(C) Same as in (B) using A715C as sensor cysteine for formation of Smc rods by the hinge proximal Smc coiled coil. In vivo crosslinking of Smc(A715C) with bismaleimidoethane (BMOE) in strains BSG1921–BSG1924, BSG1949–BSG1951, and BSG2036. T test statistics: ∗∗∗p ≤ 0.001; not significant (n.s.), p > 0.05.
See also Figure S4.
Smc head engagement must be a transient or rare phenomenon since it is barely detectable by cross-linking in wild-type Smc/ScpAB (Figure 4B). The maintenance of its association with the chromosome is thus very likely independent from continuous engagement of head domains. Smc head engagement, however, must be crucial during the establishment of chromosome association, because mutants defective in head engagement are unable to bind to the chromosome altogether. A stable association with the chromosome is likely created via the entrapment of chromosomal DNA within the Smc/ScpAB ring, a process that we have recently shown to be dependent on ATP hydrolysis (Wilhelm et al., 2015). We thus propose that Smc/ScpAB displays two distinct modes of binding to the chromosome. The first one, designated as pre-hydrolysis mode, occurring mostly or exclusively at parS sites presumably via a physical interaction with the chromosome, has a strict requirement for Smc head engagement. The second one, generated by transient head engagement and subsequent ATP hydrolysis, designated as post-hydrolysis mode, features a much more dispersed distribution on the bacterial chromosome and likely involves the entrapment of one or more DNA double helices within the Smc/ScpAB ring.
Rod-Shaped Smc Dimers Poorly Target ParB/parS
The levels of head engagement in Smc, Smc(EQ), and Smc(mH-EQ) correlate well with their efficiency of targeting to parS on the chromosome (Figures 3C and 4B). However, when scpA is deleted, both Smc(EQ) and Smc(mH-EQ) display similarly low levels of head cross-linking, whereas Smc(mH-EQ) but not Smc(EQ) exhibits strong enrichment at parS on the chromosome (Figures 3C and 4B). We conclude that hinge dimerization must have additional effects on Smc(EQ), through which it restricts chromosomal targeting, and Smc head engagement—albeit being essential—is not sufficient for targeting of Smc/ScpAB to parS sites.
Smc dimers form straight rods via the close juxtapositioning of the Smc coiled coils (Soh et al., 2015). Upon Smc head engagement and DNA binding, however, they have been proposed to undergo an extensive conformational change to a more open, possibly ring-like configuration in vitro (Soh et al., 2015). Conceivably, this structural transition might also regulate the binding of Smc/ScpAB to ParB/parS. If this were the case, then any Smc mutant that efficiently targets to parS might harbor unstable Smc rods. To investigate this, we employed in vivo cross-linking of a cysteine residue (A715C) located at the hinge-proximal interface between the two Smc coiled coils as an indicator for the formation of Smc rods (Figure 4C) (Soh et al., 2015). As reported previously, ∼35% of wild-type Smc(A715C) proteins were cross-linked into covalent dimers by BMOE (Figure 4C). The mutant hinge strongly decreased the fraction of Smc dimers displaying coiled coil rods, irrespective of the presence or absence of ScpA protein (Figure 4C). Similarly, the E1118Q mutation lead to a significant reduction in the fraction of Smc dimers with rod-shaped coiled coils providing direct evidence that the ATPase cycle affects the architecture of the Smc coiled coils near the Smc hinge in vivo. Crucially, the partial dissolution of Smc(EQ) rods was lost when the scpA gene was deleted, whereas the more pronounced opening of the coiled coils in Smc(mH, EQ) was unaffected by ΔscpA. The ScpA subunit thus facilitates the opening of the Smc rod (Figures 4B and 4C). Altogether, these data strongly support the notion that dimerization at the Smc hinge promotes Smc rod formation, which in turn opposes head engagement. ScpA is required to antagonize rod-stabilization exerted by the Smc hinge and consistent with this notion it becomes dispensable for rod opening (and chromosomal targeting) in the absence of a functional hinge. Hence, a combination of two interrelated structural features seems to be responsible for the targeting of Smc/ScpAB to parS sites on the chromosome: (1) engagement of Smc head domains, and (2) dissolution of the Smc rod. Both features appear to be rare or short-lived in wild-type Smc/ScpAB, presumably due to the inhibition by the hinge and the destabilizing action of the Smc ATPase. Nevertheless, a large fraction of cellular Smc/ScpAB must at least transiently adopt this conformation in order to localize to the replication origin region and to be able to form Smc-GFP foci in vivo (Figure 1) (Gruber and Errington, 2009, Sullivan et al., 2009).
Concomitantly, our A715C cross-linking experiments indicate that Smc coiled coils can be juxtaposed in a sizeable fraction of proteins even when dimerization at the Smc hinge is impaired, the ScpA bridge absent and Smc heads almost completely disengaged (Figure 4B). Thus, the association between Smc coiled coils contributes considerably to Smc dimerization.
