Abstract
Background:
Variations in the hepatic lipase (HL) gene are the potential candidate for coronary artery disease (CAD) especially in type 2 diabetes mellitus (T2DM) in diverse populations. We assessed the association of -514C/T and -250G/A polymorphisms in HL (LIPC) gene with CAD risk in Iranian population with type 2 diabetes.
Materials and Methods:
We evaluated 322 type 2 diabetic patients, 166 patients with normal angiograms as controls and 156 patients those identified with CAD undergoing their first coronary angiography as CAD cases. Genotyping of -514C/T and -250G/A polymorphisms in the promoter of the LIPC gene were studied by polymerase chain reaction (PCR)-restriction fragment length polymorphism technique.
Results:
Genotype distributions in CAD cases (73.7%, 20.5%, and 5.8% for −250G/A) and (62.2%, 32.7%, and 5.1% for -514C/T) were significantly different from those in controls (60.8%, 37.4%, and 1.8% for -250G/A) and (51.2%, 48.2%, and 0.6% for -514C/T). CAD cases had lower A-allele frequency than controls (0.131 vs. 0.196, P = 0.028). The odds ratio for the presence of -250 (GG + GA) genotype and A allele in CAD cases were 2.206 (95% confidence interval [CI] =1.33–3.65, P = 0.002) and 1.609 (95% CI = 1.051 −2.463, P = 0.029) respectively. Haplotype analysis demonstrated a significant association between especially LIPC double mutant (−250 A/-514 T) haplotype and presence of CAD.
Conclusion:
Our findings indicated that -250 G/A polymorphism rather than -514 C/T polymorphism of LIPC gene is more associated with the increased risk of CAD particularly in women with T2DM.
Key Words: Type 2 diabetes, coronary artery disease, LIPC gene, polymorphisms
INTRODUCTION
Coronary artery disease (CAD) is a well-known cause of morbidity and mortality in the subjects with type 2 diabetes in all nations. Recently, CAD considers a major public health problem in developing and developed countries ant its increasing incidence is a cause of substantial concern in the medical community worldwide.[1] The genetic and environmental factors and their interactions with each other have been involved in the development of CAD.[2,3] In spite of efforts to determine the molecular and genetic contributing factor that could account for variations in CAD,[3] the etiology and the complex multigenic basis of atherosclerosis is still not completely identified.
Hepatic lipase (HL), a lipolytic enzyme synthesized primarily in the liver, has an important role in the metabolism of together pro and anti-atherogenic lipoproteins.[4] HL has several metabolic functions, such as hydrolysis of triglycerides (TGs), lipolysis of phospholipids, forming of small dense atherogenic low-density lipoprotein (LDL) particles, and the catabolism of high-density lipoprotein (HDL).[4] High HL activity is related to low serum level of HDL, and HL converts large TG-rich HDL subfraction (HDL2) into small and dense HDL3.[5] It has been found that approximately 20% to 30% of the specific variation in the HL activity is due to the presence of a common polymorphism in the promoter region of the LIPC gene.[6]
The human HL gene (LIPC), located on chromosome 15q21, is consist of nine exons and eight introns, covers over up 30 kb of DNA, and encodes a protein with 449 amino acids residue.[7,8] The Four common single nucleotide polymorphisms (SNPs) in the promoter region, consist of four highly correlated polymorphism in the 5’-Flanking region of the LIPC gene (-250 G/A,-514C/T,-710T/C and -763A/G) with respect to the transcription star site, were determined to be in perfect disequilibrium.[9,10] A substitution in the promoter region of the LIPC gene (-250G/A) has been reported to be related to modifications of plasma lipid levels[11,12,13,14,15,16,17,18] and the risk of CAD[19,20] in some studies but not in others.[21,22,23,24] In addition, this SNP of LIPC gene has been shown to regulate insulin sensitivity,[15] but its effect on the risk of CAD in type 2 diabetes has not been completely understood. The C/T substitution at position-514 in the promoter region of the LIPC gene has been shown is correlated with up to 30% reduction in promoter activity in vitro,[25,26,27] obvious decrease plasma HL activity, and higher serum levels of HDL, HDL2, and large LDL particles. The association between -514C/T polymorphisms of LIPC gene with the serum HDL-C levels was demonstrated.[28,29] Thus, there is considerable evidence of associations between two common promoter polymorphisms of the HL gene and HL activity and plasma lipoprotein concentrations. In addition, result indicated that the 514C/T polymorphism of LIPC gene is not associated with coronary heart disease (CHD) susceptibility.[30] However, the association between these two common SNPs of LIPC gene on the atherosclerosis in different population has been controversial.[29] The aim of this study was to determine the association between these polymorphisms (-250G/A and -514C/T) of the LIPC gene with CAD in subjects with type 2 diabetes in a sample of Iranian population.
MATERIALS AND METHODS
Study population
We evaluated 855 type 2 diabetes mellitus (T2DM) patients, age between 40 and 60 years, genetically unrelated, and with ancestry in the state of Khuzestan, Iran. All patients performed carotid angiography at the Department of Cardiology, Ahwaz Imam Khomeini Hospital, Ahvaz Jundishapur University of Medical Sciences. Details of sample collection, processing and relevant corresponding clinical data are described elsewhere.[31] Briefly, according to the angiograms, patients were classified into two groups including patients with CAD and without CAD or controls. The control group consisted of type 2 diabetic patients older than 40 years who had a negative history of CAD, normal exercise tolerance test, and normal angiograms. Diagnosed myocardial infarction during the previous 3 months, body mass index (BMI) >30 and treatment for chronic infectious disease or malignancy were exclusion criteria. For every participant, baseline information was conducted by trained research assistants and included questionnaires related to social and medical history and physical examination. Lastly, 322 patients with complete data were included in the final analysis. Hypertension was determined as systolic blood pressure ≥140 mmHg and/or diastolic blood pressure ≥90 mmHg or treatment with oral anti-hypertension drugs. T2DM was diagnosed according to the American Diabetes Association criteria. CAD was defined as the presence of at least one significant coronary artery stenosis of more than 50% luminal diameter on coronary angiography.
