Abstract
It has become a matter of orthodoxy that among wasps, ants, bees, and other insects in the order Hymenoptera, only uniparental haploid males that arise from unfertilized eggs are capable of reproduction. This idea is of interest because the best understood and perhaps most widespread sex determination system among these insects [known as single locus complementary sex determination (sl-CSD)] does not depend on ploidy alone and, paradoxically, consistently results in small numbers of diploid biparental males. To date, the reproductive potential of diploid males has been studied in 13 of the perhaps 200,000 hymenopterans world-wide; in each of these instances, the diploid males are genetic dead ends because they are inviable or sterile. The data from these species have resulted in a general conclusion that has been invoked for virtually all species with sl-CSD and has become the basis for assumptions regarding conservation biology, sex ratio analysis, and the evolution of social behavior. Here, we report that in the solitary vespid wasp Euodynerus foraminatus, both diploid and haploid males are fertile, which documents normal fertility in diploid males of a hymenopteran with sl-CSD. This wasp has high levels of inbreeding because of frequent brother–sister mating in nature; therefore, diploid males are more frequently produced and thus more likely exposed to selection favoring their fertility. Because inbreeding and diploid male production may be important features of the population biology of many hymenopterans, we sound a cautionary note regarding ideas about the evolutionary ecology of these insects.
Nearly 20% of animal species reproduce by means of the genetic system of arrhenotokous haplodiploidy; females are diploid and arise from fertilized eggs, whereas males are haploid and develop from unfertilized eggs. Haplodiploidy has evolved independently in nematodes, rotifers, mites, scale insects, white flies, thrips, and ambrosia beetles and is characteristic of the entire insect order Hymenoptera (ants, bees, wasps, and sawflies), which may include 200,000 species. Empirical evidence indicates that a diversity of genetic mechanisms control sex determination among these animals. Among hymenopterans, the best understood of these is single-locus complementary sex determination (sl-CSD), which depends on allelic variation at a sex locus (1, 2). An individual develops as a female only when it is heterozygous at the sex locus. If an individual is hemizygous (haploid) it becomes male, but diploid individuals homozygous at the sex locus also develop as males.
Diploid males are generally considered to be a genetic dead end in the Hymenoptera (3–6). In some cases, they have low viability (1, 7–9). In a number of species, diploid males have normal viability (6, 10, 11) but fail to pass their genetic material to subsequent generations because they are unable to mate properly (12) or because they are sterile (13–16). Some diploid males have been shown to produce viable sperm, but this sperm is diploid rather than haploid and results in sterile triploid progeny (10, 12, 17–19). The picture that has emerged regarding diploid males is that they perform poorly across a variety of traits associated with fitness, and should they succeed in fathering any surviving offspring, these offspring will themselves be incapable of reproducing. Thus, diploid males present a potential cost at two levels: a cost to their parents because they will not transmit genetic material and an additional cost to females that mate with them because no fertile offspring will result from the union. This view of diploid males has virtually become dogma and has consequences for ideas about sex ratio evolution (20), the evolution of social behavior (21, 22), and conservation genetics (23).
Diploid male offspring result from so-called matched matings (24) in which the parents share an allele at the sex locus. Thus, diploid males will occur in randomly mating populations whenever a matched mating occurs by chance. The greater the diversity of sex alleles, the lower the chance will be of a matched mating. Because of this random production of diploid males, hymenopteran sex alleles are under frequency-dependent selection, with the rare allele having the greatest advantage (25). An equilibrium number of sex alleles, increasing with effective population size, is expected to result from a balance between mutation and drift. Individuals that are morphologically male but diploid, as revealed by heterozygosity for a genetic marker or by karyotype analysis, provide evidence that a species has CSD. Furthermore, a central prediction of the sl-CSD model is that inbreeding will cause an increase in the proportion of diploid males because it increases homozygosity at the sex locus. When inbreeding fails either to produce diploid males or result in sex ratio shifts, a noncomplementary form of sex determination must be functioning (26–28).