The Smc Hinge Domain Is Dispensable for Targeting to parS DNA
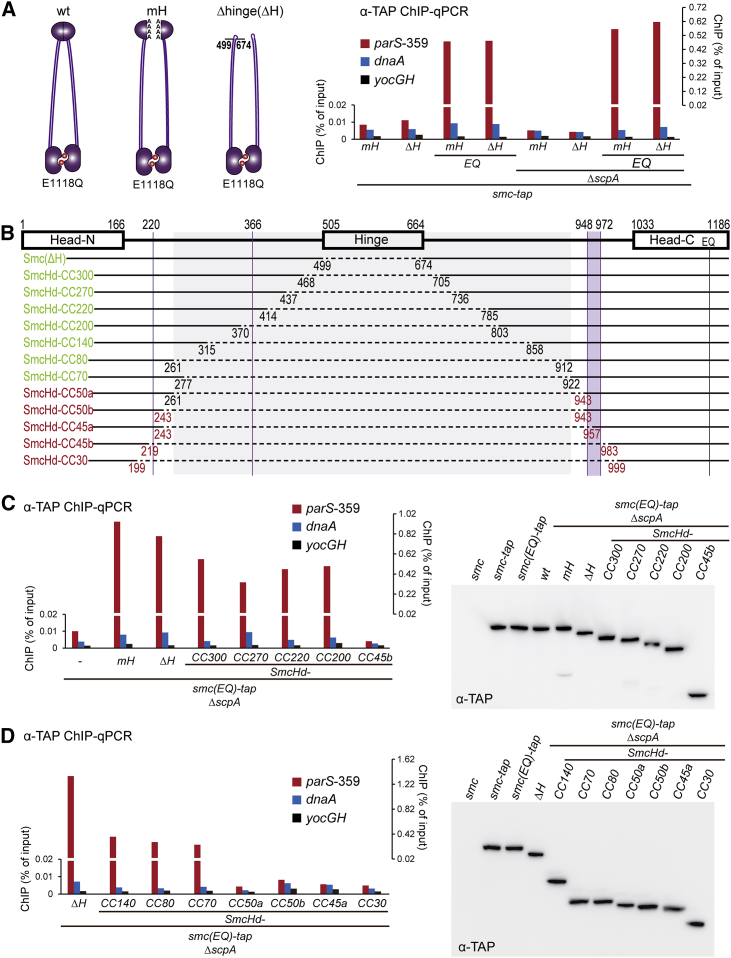
A DNA binding site has previously been mapped to the bottom surface of the Bs Smc hinge dimer (Hirano and Hirano, 2006, Soh et al., 2015). DNA binding at the coils/hinge junction appears to be sterically blocked by the juxtapositioning of the Smc coiled coils and promoted by ATP-dependent dissolution of the Smc rod (Soh et al., 2015). The same mechanism could be responsible for the targeting of Smc/ScpAB to parS sites on the chromosome. If ParB/parS—like naked DNA—were to bind to the bottom of the Smc hinge dimer, then the presence of the hinge domain would be crucial for localization of Smc(EQ) to the chromosome. To test this, we constructed an Smc fragment lacking the entire hinge domain (“ΔH” for hinge deletion) by connecting the end of Smc’s N-terminal coiled coil helix (amino acids 1–499) to the start of the C-terminal coiled coil helix (aa 674–1186) using a flexible linker peptide (-GGGSGGGSGGG-). The Smc(ΔH) construct was fused to a TAP tag at its C terminus and integrated at the endogenous smc locus. Smc(ΔH) was expressed at normal levels in B. subtilis but failed to localize to the chromosome as judged by α-TAP ChIP (Figures 5A and S5A). However, a Smc(ΔH) variant harboring the E1118Q mutation displayed robust localization to parS-359 in the presence and absence of the ScpA subunit (Figure 5A). Overall, strains harboring either a mutant Smc hinge domain or a complete deletion of the Smc hinge produced very similar results, clearly demonstrating that the Smc hinge is dispensable for the targeting of Smc(EQ) to parS (Figure 5A) and confirming that the hinge domain regulates chromosomal targeting indirectly—likely by affecting other parts of the Smc/ScpAB complex.
Figure 5.
A Large Central Part of Smc Is Dispensable for Targeting to parS
(A) ChIP-qPCR was performed with TAP-tagged alleles of Smc using IgG-coupled magnetic beads for immunoprecipitation. Strains: BSG1671–BSG1672, BSG1689, BSG1691, BSG1779–BSG1780, and BSG1895–BSG1896. The schemes on top represent modifications to the Smc hinge in Smc(mH) (left) and Smc(ΔH) (right).
(B) Schematic overview of the series of internal Smc truncation constructs. Solid and dashed horizontal lines denote the presence and absence of Smc sequences in a given truncation construct. A gray box demarcates the central portion of the Smc protein, which is dispensable for targeting to parS-359. N-terminal and C-terminal Smc sequences are fused via a short peptide linker (-GGGSGGGSGGG-). The name of a given truncation construct indicates the predicted length of its Smc coiled coil. Labels in green and red colors indicate efficient and inefficient targeting to parS. All proteins are tagged with a TAP tag at their C terminus. Purple vertical lines and boxes indicate disruptions in the Smc coiled coil (Waldman et al., 2015).