Coronary angiography
Coronary artery angiography was performed by using standard Judkins technique. Angiographic analysis was carried out by experienced cardiologists who were blinded to the investigate protocol. Angiography results were separated into sections with CAD (≥50% obstruction in ≥1 coronary artery) and without CAD. Gensini score estimates the intensity of CAD, and preliminary results are described elsewhere.[32]
Biochemical and anthropometric measurements
All study subjects, were assessed in the morning after at least 12-h fasting. Body height and weight were determined without shoes and in light clothing and BMI was calculated as weight (kg)/height2 (m2) or height squared. In each case, after an overnight fasting, 2 mL of blood was drawn into tubes treated with K3- ethylene diamine tetraacetic acid tubes for DNA extraction. Blood cell fraction, serum and plasma were frozen at -20°C. HDL-cholesterol (HDL-C), LDL-cholesterol (LDL-C), total cholester, TG, and serum glucose were measured by enzymatic methods with a clinical chemistry analyzer (Vital Scientific, Spankeren, the Netherlands), as previously described elsewhere.[31]
Genotyping
DNA was extracted from leukocytes, taken from peripheral blood, by using a salting-out procedure (Miller's method).[33] Genotyping of -250G/A (rs2070895), and -514C/T (rs1800588) SNPs, in the promoter region of the LIPC gene were performed by polymerase chain reaction (PCR)-restriction fragment length polymorphism, followed by digestion with the restriction enzymes DraI and NlaIII (New England Biolabs), respectively. Target DNA was amplified using two different primer pairs, one pair for -250G/A and the other for -514C/T polymorphism [Table 1]. PCR amplifications were performed in a total volume of 25 μL containing 12.5 μl commercially available PCR premix (AccuPower PCR PremiX; Bioneer, Daejeon, South Korea) containing (dNTP, Taq DNA polymerase, MgCl2, 10 × PCR buffer), 2.0 μl (20 pmol/μl) forward and reverse primers, 100 ng genomic DNA, and 6.5 μl sterile nuclease free water. Thermal cycling conditions were similar for two SNPs and were as follows: An initial denaturation step at 94°C for 5 min, 35 cycles at 94°C for 30 s, 59 or 57°C for 30 s (depend on primers), 72°C for 45 s and a final extension step at 72°C for 5 min. The amplification products were digested with an appropriate enzyme according to the manufacturer's protocols. The digested fragments were then separated by electrophoresis on 2% agarose gels, followed by ethidium bromide staining and visualized under ultraviolet light. For -250 G/A polymorphism, the wild type genotype doesn’t contain a DraI restriction site and yields a fragment with 584 bp; heterozygotes display three bands with 584 bp, 428 bp and 156 bp, whereas homozygotes show two bands with 428 bp and 156 bp [Figure 1]. For -514 C/T polymorphism, the wild type genotype doesn’t contain a NlaIII restriction site and yields a 459 bp fragment; heterozygotes display three bands with 459 bp, 291 bp and 168 bp, whereas homozygotes show two bands with 291 bp and 168 bp [Figure 2].
Table 1.
PCR primers, products, and restriction enzymes for the genotyping of LIPC polymorphisms

Figure 1.
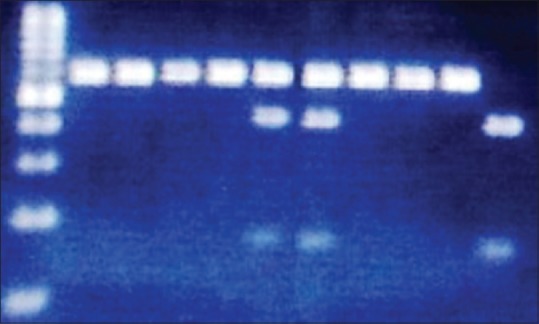
Product of polymerase chain reaction-restriction fragment length polymorphism for −250G/A genotypes of LIPC gene on the 2% agarose gel. Lane 1 is DNA size marker 100 bp, lane 2–5 and 8–10 are related to the GG genotype, lane 6, 7 are related to the GA genotype and lane 11 is related to the AA genotype
Figure 2.