Because of the relationships among inbreeding, diploid male production, and reproductive failure by diploid males, natural selection should favor outcrossing mechanisms (inbreeding avoidance) in species with sl-CSD. Among species in which inbreeding is a common part of the mating system, genetic mechanisms other than sl-CSD should exist (5), which appears to be the case. In many species with confirmed sl-CSD, individuals are spatially scattered and develop in isolation from relatives, as with some sawflies and ichneumons (12, 17, 29–31), thus promoting outbreeding. In honey bees and some parasitoids with sl-CSD, brothers and sisters develop in close proximity but disperse from their natal area (thus avoiding siblings) before mating (32, 33). In addition to dispersing before mating, some species with sl-CSD are able to recognize siblings and avoid them as sexual partners (34, 35). Many hymenopterans, however, regularly mate at their natal site before dispersal, so that mother–son or sister–brother matings are common. For those inbreeding species that have been studied, the sl-CSD hypothesis has been rejected because the expected diploid males and sex ratio shifts have not been observed in controlled breeding experiments (27). The combination of sl-CSD and frequent inbreeding in a single species would run counter to these expectations and present a paradox inviting scrutiny; Euodynerus foraminatus, a solitary hunting vespid wasp, provides such a paradox.
In E. foraminatus broods, brothers and sisters develop in the same nest. Males reach maturity first, remain at the natal nest, and mate with their emerging sisters. Behavioral observations and population genetic data indicate that up to 66% of females mate with a brother at their natal nest before dispersing, and the remaining females mate randomly in the population after dispersal (36, 37). However, laboratory breeding experiments confirm that E. foraminatus has sl-CSD and that the resulting diploid males have egg-to-adult viability comparable to haploid males (38). If diploid males in E. foraminatus are the genetic dead end that they are in other species, then this wasp should be suffering a significant sex determination load because of the production of diploid males and possibly an additional loss in fitness to any female that mates with a diploid male and produces sterile triploid offspring. In this study, we investigated the possible consequences of diploid male production in E. foraminatus through breeding experiments and microsatellite DNA genotyping. These methods enabled us to determine (i) whether diploid males are capable of both mating and fathering offspring, (ii) the ploidy of any daughters fathered by diploid males, and (iii) whether such daughters were themselves fertile.
Methods
Wasp Biology and Artificial Rearing. A nesting female of E. foraminatus finds preexisting narrow cylindrical cavities, such as hollow twigs or vacant insect tunnels in dead wood, deposits an egg, and then hunts for caterpillars, which she paralyzes by stinging and then stuffs into the hole with her egg. She then builds a mud partition enclosing the egg with its food. The female repeats this process so that the cavity becomes filled with a linear series of brood cells. Mothers control fertilization of their eggs and deposit fertilized (female-producing) eggs in the inner part of a nest and unfertilized (male-producing) eggs in the outer cells (39, 40). The sequence of sexes within a nest reveals information about whether the female had a matched mating. In unmatched matings, all of a female's fertilized eggs develop as an uninterrupted series of daughters in the innermost part of the nest, but in matched matings, half of the fertilized eggs will develop randomly as diploid males. These diploid males can sometimes be recognized by their “out-of-order” nest position if they are followed within the nest by a cell in which a female develops (38).
We collected broods of wasps from nature by using artificial nest cavities (40) placed at five locations separated by at least 5 km in Allegan and Kalamazoo counties, Michigan. We assumed that wasps reared from the same nest were siblings, and that wasps from separate field sites were unrelated. Two or three days after eclosing as adults, pairs were placed together in cages and allowed to mate. After copulating, individual females were caged and provided with nesting materials (41). Nesting females were maintained in a greenhouse at approximately ambient temperature and photoperiod.
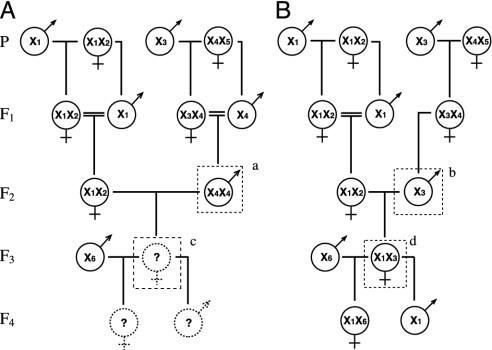
Experimental Breeding Pedigrees. We bred the wasps according to the scheme shown in Fig. 1. The parental generation consisted of free-flying wild wasps. Their offspring, reared from nests collected from the field, constituted the F1 generation. Thirty-one nest-mate pairs (siblings) of F1 wasps were mated to obtain the F2 generation containing both diploid and haploid males. According to the sl-CSD model, half of these matings will be matched and half of the diploid offspring from matched matings will be male (Fig. 1 A). The other half of the matings will be unmatched and produce only haploid males and diploid females (Fig. 1B). We used matings of F1 sibling pairs for the control as well as the experimental pedigrees so that any effects of inbreeding at loci other than the sex locus would be uniform in later generations. Sibling matings were only used among F1 adults to produce the F2 generation, which was the only generation to include diploid males (see below for methods of determining whether a particular male is haploid or diploid); all of the pairings in subsequent generations were outcrosses involving unrelated wasps derived from different field locations. To test the fertility of diploid males, diploid (Fig. 1, experimental wasp a) and haploid (Fig. 1, control wasp b) males of the F2 generation were bred to see whether they were capable of producing offspring (Fig. 1, F3 females c and d). Because diploid males did produce daughters (see below), we tested the fertility of these daughters by pairing them with unrelated males to see whether they could produce an F4 generation.