(C) ChIP-qPCR against the TAP tag of strains BSG1520, BSG1689, BSG1779, BSG1825, BSG1871–BSG1872, and BSG1874–BSG1875 (left panel). Immunoblot against the TAP tag with strains BSG1002, BSG1016, BSG1475, BSG1520, BSG1689, BSG1779, BSG1825, BSG1871–BSG1872, and BSG1874–BSG1875 (right panel).
(D) Same as in (C) with another set of Smc truncation constructs. ChIP-qPCR with strains BSG1779, BSG1824, BSG1826–BSG1830 and BSG1873 (left panel). Anti-TAP immunoblot with strains BSG1002, BSG1016, BSG1475, BSG1779, BSG1824, BSG1826–BSG1830, and BSG1873 (right panel).
See also Figure S5.
A Mini-Smc Localizes to parS Sites on the Chromosome
The above results demonstrate that neither the Smc hinge nor the ScpA and ScpB subunits are strictly required for the localization of Smc(EQ) to parS sites. In order to fine map potential binding sites for ParB/parS on the Smc(EQ) protein, we removed increasingly larger segments of the central part of a Smc(EQ)-TAP allele by fusing selected N- and C-terminal Smc sequences using a short linker peptide (Figure 5B). Twelve such Smc fragments (designated as SmcHd-CC330 to SmcHd-CC30) were integrated into the endogenous smc locus by allelic replacement in a ΔscpA strain. All these truncated Smc(EQ) proteins were expressed at normal levels in B. subtilis as judged by immunoblotting against the TAP tag (Figures 5C and 5D). Intriguingly, the seven larger fragments (SmcHd-CC330–SmcHd-CC70) yielded strong and specific enrichment at the parS-359 locus similar to the Smc(mH-EQ) and Smc(ΔH-EQ) proteins. In contrast, the five shorter constructs (SmcHd-CC50a– SmcHd-CC30) lacked any specificity for parS-359 and instead displayed background levels of enrichment at all tested loci (Figures 5C and 5D). These results demonstrate that a large, central portion of Smc—comprising its hinge domain and approximately two-thirds of the hinge proximal coiled coil—is dispensable for the specific recognition of ParB/parS. A region located within the head proximal part of the Smc coiled coil, however, appears to be critical for parS targeting because even small truncations in this region totally abolish localization to parS. An Smc moiety critical for localization to parS thus appears to be located around hundred amino acid residues away from the Smc head domain on the Smc coiled coil. Mapping of the coiled coil register demonstrates that this head-proximal third of the Smc coiled coil includes a region in which the heptate register is interrupted in the N-terminal coiled coil helix and a 24 amino acid long peptide is inserted into the C-terminal α-helix (Figure S5D), (Waldman et al., 2015). Possibly, these extra sequences protrude from the Smc coiled coil and might be involved in the interaction with ParB protein and/or parS DNA. In the rod configuration, however, the protrusions might be sterically obstructed or otherwise masked.
Smc/ScpAB Relocates from parS Loading Sites to Distant Parts of the Chromosome in an ATP Hydrolysis-Dependent Manner
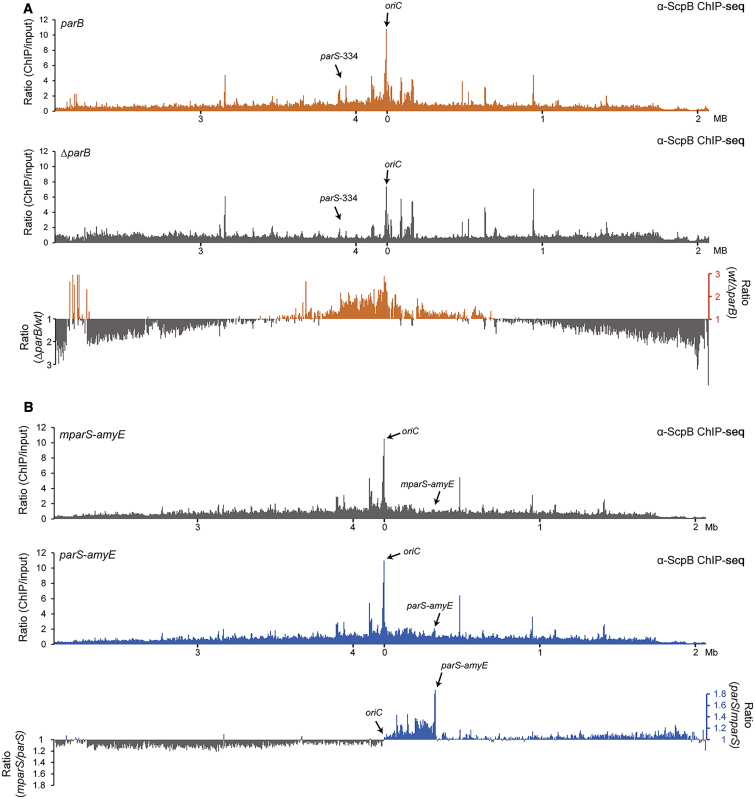
Smc/ScpAB exhibits very high specificity for parS sites on the bacterial chromosome when it is locked in its pre-hydrolysis conformation (Figures 1D and 2D). Nonetheless, only a small proportion of wild-type Smc/ScpAB actually localizes to parS sites in B. subtilis as judged from anti-ScpB and anti-Smc ChIP-seq profiles (Figures 2A and 6A). In fact, Smc/ScpAB rather displays a very broad distribution over the bacterial chromosome with a moderate peak at the replication origin and shallow gradients toward the replication terminus along both arms of the chromosome (Figures 6A and S2C) (Gruber and Errington, 2009). Formation of this long range gradient is completely abolished in the absence of ParB protein (Figure 6A). This raises the intriguing question of how a highly-localized pool of ParB protein might establish a very wide gradient of Smc/ScpAB on the chromosome. Conceivably, Smc/ScpAB might first load onto the chromosome at a parS site and then redistribute into neighboring and more distant regions of the chromosome. To test this, we modified the pattern of chromosomal recruitment of Smc/ScpAB by inserting a single additional parS site into the B. subtilis chromosome and observed changes in the chromosomal distribution of wild-type Smc/ScpAB. We inserted a 75-bp fragment of the parB gene including the parS-359 site or its non-functional variant, mparS, into the non-essential amyE gene located ∼330 kb away from the replication origin on the right arm of the chromosome. The presence of the ectopic parS site at amyE, designated as parS-amyE, had