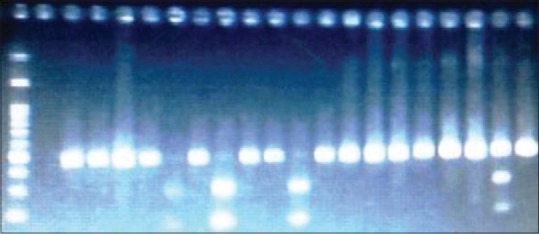
Product of polymerase chain reaction-restriction fragment length polymorphism for −514C/T genotypes of LIPC gene on the 2% agarose gel. Lane 1 is DNA size marker 100 bp, Lane 2 is related to the non-template control, lane 3–6 and 8,10,11,13-19, and 21 are related to the CC genotype, lane 21 is related to the CT genotype and lanes 7,9 and 12 are related to the TT genotype
Statistical analyses
All statistical analyses were performed by the SPSS version 15.0 software (SPSS, Inc., Chicago IL, USA). Continuous variables are expressed as mean ± standard deviation, and compared by student's t-test. All frequencies were estimated using the gene-counting method and both polymorphisms were tested for Hardy-Weinberg's equilibrium using the Chi-squared test. Categorical variables were presented as the total number (percentage) and compared by Chi-square-test. Comparisons of continuous variables between two SNPs genotypes of LIPC gene were tested by analysis of variance. Odds ratio (OR) and 95% confidence interval (CI) were estimated for CAD by logistic regression to assess the relative risk by a particular allele and genotype. The haplotypes distribution in CAD and control groups were estimated according to the two-stage iterative method named Expectation Maximization algorithm using the software SNPStats (http://www.bioinfo.iconcologia.net/SNPstats). The risk for every haplotype was compared respect to the reference category; it is the most frequent haplotype. A P < 0.05 was considered to be statistically significant for all analyses.
RESULTS
Demographic and biochemical characteristic of the studied subjects
The demographic and biochemical features of the CAD cases (T2DM patients with CAD) and controls (T2DM patients without CAD) are presented in Table 2. Among the 322 participants, 156 patients had angiographically-proven CAD and 166 had almost normal coronary arteries (without CAD group). The CAD cases had significantly higher of fasting blood sugar (FBS) (P = 0.014), BMI (P = 0.011), systolic blood pressure (P = 0.001), diastolic blood pressure (P < 0.001), total cholesterol (P = 0.020), TG (P = 0.016), LDL-C (P < 0.001), and lower HDL-C (P < 0.001) than controls. For the demographic data the age of CAD cases were significantly higher than that in control group (P < 0001). In general, an elevated level of LDL-C, TG and lower level of HDL-C have been considered to increase the risk of CAD.
Table 2.
Demographic and biochemical characteristics of CAD cases and controls
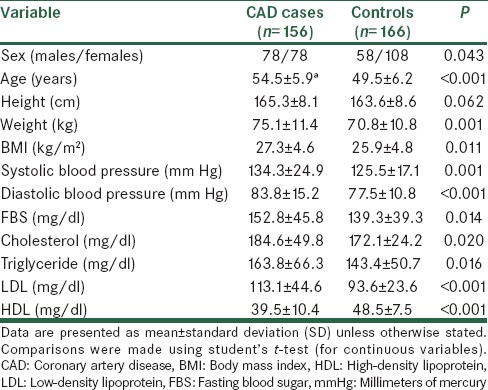
The genotype and allele frequencies of -250G/A and -514 C/T polymorphisms in hepatic lipase gene
The allele and genotype frequencies of the two LIPC loci among our controls and CAD cases were in Hardy-Weinberg equilibrium. CAD cases for -250G/A and controls for -514C/T showed a distribution that deviated significantly from a distribution expected in a population with genotype distribution in Hardy-Weinberg equilibrium. These deviations may be due to the small sample size. However, it was not likely that any of the assumptions for Hardy-Weinberg equilibrium was violated, and we attribute such departure to chance. The distributions of the genotype and allele frequency in CAD cases and controls are shown in Table 3. Distribution of GG, GA and AA genotype for 250G/A polymorphism are significantly different between controls and CAD cases (P = 0.001). For -514 C/T polymorphism the distribution of CC, CT and TT genotype are significantly different between controls and CAD cases (P = 0.002). The frequencies of G and A alleles for -250G/A SNP are shown significant different between controls and CAD cases (P = 0.028). However, the frequencies of C and T alleles for -514C/T SNP are shown not significant different between controls and CAD cases (P = 0.092) [Table 3].
Table 3.
Genotype and allele distribution of two SNPs in CAD cases and controls and their association with CAD
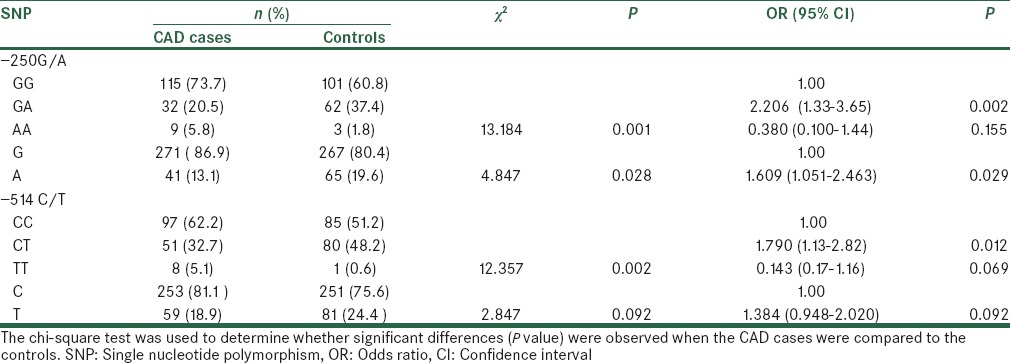
Genotype distribution of -514C/T and -250G/A polymorphism according to the gender in study population
Genotype frequencies in overall study population according to the gender are shown in Table 4.
Table 4.