Fig. 1.
Experimental (A) and control (B) breeding protocols. X1, X2,..., X5 represent hypothetical distinct alleles at the sex locus. The experimental and control pedigrees differ in that biparental diploid F2 males (male a) were used in the experimental lineage and uniparental haploid males (male b) were used in the control lineage. According to the current understanding of sl-CSD, if diploid males are fertile, their offspring (F3 female c) should be triploid and sterile and no F4 offspring should be produced.
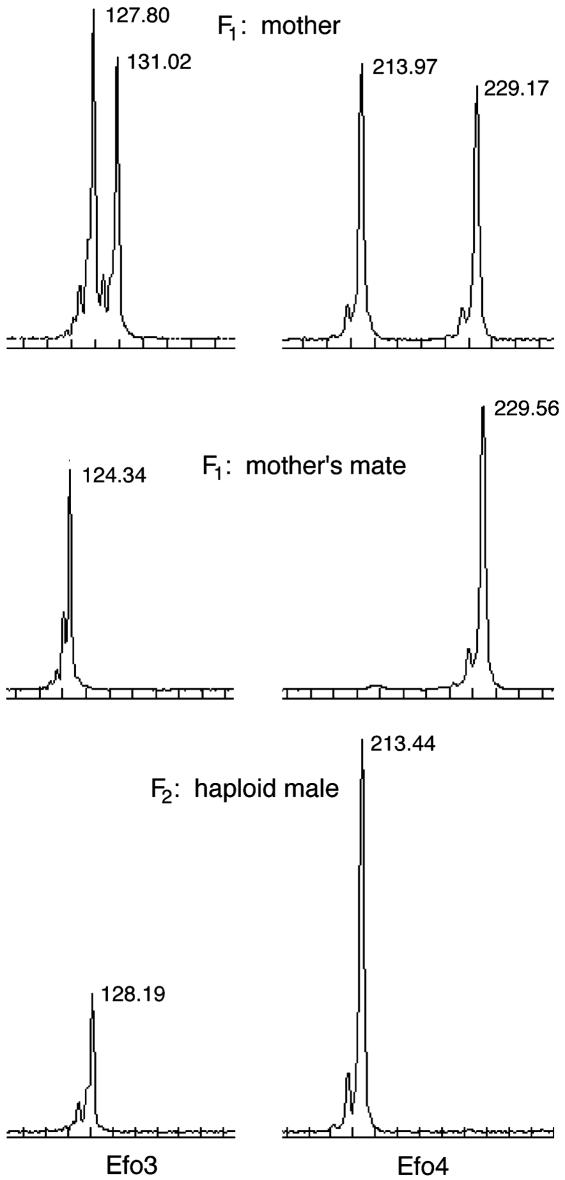
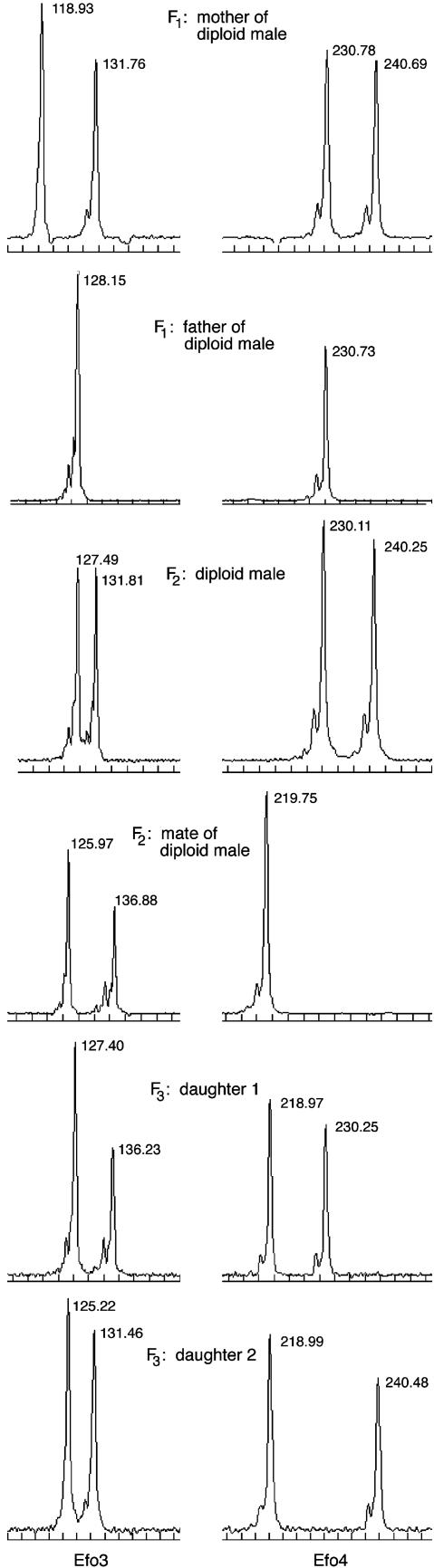
Genetic Analysis: Determination of Males as Haploid or Diploid. Because our distinction between experimental and control groups is based on the genotypes of males at the sex locus and because at this time the precise nature of the sex locus in E. foraminatus is unknown, an individual's sex locus genotype must be inferred based on other genotypic or phenotypic characteristics. To determine whether the male wasps we used for breeding in the F2 generation were haploid or diploid, we genotyped them, along with their parents, at five microsatellite loci (42). Using microsatellites allowed us not only to determine the ploidy of individual wasps but also to track the segregation of alleles from one generation to the next. Because all male wasps in the F2 generation were sons of sib-mated parents, there is an increased chance that diploid males could be homozygous at all markers and appear to be haploid. We categorized males as haploid (uniparental) only if they were not heterozygous at any locus, had exclusively maternal alleles, and lacked any distinctive paternal alleles observed in their putative “father” (mother's mate) (Fig. 2). We categorized males as diploid (biparental) only if they were heterozygous at one or more loci and carried both paternal and maternal alleles (Fig. 3).