no discernible effects on the growth of B. subtilis. We then tested whether the artificial parS-amyE locus serves as chromosomal landing site for Smc/ScpAB using ScpB ChIP-seq experiments in strains harboring the smc(EQ) gene (Figure S6). Efficient targeting of Smc(EQ) to parS-amyE suggests that also a significant fraction of wild-type Smc/ScpAB is loaded onto the chromosome at the synthetic parS-amyE locus (Figure S6). The pattern of wild-type Smc/ScpAB localization measured by ScpB ChIP-seq was superficially similar in cells harboring the ectoptic parS or mparS site (Figure 6B). However, the levels of ScpB enrichment were moderately—but consistently—higher for example in a region between the replication origin and the amyE locus when the additional parS site was present. In order to get a global and more quantitative picture of the differences between the two ChIP samples, we calculated enrichment ratios for each window along the chromosome (Figure 6B). Strikingly, the differences between the two samples followed a clear pattern with opposite trends on the two arms of the chromosome. Almost all loci on the right arm of the chromosome, which includes the ectopic parS site, were more highly enriched (on average by ∼20%) in the parS-amyE sample, whereas DNA from the left arm of the chromosome was generally more enriched (∼20%) in the mparS sample. Clearly, the addition of a single parS site at a defined position on the chromosome affects Smc distribution in a chromosome arm-specific manner. What might be the underlying molecular mechanism? Smc/ScpAB could relocate from parS by three-dimensional (3D) diffusion within a chromosomal domain or by one-dimensional (1D) translocation along the DNA backbone. 3D diffusion seems a highly unlikely explanation for intra-arm-specific relocation because loci on opposite chromosome arms are thought to be in close proximity in B. subtilis (Le et al., 2013, Marbouty et al., 2014, Marbouty et al., 2015, Umbarger et al., 2011, Wang et al., 2015). Our data is much more consistent with 1D translocation of Smc/ScpAB along a DNA double helix within a chromosome arm. According to this hypothesis, loading of Smc/ScpAB at the ectopic parS site might titrate condensin away from endogenous parS sites and thereby reduce loading on one arm of the chromosome, while increasing the fraction loaded onto the other. Curiously, the re-distribution of Smc complexes loaded onto the chromosome at the ectopic parS site appears to occur differently toward the replication origin and the terminus. In B. subtilis, most genes are co-oriented with respect to DNA replication. Thus, the apparent difference in relocation toward and away from the replication origin might be due to head-on encounters with transcription or replication complexes (Wang et al., 2015). Overall, these experiments provide evidence that Smc/ScpAB is able to translocate along a DNA double helix over large distances on the bacterial chromosome after its release from parS sites.
Figure 6.
Smc/ScpAB Relocates from parS Loading Sites to Distant Parts of the Chromosome upon ATP Hydrolysis
(A) ChIP-seq using α-ScpB antiserum on strains BSG1002 (parB) (top panel) and BSG1052 (ΔparB) (middle panel). Reads were mapped to 5-kb bins. Signals for IP samples were divided by the signals of the normalized input. Ratios were calculated by dividing the values obtained for the wild-type strain by the numbers of the ΔparB strain (bottom panel). All values above one are shown in orange colors. For all other windows the inverse ratio was calculated and displayed in gray colors.
(B) ChIP-seq using α-ScpB antiserum on strains BSG1470 (mparS-amyE) (top panel) and BSG1469 (parS-amyE) (middle panel). Reads were mapped to 5-kb bins. Signals for IP samples were divided by the signals of the normalized input. Ratios were calculated by dividing the values of the parS-amyE strain by the mparS-amyE strain (bottom panel). A number above one indicates more reads in the parS-amyE sample (shown in the blue colors), for all other windows, the inverse ratios were calculated and displayed in gray colors.
See also Figure S6.
Discussion
The mechanistic bases for SMC’s dynamic association with chromosomes are in many ways mysterious. Here, we reveal that the Smc ATPase cycle defines two different configurations of Smc/ScpAB, which distinctly interact with the bacterial chromosome. The pre-ATP hydrolysis state displays high specificity for parS proximal DNA, whereas the specificity for parS is lost upon ATP hydrolysis leading to the redistribution of Smc/ScpAB within the chromosome.