Genotype distribution of two SNPs in CAD cases and controls and their association with CAD according to the gender
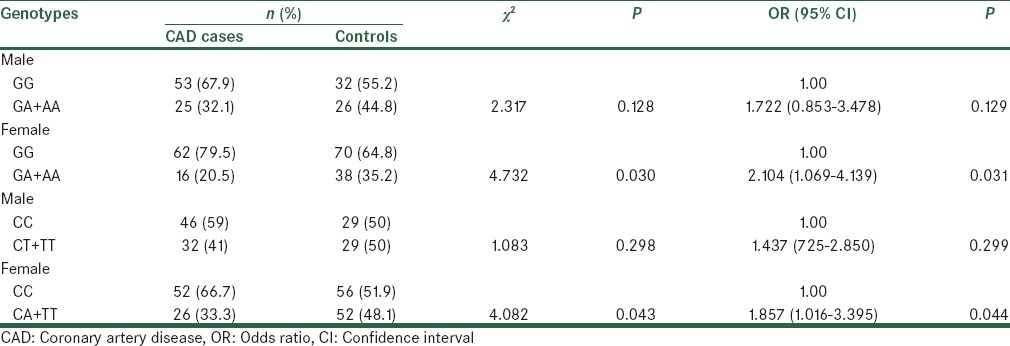
For -514C/T polymorphism distribution of CC, and CT + TT genotype in females are significantly different between CAD cases and controls (p =0.043). However, distribution of CC, and CT + TT genotype in males are not significantly different between CAD cases and controls (P = 0.298). Similarly, for -250 A/G polymorphism distribution of GG, and GA + AA genotype, in females are significantly different between CAD cases and controls (P = 0.030). While, distribution of GG, and GA + AA genotype, in males are not significantly different between CAD cases and controls (P = 0.128). Among females, comparing to the subjects with GG reference genotype of -250 G/A polymorphism, those carrying the GA genotype (OR = 2.104, 95% CI = 1.069-4.139, P = 0.031) had significant increased risk for CAD. Similarly, Among females, comparing to the subjects with CC reference genotype of -514 C/T polymorphism, those carrying the CT genotype (OR = 1.857, 95% CI = 1.016-3.395, P = 0.044) had significant increased risk for CAD. In fact, our results indicated that the female might be at higher increased risk of CAD than the male with type 2 diabetes.
The risk assessment of genotype and allele frequencies of two polymorphisms for coronary artery disease
The odds ratio of genotype and allele frequencies of -250 G/A and -514 C/T polymorphisms for CAD are presented in Table 3. The OR for genotypes and alleles of -250 A/G polymorphism are 2.206 (95% CI = 1.333-3.650, P = 0.002) for GA genotype and 0.380 (95% CI = 0.100-1.440, P = 0.155) for AA genotype comparing to the GG reference genotype. The OR for A allele of -250 polymorphism comparing to the reference G allele was 1.609 (95% CI = 1.051-2.463, P = 0.029). In addition, the odds ratio for genotypes and alleles of -514 C/T polymorphism are 1.790 (95% CI = 1.134-2.825, P =0.012) for CT genotype and 0.143 (95% CI = 0.017-1.164, P = 0.069) for TT genotype comparing to the CC reference genotype. The OR for T allele of -514C/T polymorphism comparing to the reference C allele is 1.384 (95% CI = 0.984-2.020, P = 0.092). In addition, in the entire population study the influence of -514C/T and -250G/A polymorphisms on the studied variables are not significant only in CC genotype which is decrease serum level of HDL-C (data not shown here).
Haplotype distribution in study population
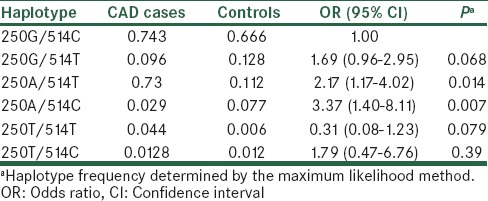
The haplotype analysis stratified by study subjects of two studied SNPs is presented in Table 5. Using the software SNPStats, we estimated all possible haplotypes from the observed genotypes of two SNPs, -514C/T and -250G/A. Then, we compared haplotype frequency of LIPC between CAD cases and controls. Six haplotypes derived from the two SNPs accounted for about 95% of haplotype variations (ORs and P values for the associations are listed in Table 5). Among these six haplotypes, using the haplotype consisting of wild allele of each SNP (250G -514C) as reference, two haplotype (-514T/-250A) and (-514C/-250A) are found to be significantly associated with the increased risk of CAD, OR = 2.17 (95% CI = 1.17-4.02, P = 0.014) and OR = 3.37 (95% CI = 1.40-8.11, P = 0.007), respectively.
Table 5.
Haplotype frequencies estimation and haplotype association with CAD
DISCUSSION
In this study we reported significant difference in the distribution of allele or genotype frequencies of -250G/A and -514C/T polymorphisms in LIPC gene between CAD cases and controls. Additionally, we found the association between these two polymorphisms and anthropometric indices and metabolic characteristics, such as HDL-C. This is in agreement with Pihlajamäki et al.[15] However, Hegel et al.[23] have reported an absence of association between the genetic variation in the promoter of LIPC gene and plasma lipoproteins in the Canadian population.