Fig. 2.
Electropherograms of microsatellite fragments showing the parentage of a haploid male. The numbers next to each peak are the fragment lengths as measured by Beckman Coulter ceq 2000 fragment analysis software. Results for two loci in each individual are shown; diagrams for the Efo3 locus are on the left, and diagrams for the Efo4 locus are on the right. The mother is heterozygous at both the Efo3 and Efo4 loci. In this case, the haploid male has inherited allele 128 at the Efo3 locus and allele 213 at the Efo4 locus from his mother. The mother's mate's alleles are absent, indicating that the son is uniparental and haploid.
Fig. 3.
Electropherograms for the family of a diploid male showing (from top to bottom) the diploid male's mother, his father, the diploid male, the diploid male's mate, and two daughters of the diploid male. The Efo3 locus is on the left, and the Efo4 locus is on the right. In this case, the male is heterozygous (diploid/biparental) at both loci, having inherited allele 127 at the Efo3 locus and allele 230 at the Efo4 locus from his father. The electropherograms for the daughters show that they are heterozygous (diploid, not triploid), having inherited one distinctive allele from their father and one from their mother. Note that each daughter carries a different paternal allele.
We timed the duration of the stages of mating for haploid and diploid males and for daughters of diploid males and daughters of haploid males. We performed statistical analyses according to Zar (43). χ2 tests are with Yates correction, and all t tests are two-tailed.