Pre-ATP Hydrolysis Smc/ScpAB: A Tightly Regulated Configuration for ParB/parS Targeting
Our findings define the ATP engaged form of SmcHd(EQ)-CC80 as minimal structure for the specific recognition of Smc/ScpAB’s chromosomal target (Figure 5). They highlight the importance of a head proximal segment of the Smc coiled coil in parS-DNA targeting and raise intriguing questions. How is binding of Smc/ScpAB to ParB/parS-DNA enabled by Smc head engagement? And conversely, how is physical association with the chromosome blocked when Smc heads are disengaged? Based on the strict dependence of parS targeting on Smc head engagement and its inverse correlation with Smc rod formation, we propose that a putative interface for ParB/parS on the head-proximal Smc coiled coil is concealed or distorted within the Smc rod. Smc head engagement might simply trigger the opening of the Smc rod and thereby unmask an interfaces for binding to ParB/parS. However, dimerization defective Smc proteins only very poorly localize to parS sites, despite featuring mostly “disengaged” Smc coiled coils (Figures 4 and 5). Conceivably, the interaction via a single interface on monomeric Smc is too transient for significant levels of targeting to parS. If so, then Smc head engagement might arrange the two interfaces on a given Smc dimer in a way that allows them to co-operatively and thus more stably bind a ParB protein/parS DNA complex. In this regard, it is tempting to speculate that DNA passes between the Smc coiled coils, because in this instance the 2-fold symmetry axis in the Smc dimer is matched to the one in ParB dimers. Binding of Smc to ParB/parS-DNA would therefore be restricted to the rare occasions when Smc head domains engage with one another to dissolve the Smc rod.
Although we have not been able to directly detect the recruitment of wild-type Smc to parS sites, at least three observations strongly suggest that wild-type Smc (like Smc(EQ)) is targeted to parS on the chromosome. First of all, the formation of chromosomal foci by wild-type Smc/ScpAB as well as its localization to the replication origin depend on ParB protein and parS sites (Gruber and Errington, 2009, Sullivan et al., 2009). Second, the efficiency of DNA entrapment by Smc/ScpAB is strongly decreased by the absence of ParB, indicating that most Smc/ScpAB is loaded onto the chromosome at parS sites (Wilhelm et al., 2015). Third, the chromosomal distribution of Smc is changed by an additional parS site. We propose that transient head engagement in Smc/ScpAB governs its brief encounters with chromosomal parS sites. It is important to note that the Smc(EQ) protein might display residual levels of ATP hydrolysis activity (Hirano and Hirano, 2004). It is thus conceivable that some observations made with Smc(EQ) such as its dependence on ScpAB during head engagement and parS localization and the inhibition by hinge dimerization might be specific to this partially defective ATP hydrolysis mutant. Furthermore, it is possible that the association of Smc(mH-EQ) (and Smc(EQ)/ScpAB) with ParB/parS might be structurally somewhat different from wild-type Smc/ScpAB.
Smc/ScpAB Relocation on the Bacterial Chromosome
The chromosomal distribution of Smc/ScpAB displays a single, broad peak centered on the replication origin and extending all the way to the replication terminus region (Figure 6A). Formation of such molecular gradients can be explained by a localized source of molecules and their random/diffusional or directed motion away from the source. Loading at parS establishes a tightly localized source of chromosomal Smc/ScpAB. Here, we present evidence for the subsequent relocation of Smc/ScpAB from its loading sites into flanking DNA. Our findings are consistent with the idea that Smc/ScpAB is able to translocate on the bacterial chromosome over hundreds of kilobases. Interestingly, cohesin has also been suggested to move away from its loading sites upon ATP hydrolysis possibly over several tens of kilobases (Hinshaw et al., 2015, Hu et al., 2011). What might be the purpose of such a striking and apparently conserved propensity for chromosomal relocation and what could be the molecular driving force?
The association of SMC complexes with chromosomes by DNA entrapment provides an obvious basis for a DNA sliding mechanism. Sliding of Smc/ScpAB rings along a single DNA molecule could help to identify and eliminate tangles within chromosomal DNA or between sister DNA molecules and thus promote chromosome segregation. However, this simple mechanism by itself fails to explain how Smc/ScpAB could organize the chromosome. Conceivably, Smc/ScpAB (like cohesin in eukaryotes) acts as a DNA clamp by capturing two or more DNA double helices within a single Smc/ScpAB ring or through the association of two or more Smc/ScpAB rings each entrapping a single DNA double helix. Taking into account the proposed DNA relocation activity, DNA loops could be formed by Smc/ScpAB and continuously expanded by the translocation of DNA through Smc/ScpAB rings. Extrusion of DNA loops—created at a parS site—explains how the left arm of the chromosome might be brought together with its right counterpart to establish the longitudinal organization of the chromosome observed in Caulobacter crescentus and B. subtilis (Le et al., 2013, Marbouty et al., 2014, Marbouty et al., 2015, Umbarger et al., 2011, Wang et al., 2015). Loop extrusion by SMC complexes also provides a simple solution for the formation of linearly condensed rod-shaped chromosomes during mitosis (Alipour and Marko, 2012, Bürmann and Gruber, 2015, Nasmyth, 2001). However, the driving force for any proposed relocation and loop extrusion mechanisms remains enigmatic. Smc/ScpAB appears to be able to translocate on the bacterial chromosome against the flow of replication forks and active transcription units, making a role of RNA polymerase and replication fork proteins in translocation unlikely (Figure 6) (Wang et al., 2015). Smc itself is an enzyme that could harbor energy from the hydrolysis of ATP to perform work. In principle, it could act as a motor protein for example by using its head engagement/disengagement cycle to progressively move DNA through its ring in a directional manner (Figure 7). For example, repetitive transitions between Smc rod and Smc ring states might allow continuous expansion of loops of chromosomal DNA formed at parS (Figure 7) (Alipour and Marko, 2012, Nasmyth, 2001). Our data indeed suggest that ATP hydrolysis by Smc/ScpAB is involved in its chromosomal redistribution after loading. However, it remains to be determined whether continuous ATP hydrolysis by Smc is needed for relocation to distant positions on the chromosome or whether a single round of ATP hydrolysis is sufficient to trigger the relocation process.