The association between LIPC –250G/A polymorphism and CAD is consistent with other studies, and confirmed the results of Zambon et al. reported that the allele frequency of the -250G/A polymorphism is 47% in a small group of Japanese Americans.[21] De Andrade et al. in a study conducted on Brazilian population showed the prevalence of the -250 A allele in the population-based sample was 32%. Overall, the -250 A allele was somewhat lower in CAD patients than in the controls, however, when the CAD+ sample was compared with the control sample, a significant association was observed between the -250 A allele and severe CAD (P = 0.02). The same studies confirmed that the allelic distribution is different according to the geographical origin of the study group.[19] In our study the genotype frequencies based on gender showed a significant difference between women of two groups. But this difference was not seen in men. The finding of a positive and significant association between female and risk of CAD is a novel finding in Iranian subjects with type 2 diabetes. However, Ko et al.[13] found significant associations between the -250G/A polymorphisms of the HL gene and HDL-C levels in obese men. The result of this study showed that there were no significant differences for FBS based on both polymorphic sites of HL gene in CAD cases and controls. However, the HDL-C levels have significantly increased in GG genotype of -250G/A polymorphism compared to the other genotype. Jimenez et al, demonstrated no significant associations between the -250G/A polymorphism and plasma HDL-C levels.[12] Zambon et al, demonstrated that not only its associated with HDL-C levels, but also -250G/A polymorphism related to small dense LDL particles.[21] Although low HDL-C is a well-established risk factor for CAD, the association of LIPC promoter variants with the risk of CAD appears to be a paradox showing the highest CAD risk in minor allele carriers also associated with elevated HDL-C.[34,35] This contradiction may be due to a HL-induced modulation of the HDL particle,[36] a balance that is actually influenced by physical activity.[37] De Andrade in a study on Brazilian population a significant association was observed between the -250 A allele and severe CAD. Also, the multiple logistic regression analyses that included the conventional risk factors showed that the -250 GG homozygote genotype was an independent predictor of CAD (OR = 1.79; P = 0.025).[23] In this study, we found that GG, GA genotypes and A allele of the LIPC -250G/A polymorphism were the high risk of CAD in T2DM patients. A study conducted by Valdivielso et al., showed that the -250A allele was significantly associated with peripheral arterial disease (PAD), and suggest that the presence of the -250 A allele in the promoter of the LIPC gene confers susceptibility to PAD in subjects with type 2 diabetes.[38]
Our results showed there was a significant association between CT genotype of – 514 C/T polymorphism and CAD risk in T2DM subjects. The effect of the -514C/T polymorphism on the presence of CAD is controversial. Jansen et al.[39] observed a higher frequency of the T allele in patients with CAD; however these results couldn’t be confirmed by the other studies.[40,41] A study was presented by Andersen et al., on over than 10,000 subjects of the kobenhavn town Heart Study showed homozygous presence of the rare T allele in the LIPC gene was associated with a risk of CAD. Jansen et al.[39] observed that the rare -514 T allele of the HL promoter was associated with CAD in Dutch patients has so far been without a good explanation. Zhang et al.[40] in a study on the US diabetic men, the association between the LIPC -514C/T polymorphism and CHD risk was modulated by both overall adiposity and central obesity. Among obese men, the CT or TT genotype was associated with significantly increased risk of CHD compared with those with the CC genotype. However, among lean men this polymorphism was associated with a reduction in CHD risk. But in this study, we reported no significant difference for genotype distribution of both polymorphisms between men of two groups. Several studies suggested a pro-atherogenic role of -514 C/T polymorphism.[42,43] For instance, in a Danish cohort study, participants with the TT genotype were observed to have a 1.5-fold (95% CI = 1.0- 2.2) increased risk of ischemic heart disease compared with those with the CC genotype.[34] The extent of coronary atherosclerosis determined by angiography was reported to be higher in participants with the T allele.[35] The LIPC − 514 T allele was also reported to be associated with a decrease in coronary flow reserve[44] and the presence of subclinical CHD. Conversely, no association of the LIPC -514C/T polymorphism with the risk for CHD or CAD was observed in a study of Finnish men.[45] Also, in this study we observed no significant association of LIPC -514T TT genotype and T allele with the risk for CAD. However, the observed association with HDL-C is in line with the results of a meta-analysis on the -514C/T variant, showed an increase in plasma levels of HDL-C in homozygous minor T-allele carriers compared with homozygous major C allele carriers.[46]
The association between two LIPC loci and CAD was also studied by haplotype analysis. Of the possible four haplotypes, the double mutant–250A/–514T haplotype was under-represented in both groups, and significant preferential association with incident of CAD in T2DM patients was seen. In addition, the haplotype -250A/-514C compare to reference haplotype has increased 3-fold risk of CAD in type 2 diabetes. In several studies, the rare allele, or the rare haplotype, has been associated with low HL activity and with high TG levels,[34,47] high HDL cholesterol levels,[35,39] buoyant LDL particles,[47] and CHD.[33] The observation of Jansen et al.[39] that the rare-514 allele of the HL promoter was associated with CAD in Dutch patients has so far been without a good explanation. subjects with this rare, or the respective haplotype, have low HL activity,[47] high HDL cholesterol levels,[39] and buoyant LDL particle,[47] which have all been associated with low risk of CHD.
Study strengths
To the best of our knowledge, this study is the first report of a gender-specific association between two common promoter SNPs (-250G/A and -514C/T) of LIPC gene and the risk of CAD in Iranian subjects with type 2 diabetes. In addition, we find the LIPC double mutant (-250 A/-514 T) haplotype is associated with the presence of CAD in diabetic patients in a sample of Iranian population.