Results
Mating Ability and Fertility of Diploid Males. We established 39 F2 pairings with haploid or diploid males. On the basis of the microsatellite criteria described above, we identified 19 males as diploids and 18 as haploids. Two of the males had pedigrees lacking adequate allelic variation to confirm a diploid or haploid status; their families were not included in our analysis. The mating abilities of haploid and diploid males were comparable with regard to their ability to mount females and engage in copulation. All 19 diploid males and 16 of 18 haploid males mated, and for the males that mated, we found no differences between the groups in the time required for courtship (t = 1.30, P = 0.20) or to complete copulation (t = 0.02, P = 0.98). Of the 19 females that mated with diploid males, three never attempted to nest, and similarly, four of the 16 females mated to haploid males failed to nest (χ2 = 0.27, P = 0.60). These nonnesting wasps are not included in further analyses.
Data comparing the reproductive output of nesting females mated to diploid versus haploid males are presented in Table 1. Females mated to diploid males and females mated to haploids provisioned comparable numbers of nest cells (x̄ = 37.3 vs. x̄ = 32.3, t = 1.61, P = 0.12). The proportion of immature mortality in the two groups was the same: 0.25. Thus, regardless of their mate's ploidy, females produced similar numbers of offspring.
Table 1. Numbers of offspring and sex ratios produced by female wasps mated to diploid males or haploid males.
| Female | Sons | Daughters | Dead | Total | Sex ratio |
|---|---|---|---|---|---|
| Broods with diploid fathers | |||||
| 1 | 40 | 0 | 8 | 48 | 1.00 |
| 2 | 4 | 27 | 9 | 40 | 0.13 |
| 3 | 9 | 36 | 7 | 52 | 0.20 |
| 4 | 2 | 20 | 14 | 36 | 0.09 |
| 5 | 2 | 15 | 12 | 29 | 0.12 |
| 6 | 8 | 13 | 14 | 35 | 0.38 |
| 7 | 33 | 2 | 8 | 43 | 0.94 |
| 8 | 2 | 10 | 21 | 33 | 0.17 |
| 9 | 11 | 14 | 5 | 30 | 0.44 |
| 10 | 8 | 28 | 8 | 44 | 0.22 |
| 11 | 13 | 15 | 5 | 33 | 0.46 |
| 12 | 1 | 22 | 12 | 35 | 0.04 |
| 13 | 16 | 16 | 8 | 40 | 0.50 |
| 14 | 10 | 12 | 8 | 30 | 0.45 |
| 15 | 21 | 16 | 6 | 43 | 0.57 |
| 16 | 7 | 12 | 6 | 25 | 0.37 |
| Total | 187 | 258 | 151 | 596 | — |
| Mean | 11.7 | 16.1 | 9.4 | 37.3 | 0.38 |
| Broods with haploid fathers | |||||
| 17 | 0 | 7 | 11 | 18 | 0.00 |
| 18 | 6 | 23 | 13 | 42 | 0.21 |
| 19 | 7 | 32 | 10 | 49 | 0.18 |
| 20 | 2 | 17 | 6 | 25 | 0.11 |
| 21 | 5 | 13 | 10 | 28 | 0.28 |
| 22 | 2 | 22 | 12 | 36 | 0.08 |
| 23 | 3 | 20 | 9 | 32 | 0.13 |
| 24 | 2 | 21 | 8 | 31 | 0.09 |
| 25 | 0 | 21 | 5 | 26 | 0.00 |
| 26 | 5 | 19 | 1 | 25 | 0.21 |
| 27 | 2 | 30 | 7 | 39 | 0.06 |
| 28 | 4 | 28 | 4 | 36 | 0.13 |
| Total | 38 | 253 | 96 | 387 | — |
| Mean | 3.2 | 21.1 | 8.0 | 32.3 | 0.12 |
There were however significant differences between the groups in the numbers of male offspring. Females mated to diploids averaged significantly more sons (x̄ = 11.7) than females mated to haploids (x̄ = 3.2, t = 2.97, P = 0.01). However, for male reproductive success, the critical factor is whether their sperm are used in fertilizations to make daughters. Diploid males produced, on average, 16.1 daughters versus the 21.1 daughters achieved by haploid males; diploid males thus have 76% the fertility of haploids. Even so, the two-tailed test does not indicate a significant difference (t = 1.60, P = 0.12). The range in the number of daughters for diploid males (0–36) is greater than for haploids (7–32); the fertility of diploid males spans the range from zero to levels indistinguishable from that of normal haploid males. Because of the high variability in our sample, there may be undetected differences between diploid and haploid male reproduction. However, clearly some diploid males have fertility comparable to or exceeding that of some haploid males.