The Mechanics of Smc Rod Making and Rod Breaking
We discovered an unexpected antagonistic functional relationship between the two globular domains located at distal ends of the long Smc coiled coil: dimerization at the Smc hinge has a clear inhibitory activity on the engagement of Smc head domains. Conversely, head engagement reduces the level of Smc coiled coil alignment. How might this long-distance communication happen mechanistically? We propose that the Smc coiled coil acts as rather stiff rod, which positions the Smc head in a way that is incompatible with ATP-dependent head engagement when the two Smc coiled coils are being aligned side-by-side. Dimerization of Smc hinge domains (“rod maker”) promotes Smc rod formation presumably by simply bringing the ends of two Smc coiled coils in close proximity in a way that allows them to zip up. Head engagement with the help of ScpAB (“rod breakers”), however, positions the two other ends of the Smc coiled coils at a distance to each other, thus favoring the unzipping of the Smc rod. In analogy, to the role of NBDs in ABC transporters, engagement and disengagement of Smc head domains might transform the Smc coiled coil between a rod-like state and a more open ring-like state. Only the open state appears to be able to contact DNA and the ParB/parS substrates via two separate interfaces located at the Smc hinge and within the head proximal coiled coil, respectively. Substrate binding is then likely triggering ATP hydrolysis, thereby driven head disengagement and rod re-formation, which in turn releases DNA and ParB/parS from its binding sites (Figure 7). We propose that transitions between open and closed states are central aspects of SMC biochemistry – conceivably regulating substrate binding to many or all SMC/kleisin complexes.
Experimental Procedures
Strain Construction
Genetic modifications were introduced via double cross-over recombination into the genome of B. subtilis 1A700. Cells were made competent and grown on SMG or NA medium supplied with antibiotics as previously described (Bürmann et al., 2013). Relevant genotypes are given in Table S1.
ChIP-qPCR
ChIP-qPCR experiments were essentially performed as previously described (Gruber and Errington, 2009). Detailed information is available in the Supplemental Information.
ChIP-Seq
ChIP samples were prepared as described above with the exception that several immunoprecipitate (IP) samples were loaded onto the same PCR purification column to obtain sufficient DNA material. DNA (1–5 ng) was analyzed by Illumina sequencing at the Max Planck Genome Centre in Cologne. Briefly, DNA was fragmented by sonication (Covaris S2) to fragment sizes ranging from 220–280 bp with a main peak of ∼250 bp. DNA libraries were prepared using the Ovation Ultralow Library System (NuGEN) kit (version V1) including 15 cycles of PCR amplification. Five to ten million sequence reads were obtained on a HiSeq2500 (Illumina) with 100-bp read length. The obtained reads were mapped to the genome with Bowtie (http://GalaxyProject.org) using default settings and randomly assigning sequencing reads from repetitive DNA elements to a single location. Subsequent data analysis was performed using Seqmonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) and Microsoft Excel.
BMOE Cross-Linking
In vivo cross-linking of cysteine-modified Smc protein was performed as described previously (Soh et al., 2015).
Microscopy
Overnight cultures in SMG medium were diluted to an OD600 of 0.005 and grown to OD600 0.02–0.03 in SMG medium. Cells were mounted on agarose pads and visualized on an Applied Precision DeltaVision RT system equipped with an Olympus IX-71 inverted base microscope, an Olympus UPlanApo 100×/NA1.35 oil immersion objective and a Photometrics CoolSNAP HQ 12 bit monochrome camera at the Imaging Facility of the Max Planck Institute of Biochemistry, Martinsried.
Author Contributions
Strain Construction, A.M., L.W., and F.B.; Live-Cell Imaging, A.M. and L.W.; ChIP-qPCR, A.M. and L.W.; ChIP-Seq Experiments, A.M.; Cys Cross-Linking Experiments, F.B.; Exploratory ChIP-qPCR Experiments, A.A.; Mapping of the Smc Coiled Coil Register and Protein Purification, M.-L.D.-D; Conception and Interpretation of Experiments, A.M., F.B., and S.G.; Manuscript Preparation, A.M., F.B., L.W., and S.G.