Study limitations
First, because of the cross-sectional nature of the present study we cannot elucidate the mechanism or determine the direction of causality. Thus, further longitudinal studies are necessary to determine whether our results came from simple associations or indicate to causal relationships. Second, distribution of -250 A/G genotype in diabetic patients with CAD and distribution of -514C/T in controls were significantly deviated from a distribution expected in a population according to the Hardy-Weinberg equilibrium.
CONCLUSION
Our results indicated that -250 G/A polymorphism rather than -514 C/T polymorphism of LIPC gene is more associated with the increased risk of CAD particularly in women with T2DM. To the best of our knowledge, there was no report to provide information about the role of two common SNPs of LIPC gene on the risk of CAD in type 2 diabetic patients of Iranian population.
ACKNOWLEDGMENTS
This study is part of M.SC thesis for Mohammad Bazyar which has been kindly supported by Cellular and Molecular Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. (Grant No. CMRC-95). Since in this study blood samples were derived from participated patients in another research project with grant number of CMRC-66; hence we would like to appreciate a lot all researchers of that project. Also, we would like to thank the staffs of Department of Cardiology, Ahwaz Imam Khomeini Hospital for their friendly cooperation. We have extended our thanks to all the participants and their relatives for their kind cooperation with the researchers as well.
Footnotes
Source of Support: We thank the financial support of research deputy, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. We are also grateful to the head and staffs of department of clinical biochemistry of Ahvaz Jundishapur University of Medical Sciences
Conflict of Interest: None declared.
REFERENCES
- 1.Tradif JC. Coronary artery disease in 2010. European Heart Journal Supplements. 2010;12:C2–10. [Google Scholar]
- 2.Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97:1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 3.Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, De Faire U. Heritability of death from coronary heart disease: A 36-year follow-up of 20,966 Swedish twins. J Intern Med. 2002;252:247–254. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 4.Jansen H, Verhoeven AJ, Sijbrands EJ. Hepatic lipase: A pro- or anti-atherogenic protein? J Lipid Res. 2002;43:1352–1362. doi: 10.1194/jlr.r200008-jlr200. [DOI] [PubMed] [Google Scholar]
- 5.Zambon A, Austin MA, Brown BG, Hokanson JE, Brunzell JD. Effect of hepatic lipase on LDL in normal men and those with coronary artery disease. Arterioscler Thromb. 1993;13:147–53. doi: 10.1161/01.atv.13.2.147. [DOI] [PubMed] [Google Scholar]
- 6.Eller P, Schgoer W, Mueller T, Tancevski I, Wehinger A, Ulmer H, et al. Hepatic lipase polymorphism and increased risk of peripheral arterial disease. J Intern Med. 2005;258:344–8. doi: 10.1111/j.1365-2796.2005.01549.x. [DOI] [PubMed] [Google Scholar]
- 7.Cai SJ, Wong DM, Chen SH, Chan L. Structure of the human hepatic triglyceride lipase gene. Biochem. 1989;28:8966–71. doi: 10.1021/bi00449a002. [DOI] [PubMed] [Google Scholar]
- 8.Ameis D, Stahnke G, Kobayashi J, McLean J, Lee G, Büscher M, et al. Isolation and characterization of the human hepatic lipase gene. J Biol Chem. 1990;265:6552–5. [PubMed] [Google Scholar]
- 9.Guerra R, Wang J, Grundy SM, Cohen JC. A hepatic lipase (LIPC) allele associated with high plasma concentrations of high density lipoprotein cholesterol. Proc Natl Acad Sci USA. 1997;94:4532–7. doi: 10.1073/pnas.94.9.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murtomaki S, Tahvanainen E, Antikainen M, Tiret L, Nicaud V, Jansen H, et al. Hepatic lipase gene polymorphisms influence plasma HDL levels. Results from Finnish EARS participants European Atherosclerosis Research Study. Arterioscler Thromb Vasc Biol. 1997;17:1879–4. doi: 10.1161/01.atv.17.10.1879. [DOI] [PubMed] [Google Scholar]
- 11.Bertolini S, Pisciotta L, Di Scala L, Langheim S, Bellocchio A, Masturzo P, et al. Genetic polymorphisms affecting the phenotypic expression of familial hypercholesterolemia. Atherosclerosis. 2004;174:57–65. doi: 10.1016/j.atherosclerosis.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 12.Jiménez-Gómez Y, Pérez-Jiménez F, Marín C, Gómez P, Moreno R, Delgado J, et al. The -250G/A polymorphism in the hepatic lipase gene promoter influence the postprandial lipemic response in healthy men. Nutr Metab Cardiovasc Dis. 2008;18:173–81. doi: 10.1016/j.numecd.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 13.Ko YL, Hsu LA, Hsu KH, Ko YH, Lee YS. The interactive effects of hepatic lipase gene promoter polymorphisms with sex and obesity on high density- lipoprotein cholesterol levels in Taiwanese-Chinese. Atherosclerosis. 2004;172:135–42. doi: 10.1016/j.atherosclerosis.