Reproductive Capabilities and Ploidy of Daughters of Diploid Males. The behavior of daughters of diploid males did not differ from that of daughters of haploid males with regard to courtship (t = 0.53, P = 0.60) or total time required for mating (t = 1.76, P = 0.09), nor did daughters of diploid and haploid males show any differences in nesting and reproduction. Of the 30 F3 females, 11 of 18 that had diploid fathers nested, and 10 of 12 with haploid fathers nested (χ2 = 0.80, P = 0.37). Nesting females with diploid versus haploid fathers provisioned an average of 23.8 versus 24.8 cells respectively (t = 0.25, P = 0.81), and mortality among the offspring of the two kinds of females was also similar (0.41 versus 0.43, χ2 = 0.01, P = 0.93) (Table 2). Because the daughters of diploid males had normal fertility, we would expect them to be diploid rather than triploid. By using microsatellites, we were able to test the ploidy of these females. Thirteen diploid males and their mates had microsatellite allelic combinations that allowed us to unequivocally determine whether their daughters were diploid or triploid. We genotyped 47 daughters from these crosses. In all cases, the daughters were diploid with one distinctive allele from each parent, and their diploid fathers could pass either allele at a locus to these daughters (Fig. 3).
Table 2. Numbers of offspring produced by female wasps that had diploid or haploid fathers.
| Female | Sons | Daughters | Dead | Total |
|---|---|---|---|---|
| Daughters of diploid fathers | ||||
| A | 5 | 20 | 15 | 40 |
| B | 5 | 8 | 7 | 20 |
| C | 1 | 6 | 9 | 16 |
| D | 1 | 2 | 6 | 9 |
| E | 5 | 12 | 8 | 25 |
| F | 2 | 19 | 11 | 32 |
| G | 2 | 5 | 10 | 17 |
| H | 5 | 10 | 15 | 30 |
| I | 1 | 5 | 6 | 12 |
| J | 4 | 16 | 12 | 32 |
| K | 5 | 12 | 11 | 28 |
| Mean | 3.3 | 10.6 | 10.1 | 23.8 |
| Daughters of haploid fathers | ||||
| L | 13 | 9 | 7 | 29 |
| M | 4 | 15 | 11 | 30 |
| N | 2 | 8 | 14 | 24 |
| O | 2 | 16 | 9 | 27 |
| P | 2 | 4 | 11 | 17 |
| Q | 0 | 0 | 5 | 5 |
| R | 7 | 9 | 12 | 28 |
| S | 8 | 7 | 13 | 28 |
| T | 4 | 8 | 17 | 29 |
| U | 8 | 17 | 8 | 33 |
| Mean | 5.0 | 9.3 | 10.5 | 24.8 |
Discussion
Our data show that diploid males in E. foraminatus are sexually competent, father diploid daughters, and can transmit either allele at a locus through their sperm. Furthermore, daughters of these diploid males also mate normally and are fully fertile. Previous studies of diploid male reproductive abilities among hymenopterans with sl-CSD provided a sample based on only 13 species from nine genera. Our knowledge of these species indicates that they all have life cycles or behaviors that promote random mating, thus minimizing homozygosity at the sex locus and the frequency of diploid males, and that when diploid males are produced, they are nonfunctional. The information from this handful of examples has been expanded to a general assumption regarding many other species for which data are lacking. The wasp E. foraminatus stands in contrast because it has sl-CSD, high natural levels of inbreeding, and diploid males with normal viability (37, 38). Furthermore, in this study we have found that diploid E. foraminatus males have near-normal fertility. Consequently, our results bring into question widespread assumptions about the inability of diploid males to reproduce.
Investigations of sperm production in haploid male hymenopterans have shown that spermatogenesis begins with a reductional division (meiosis I) that is aborted during metaphase I (18, 44, 45). The diploid sperm made by diploid males of other species (10, 16, 17, 19) is presumably the result of these same events occurring even when two complementary sets of chromosomes are present in the spermatogonium. Spermatogenesis in these species follows the same path regardless of whether it occurs in a haploid or a diploid male. However, our observations that a diploid male of E. foraminatus passes either, but not both, of his alleles to his daughters suggests that a complete reductional division occurs during spermatogenesis. An alternative possibility is that diploid males make diploid sperm, but one chromosome set from the sperm cell is eliminated at some point in the fertilization process, resulting in a diploid zygote. Either of these possibilities implies a significant and previously unknown polymorphism that involves alteration of developmental processes according to ploidy, either during meiosis or in the zygote, so that offspring sired by diploid males are always biparental diploids.