Acknowledgments
Next-generation sequencing (NGS) library preparation and sequencing was performed at the Max Planck-Genome-Centre Cologne. We thank Thomas Gerland and Verena Kuttenberger for strain construction and exploratory experiments. We are grateful to Stefan Jentsch for sharing equipment and the Max Planck Institute of Biochemistry core facility for SEC-MALS analysis. This work was supported by a European Research Council Starting Grant to S.G. (DiseNtAngle 260853) and the Max Planck Society. M.-L.D.-D. is supported by an EMBO long-term fellowship.
Published: February 18, 2016
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Supplemental Information includes Supplemental Experimental Procedures, six figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.01.066.
Accession Numbers
The accession number for ChIP-seq data reported in this paper is GEO: GSE76949.
Supplemental Information
References
- Alipour E., Marko J.F. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–11212. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badrinarayanan A., Reyes-Lamothe R., Uphoff S., Leake M.C., Sherratt D.J. In vivo architecture and action of bacterial structural maintenance of chromosome proteins. Science. 2012;338:528–531. doi: 10.1126/science.1227126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton R.A., Lin D.C., Grossman A.D. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürmann F., Gruber S. SMC condensin: promoting cohesion of replicon arms. Nat. Struct. Mol. Biol. 2015;22:653–655. doi: 10.1038/nsmb.3082. [DOI] [PubMed] [Google Scholar]
- Bürmann F., Shin H.C., Basquin J., Soh Y.M., Giménez-Oya V., Kim Y.G., Oh B.H., Gruber S. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat. Struct. Mol. Biol. 2013;20:371–379. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- Cuylen S., Metz J., Haering C.H. Condensin structures chromosomal DNA through topological links. Nat. Struct. Mol. Biol. 2011;18:894–901. doi: 10.1038/nsmb.2087. [DOI] [PubMed] [Google Scholar]
- Danilova O., Reyes-Lamothe R., Pinskaya M., Sherratt D., Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 2007;65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gligoris T.G., Scheinost J.C., Bürmann F., Petela N., Chan K.L., Uluocak P., Beckouët F., Gruber S., Nasmyth K., Löwe J. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science. 2014;346:963–967. doi: 10.1126/science.1256917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P.L., Losick R., Strunnikov A.V. Subcellular localization of Bacillus subtilis SMC, a protein involved in chromosome condensation and segregation. J. Bacteriol. 1998;180:5749–5755. doi: 10.1128/jb.180.21.5749-5755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber S., Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- Gruber S., Haering C.H., Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–777. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Gruber S., Veening J.W., Bach J., Blettinger M., Bramkamp M., Errington J. Interlinked sister chromosomes arise in the absence of condensin during fast replication in B. subtilis. Curr. Biol. 2014;24:293–298. doi: 10.1016/j.cub.2013.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering C.H., Löwe J., Hochwagen A., Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol. Cell. 2002;9:773–788. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- Haering C.H., Schoffnegger D., Nishino T., Helmhart W., Nasmyth K., Löwe J. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol. Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Hinshaw S.M., Makrantoni V., Kerr A., Marston A.L., Harrison S.C. Structural evidence for Scc4-dependent localization of cohesin loading. eLife. 2015;4:e06057. doi: 10.7554/eLife.06057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M., Hirano T. Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J. 2002;21:5733–5744. doi: 10.1093/emboj/cdf575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M., Hirano T. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. EMBO J. 2004;23:2664–2673. doi: 10.1038/sj.emboj.7600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M., Hirano T. Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol. Cell. 2006;21:175–186. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Hirano M., Anderson D.E., Erickson H.P., Hirano T. Bimodal activation of SMC ATPase by intra- and inter-molecular interactions. EMBO J. 2001;20:3238–3250. doi: 10.1093/emboj/20.12.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlard M., Godwin J., Metson J., Lee J., Hirano T., Nasmyth K. Condensin confers the longitudinal rigidity of chromosomes. Nat. Cell Biol. 2015;17:771–781. doi: 10.1038/ncb3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Itoh T., Mishra A., Katoh Y., Chan K.L., Upcher W., Godlee C., Roig M.B., Shirahige K., Nasmyth K. ATP hydrolysis is required for relocating cohesin from sites occupied by its Scc2/4 loading complex. Curr. Biol. 2011;21:12–24. doi: 10.1016/j.cub.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada K., Miyata M., Hirano T. Molecular basis of SMC ATPase activation: role of internal structural changes of the regulatory subcomplex ScpAB. Structure. 2013;21:581–594. doi: 10.1016/j.str.2013.02.016. [DOI] [PubMed] [Google Scholar]