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Lindi V, Schwab U, Louheranta A, Vessby B, Hermansen K, Tapsell L, et al. The G-250A polymorphism in the hepatic lipase gene promoter is associated with changes in hepatic lipase activity and LDL cholesterol: The KANWU Study. Nutr Metab Cardiovasc Dis. 2008;18:88–95. doi: 10.1016/j.numecd.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Pihlajamaki J, Karjalainen L, Karhapaa P, Vauhkonen I, Taskinen MR, Deeb SS, et al. G-250A substitution in promoter of hepatic lipase gene is associated with dyslipidemia and insulin resistance in healthy control subjects and in members of families with familial combined hyperlipidemia. Arterioscler Thromb Vasc Biol. 2000;20:1789–95. doi: 10.1161/01.atv.20.7.1789. [DOI] [PubMed] [Google Scholar]
- 16.Stancakova A, Baldaufova L, Javorsky M, Kozarova M, Salagovic J, Tkac I. Effect of gene polymorphisms on lipoprotein levels in patients with dyslipidemia of metabolic syndrome. Physiol Res. 2006;55:483–90. doi: 10.33549/physiolres.930836. [DOI] [PubMed] [Google Scholar]
- 17.Wood KC, Fullerton MD, El-Sohemy A, Bakovic M. Interactions between hepatic lipase and apolipoprotein E gene polymorphisms affect serum lipid profiles of healthy Canadian adults. Appl Physiol Nutr Metab. 2008;33:761–8. doi: 10.1139/H08-054. [DOI] [PubMed] [Google Scholar]
- 18.Zhao S, Xie X, Nie S. The -250G/A polymorphism in the human hepatic lipase gene promoter affects blood lipids in Chinese. Clin Chim Acta. 2006;365:149–52. doi: 10.1016/j.cca.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 19.de Andrade FM, Silveira FR, Arsand M, Antunes AL, Torres MR, Zago AJ, et al. Association between -250G/A polymorphism of the hepatic lipase gene promoter and coronary artery disease and HDL-C levels in a Southern Brazilian population. Clin Genet. 2004;65:390–5. doi: 10.1111/j.0009-9163.2004.00243.x. [DOI] [PubMed] [Google Scholar]
- 20.Wei M, Lu YS, Li PP. Association of the hepatic lipase gene -250G/A promoter polymorphism with the susceptibility to type 2 diabetes mellitus combining with coronary heart disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2009;26:219–22. doi: 10.3760/cma.j.issn.1003-9406.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 21.Zambon A, Deeb SS, Hokanson JE, Brown BG, Brunzell JD. Common variants in the promoter of the hepatic lipase gene are associated with lower levels of hepatic lipase activity, buoyant LDL, and higher HDL 2 cholesterol. Arterioscler Thromb Vasc Biol. 1998;18:1723–9. doi: 10.1161/01.atv.18.11.1723. [DOI] [PubMed] [Google Scholar]
- 22.Tahvanainen E, Syvanne M, Frick MH, Murtomaki-Repo S, Antikainen M, Kesaniemi YA, et al. Association of variation in hepatic lipase activity with promoter variation in the hepatic lipase gene. The LOCAT Study Investigators. J Clin Invest. 1998;101:956–60. doi: 10.1172/JCI1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hegele RA, Harris SB, Brunt JH, Young TK, Hanley AJ, Zinman B, et al. Absence of association between genetic variation in the LIPC gene promoter and plasma lipoproteins in three Canadian populations. Atherosclerosis. 1999;146:153–60. doi: 10.1016/s0021-9150(99)00113-6. [DOI] [PubMed] [Google Scholar]
- 24.Shohet RV, Vega GL, Anwar A, Cigarroa JE, Grundy SM, Cohen JC. Hepatic lipase (LIPC) promoter polymorphism in men with coronary artery disease. Allele frequency and effects on hepatic lipase activity and plasma HDL-C concentrations. Arterioscler Thromb Vasc Biol. 1999;19:1975–8. doi: 10.1161/01.atv.19.8.1975. [DOI] [PubMed] [Google Scholar]
- 25.Deeb SS, Zambon SS, Carr MC, Ayyobi AF, Brunzell JD. Hepatic lipase and dyslipidemia: Interactions among genetic variants, obesity, gender, and diet. J Lipid Res. 2003;44:1279–86. doi: 10.1194/jlr.R200017-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Botma GJ, Verhoeven AJ, Jansen H. Hepatic lipase promoter activity is reduced by the C-480T and G-216A substitutions present in the common LIPC gene variant, and is increased by upstream stimulatory factor. Atherosclerosis. 2001;154:625–32. doi: 10.1016/s0021-9150(00)00478-0. [DOI] [PubMed] [Google Scholar]
- 27.Deeb SS, Peng R. The C-514T polymorphism in the human hepatic lipase gene promoter diminishes its activity. J Lipid Res. 2000;41:155–8. [PubMed] [Google Scholar]
- 28.Kashani MA, Azizi F, Hedayati M, Daneshpour MS, Shamshiri AR, Siassi F. Association between CETP Taq1B and LIPC -514C/T polymorphisms with the serum lipid levels in a group of Tehran's population: A cross sectional study. Lipids in Health and Disease. 2010;9:96–103. doi: 10.1186/1476-511X-9-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang C, Ridaura RL, Rimm EB, Rifai N, Hunter DJ, Hu FB. Interactions between the -514C/T polymorphism of the hepatic lipase gene and life style factors in relation to HDL concentrations among US diabetic men. AmJ Clin Nut. 2005;81:1429–35. doi: 10.1093/ajcn/81.6.1429. [DOI] [PubMed] [Google Scholar]