Similar variation in the functionality of diploid males may be present in other sl-CSD species. El Agoze et al. (13) present evidence that diploid males of the ichneumonid Diadromus pulchellus have fertility that is only 1% that of haploid males; however, the rare daughters produced by diploid males that they observed are apparently normal diploids rather than triploids. If species with this variability were to start inbreeding, the frequency of diploid males would increase and there would be enhanced selection for increased diploid male fertility. Other studies of natural hymenopteran populations have revealed significant numbers of diploid males and attributed their presence to habitat fragmentation and loss of sex allele diversity by drift in small, isolated populations, or else to consanguineous matings (23, 46–48). Because either of these circumstances can result in high diploid male production, they can also strengthen selection for an increase in diploid male fertility. Consequently, these species may be of interest as candidates for having functional diploid males.
The paradox that motivated this study, that a species with sl-CSD engages in frequent inbreeding, is partly resolved by the discovery that diploid males are viable, fertile, and capable of producing normal offspring. But why does E. foraminatus inbreed in the first place? Because this is a common species and individuals are strong fliers, difficulty obtaining mates seems an unlikely explanation. Perhaps the advantage to a female of mating with a brother and achieving greater genetic representation in offspring outweighs the costs of lower genetic variation among offspring. Even though diploid males are not a complete loss, their full significance in natural populations remains to be investigated. Because most males are normal haploids and the preponderance of fertilized eggs develop as females and because nesting females arrange these offspring within nests in a stereotypical female-first order, it seems that diploid males are still not a “normal” or “preferred” mode of reproduction. From a maternal point of view, diploid sons constitute a loss of the precise control over offspring sex that is a major theme in the behavioral ecology of the Hymenoptera, and in E. foraminatus this also means a loss of a female's ability to adjust the amount of provisions given to each offspring according to its sex. Furthermore, compared to having haploid sons, females also incur a loss with diploid sons because genetic representation in grandchildren through diploid sons is half of that achieved through haploid sons. In these regards, it is always better to make haploid sons.
A more efficient solution to problems with sex determination and inbreeding would seem to be the evolution of noncomplementary sex determination mechanisms, such as those observed among other inbreeding hymenopterans (26, 28, 49–52). Because we do not know all of the molecular details of CSD (2), it is not possible to make inferences about the difficulty of the evolutionary transition from complementary to noncomplementary sex determination. If sl-CSD is ancestral, then the transition to noncomplementary sex determination systems has evolved multiple times: in the chalcidoid clade (50–52), in the Ichneumonoidea (53), and in the Aculeata (26). The finding that sl-CSD is the mechanism in E. foraminatus but that diploid males have significant fertility and fertile offspring indicates an additional adaptive solution to a shared inbreeding contingency.
To the extent that reproductive diploid males may be present in other species, some ideas about the evolutionary ecology of wasps, ants, and bees will need to be reconsidered. For example, in sex ratio theory, the relative allocation of parental resources between sons and daughters is a fundamental parameter (54). If diploid males are nonfunctional, then they would be charged to the allocation toward daughters because they are an unavoidable component of the cost of making daughters. But if diploid males can reproduce, their cost to parents would become part of the effort devoted to the production of sons. The issue is further complicated because, from the maternal point of view, diploid sons have only half the value of haploids. Because haploid “sons” have no value to hymenopteran fathers, there is also a potential conflict between mates over the kind of sons that should be produced.
The prospect of fertile diploid males has intriguing implications for social hymenopterans. As a member of the family Vespidae, E. foraminatus is a close relative of the social wasps, and the vespids are closely related to the ants and bees (55, 56), so these other taxa might have similar potential regarding male fertility. Among social insects, sl-CSD presents additional costs because some fertilized eggs intended to produce workers (which are all female) would develop as nonworking diploid males and severely limit colony viability (57), even though the cost of a reduction in worker force because of sl-CSD can be ameliorated by the destruction of diploid male eggs before significant energy is devoted to their care (58, 59). The infertility of diploid males is key to some ideas about how often queens should mate (60, 61) or how many queens are best for a colony (21, 22). The cost of producing diploid males instead of workers is not incurred by socially parasitic ant species that have lost the worker caste. This is a life history that has evolved independently many times among ants and is accompanied by a correlated suite of characteristics that have been referred to as the “inquiline syndrome” (62, 63), which, in addition to loss of workers, includes mating within the natal nest, probably between siblings, before female dispersal. However, even if diploid males are functional, retention of sl-CSD would mean significant loss of control over sex ratio by queens and, consequently, loss of a major advantage of sex ratio control under inbreeding and local competition for mates (64). Ants in the genus Epimyrma exhibit the inquiline syndrome but appear to have evolved a noncomplementary sex determination mechanism (64).