- Kleine Borgmann L.A., Ries J., Ewers H., Ulbrich M.H., Graumann P.L. The bacterial SMC complex displays two distinct modes of interaction with the chromosome. Cell Rep. 2013;3:1483–1492. doi: 10.1016/j.celrep.2013.04.005. [DOI] [PubMed] [Google Scholar]
- Lammens A., Schele A., Hopfner K.P. Structural biochemistry of ATP-driven dimerization and DNA-stimulated activation of SMC ATPases. Curr. Biol. 2004;14:1778–1782. doi: 10.1016/j.cub.2004.09.044. [DOI] [PubMed] [Google Scholar]
- Le T.B., Imakaev M.V., Mirny L.A., Laub M.T. High-resolution mapping of the spatial organization of a bacterial chromosome. Science. 2013;342:731–734. doi: 10.1126/science.1242059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow J.C., Kuwano M., Moriya S., Grossman A.D. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol. Microbiol. 2002;46:997–1009. doi: 10.1046/j.1365-2958.2002.03235.x. [DOI] [PubMed] [Google Scholar]
- Marbouty M., Cournac A., Flot J.F., Marie-Nelly H., Mozziconacci J., Koszul R. Metagenomic chromosome conformation capture (meta3C) unveils the diversity of chromosome organization in microorganisms. eLife. 2014;3:e03318. doi: 10.7554/eLife.03318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbouty M., Le Gall A., Cattoni D.I., Cournac A., Koh A., Fiche J.B., Mozziconacci J., Murray H., Koszul R., Nollmann M. Condensin- and replication-mediated bacterial chromosome folding and origin condensation revealed by Hi-C and super-resolution imaging. Mol. Cell. 2015;59:588–602. doi: 10.1016/j.molcel.2015.07.020. [DOI] [PubMed] [Google Scholar]
- Mascarenhas J., Soppa J., Strunnikov A.V., Graumann P.L. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 2002;21:3108–3118. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J., Volkov A.V., Rinn C., Schiener J., Guckenberger R., Graumann P.L. Dynamic assembly, localization and proteolysis of the Bacillus subtilis SMC complex. BMC Cell Biol. 2005;6:28. doi: 10.1186/1471-2121-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby T.E., Ciampaglio C.N., Briscoe G., Erickson H.P. The symmetrical structure of structural maintenance of chromosomes (SMC) and MukB proteins: long, antiparallel coiled coils, folded at a flexible hinge. J. Cell Biol. 1998;142:1595–1604. doi: 10.1083/jcb.142.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnen A., Attaiech L., Thon M., Gruber S., Veening J.W. SMC is recruited to oriC by ParB and promotes chromosome segregation in Streptococcus pneumoniae. Mol. Microbiol. 2011;81:676–688. doi: 10.1111/j.1365-2958.2011.07722.x. [DOI] [PubMed] [Google Scholar]
- Moriya S., Tsujikawa E., Hassan A.K., Asai K., Kodama T., Ogasawara N. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol. Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- Schleiffer A., Kaitna S., Maurer-Stroh S., Glotzer M., Nasmyth K., Eisenhaber F. Kleisins: a superfamily of bacterial and eukaryotic SMC protein partners. Mol. Cell. 2003;11:571–575. doi: 10.1016/s1097-2765(03)00108-4. [DOI] [PubMed] [Google Scholar]
- Schwartz M.A., Shapiro L. An SMC ATPase mutant disrupts chromosome segregation in Caulobacter. Mol. Microbiol. 2011;82:1359–1374. doi: 10.1111/j.1365-2958.2011.07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintomi K., Takahashi T.S., Hirano T. Reconstitution of mitotic chromatids with a minimum set of purified factors. Nat. Cell Biol. 2015;17:1014–1023. doi: 10.1038/ncb3187. [DOI] [PubMed] [Google Scholar]
- Soh Y.M., Bürmann F., Shin H.C., Oda T., Jin K.S., Toseland C.P., Kim C., Lee H., Kim S.J., Kong M.S. Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol. Cell. 2015;57:290–303. doi: 10.1016/j.molcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J., Kobayashi K., Noirot-Gros M.F., Oesterhelt D., Ehrlich S.D., Dervyn E., Ogasawara N., Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- Sullivan N.L., Marquis K.A., Rudner D.Z. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbarger M.A., Toro E., Wright M.A., Porreca G.J., Baù D., Hong S.H., Fero M.J., Zhu L.J., Marti-Renom M.A., McAdams H.H. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol. Cell. 2011;44:252–264. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallet-Gely I., Boccard F. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 2013;9:e1003492. doi: 10.1371/journal.pgen.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman V.M., Stanage T.H., Mims A., Norden I.S., Oakley M.G. Structural mapping of the coiled-coil domain of a bacterial condensin and comparative analyses across all domains of life suggest conserved features of SMC proteins. Proteins. 2015;83:1027–1045. doi: 10.1002/prot.24778. [DOI] [PubMed] [Google Scholar]
- Wang X., Tang O.W., Riley E.P., Rudner D.Z. The SMC condensin complex is required for origin segregation in Bacillus subtilis. Curr. Biol. 2014;24:287–292. doi: 10.1016/j.cub.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Le T.B., Lajoie B.R., Dekker J., Laub M.T., Rudner D.Z. Condensin promotes the juxtaposition of DNA flanking its loading site in Bacillus subtilis. Genes Dev. 2015;29:1661–1675. doi: 10.1101/gad.265876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm L., Bürmann F., Minnen A., Shin H.C., Toseland C.P., Oh B.H., Gruber S. SMC condensin entraps chromosomal DNA by an ATP hydrolysis dependent loading mechanism in Bacillus subtilis. eLife. 2015;4:4. doi: 10.7554/eLife.06659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.