- 30.Wang HR, Jiang M, Qiu JP. Quantitative Assessment of the Effect of Hepatic Lipase Gene Polymorphism on the Risk of Coronary Heart Disease. Archives of Medical Research. 2010;41:383–390. doi: 10.1016/j.arcmed.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 31.Ghaffari MA, Askari Sede S, Rashtchizadeh N, Mohammadzadeh G, Majidi S. Association of CRP gene polymorphism with CRP levels and Coronary Artery Disease in Type 2 Diabetes in Ahvaz, southwest of Iran. BioImpacts. 2014;4:133–139. doi: 10.15171/bi.2014.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rashtchizadeh N, Askari Sede S, Ghaffari MA, Mohammadzadeh G, Majidi S. Associations of pentraxin 3 with presence and severity of coronary artery disease in type 2 diabetes patients. Turk J Biochem. 2015;40:37–43. [Google Scholar]
- 33.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Research. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersenn RV, Wittrup HH, Tybaerg-Hansen A, Steffensen R, Schnohr P, Nor destgaard BG. Hepatic lipase mutations, elevated high-density lipoprotein cholesterol, and increased risk of ischemic heart disease the Copenhagen City Heart Study. J Am Coll Cardiol. 2003;41:1972–82. doi: 10.1016/s0735-1097(03)00407-8. [DOI] [PubMed] [Google Scholar]
- 35.Dugi KA, Brandauer K, Schmidt N, Nau B, Schneider JG, Mentz S, et al. Low hepatic lipase activity is a novel risk factor for coronary artery disease. Circulation. 2001;104:3057–62. doi: 10.1161/hc5001.100795. [DOI] [PubMed] [Google Scholar]
- 36.Roberts CK, Ng C, Hama S, Eliseo AJ, Barnard RJ. Effect of a short-term diet and exercise intervention on inflammatory/anti-inflammatory properties of HDL in overweight/obese men with cardiovascular risk factors. J Appl Physiol. 2006;101:1727–32. doi: 10.1152/japplphysiol.00345.2006. [DOI] [PubMed] [Google Scholar]
- 37.Ansell BJ, Fonarow GC, Fogelman AM. The paradox of dysfunctional high-density lipoprotein. Curr Opin Lipidol. 2007;18:427–34. doi: 10.1097/MOL.0b013e3282364a17. [DOI] [PubMed] [Google Scholar]
- 38.Valdivielso P, José Ariza M, Vega-Román C, González-Alegre T, Rioja J, Ulzurrun E, et al. Association of the -250G/A promoter polymorphism of the hepatic lipase gene with the risk of peripheral arterial disease in type 2 diabetic patients. Journal of Diabetes and Its Complications. 2008;22:273–77. doi: 10.1016/j.jdiacomp.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 39.Jansen H, Verhoeven A, Weeks L, Kastelein J, Halley D, van den Ouweland A, et al. Common C-to-T substitution at position -480 of the hepatic lipase promoter associated with a lowered lipase activity in coronary artery disease patients. Arterioscler Thromb Vasc Biol. 1997;17:2837–42. doi: 10.1161/01.atv.17.11.2837. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C, Lopez-Ridaura R, Rimm EB, Hunter T, Hu FB. Genetic variation in the hepatic lipase gene and the risk of coronary heart disease among US diabetic men: Potential interaction with obesity. Diabetologia. 2006;49:1552–9. doi: 10.1007/s00125-006-0235-2. [DOI] [PubMed] [Google Scholar]
- 41.Ji J, Herbison CE, Mamotte CD, Burke V, Taylor RR, van Bockxmeer FM. Hepatic lipase gene − 514 C/T polymorphism and premature coronary heart disease. J Cardiovasc Risk. 2002;9:105–13. doi: 10.1177/174182670200900206. [DOI] [PubMed] [Google Scholar]
- 42.Fan Y, Laaksonen R, Janatuinen T, Vesalainen R, Nuutila P, Koivula T, et al. Hepatic lipase gene variation is related to coronary reactivity in healthy young men. Eur J Clin Invest. 2001;31:574–80. doi: 10.1046/j.1365-2362.2001.00858.x. [DOI] [PubMed] [Google Scholar]
- 43.Isaacs A, Sayed-Tabatabaei FA, Njajouo T, Witteman JC, vanDuijn CM. The -514 C/T hepatic lipase promoter region polymorphism and plasma lipids: A meta-analysis. J Clin Endocrinol Metab. 2004;89:3858–63. doi: 10.1210/jc.2004-0188. [DOI] [PubMed] [Google Scholar]
- 44.Castelli WP, Garrison RJ, Wilson PW, Abbott RD, Kalousdian S, KannelWB Incidence of coronary heart disease and lipoprotein cholesterol levels. The Framingham Study. JAMA. 1986;256:2835–8. [PubMed] [Google Scholar]
- 45.Sharrett AR, Ballantyne CM, Coady SA, Heiss G, Sorlie PD, Catellier D, et al. Atherosclerosis Risk in Communities Study Group. Coronary heart disease prediction from lipoprotein cholesterol levels, triglycerides, lipoprotein (a), apolipoproteins A-I and B, and HDL density sub fractions: The Atherosclerosis Risk in Communities (ARIC) Study. Circulation. 2001;104:1108–13. doi: 10.1161/hc3501.095214. [DOI] [PubMed] [Google Scholar]
- 46.Grarup N, Andersen G. Gene-environment interactions in the pathogenesis of type 2 diabetes and metabolism. Curr Opin Clin Nutr Metab Care. 2007;10:420–6. doi: 10.1097/MCO.0b013e3281e2c9ab. [DOI] [PubMed] [Google Scholar]
- 47.Austin M, Breslow J, Hennekens C, Buring J, Willett W, Krauss R. Low-density lipoprotein subclass patterns and risk of myocardial infarction. JAMA. 1988;13:1917–21. [PubMed] [Google Scholar]