Whether the fertility of diploid males in E. foraminatus is an unusual exception to the commonly held view that diploid males are a complete loss or whether this is an example of a more widespread phenomenon remains to be determined. Until we have data from more species, particularly from species with population structure or breeding systems that are likely to produce high frequencies of diploid males, E. foraminatus serves as a cautionary example.
Acknowledgments
We thank Todd Barkman, DeWayne Shoemaker, and Karim Essani for interactions and comments on previous versions of the manuscript. This work was supported by funds from the Western Michigan University College of Arts and Sciences and Department of Biological Sciences.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: sl-CSD, single locus complementary sex determination.
References
- 1.Whiting, P. W. (1943) Genetics 28, 365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beye, M., Hasselmann, M., Fondrk, M. K., Page, R. E. J. & Omholt, S. W. (2003) Cell 114, 419–429. [DOI] [PubMed] [Google Scholar]
- 3.Crozier, R. H. (1977) Annu. Rev. Entomol. 22, 263–288. [Google Scholar]
- 4.Stouthamer, R., Luck, R. F. & Werren, J. H. (1992) Environ. Entomol. 21, 427–435. [Google Scholar]
- 5.Cook, J. M. & Crozier, R. H. (1995) Trends Ecol. Evol. 10, 281–286. [DOI] [PubMed] [Google Scholar]
- 6.Beukeboom, L. W. (2001) Neth. J. Zool. 51, 1–15. [Google Scholar]
- 7.Bostian, C. H. (1935) Genetics 20, 280–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiting, A. R. (1961) Adv. Genet. 10, 295–348. [DOI] [PubMed] [Google Scholar]
- 9.Petters, R. M. & Mettus, R. V. (1980) J. Hered. 71, 353–356. [Google Scholar]
- 10.Inaba, F. (1939) Cytologia 9, 517–523. [Google Scholar]
- 11.Duchateau, M. J., Hoshiba, H. & Velthuis, H. H. (1994) Entomol. Exp. Appl. 71, 263–269. [Google Scholar]
- 12.Smith, S. G. & Wallace, D. R. (1971) Can. J. Genet. Cytol. 13, 617–621. [Google Scholar]
- 13.El Agoze, M., Drezen, J. M., Renault, S. & Periquet, G. (1994) Bull. Entomol. Res. 84, 213–218. [Google Scholar]
- 14.Duchateau, M. J. & Mariën, J. (1995) Insectes. Soc. 42, 255–266. [Google Scholar]
- 15.Krieger, M. J. B., Ross, K. G., Chang, C. W. Y. & Keller, L. (1999) Heredity 82, 142–150. [Google Scholar]
- 16.MacBride, D. H. (1946) Genetics 31, 224. [PubMed] [Google Scholar]
- 17.Naito, T. & Suzuki, H. (1991) J. Hered. 82, 101–104. [Google Scholar]
- 18.Woyke, J. & Skowronek, W. (1974) J. Apic. Res. 13, 183–190. [Google Scholar]
- 19.Yamauchi, K., Yoshida, T., Ogawa, T., Itoh, S., Ogawa, Y., Jimbo, S. & Imai, H. T. (2001) Insectes. Soc. 48, 28–32. [Google Scholar]
- 20.Cook, J. M. (2002) in Sex Ratios, Concepts and Research Methods, ed. Hardy, I. C. W. (Cambridge Univ. Press, Cambridge, U.K.).
- 21.Crozier, R. H. & Pamilo, P. (1996) Evolution of Social Insect Colonies (Oxford Univ. Press, Oxford).
- 22.Bourke, A. F. G. & Franks, N. R. (1995) Social Evolution in Ants (Princeton Univ. Press, Princeton).
- 23.Zayed, A. & Packer, L. (2001) Heredity 87, 631–636. [DOI] [PubMed] [Google Scholar]
- 24.Adams, J., Rothman, E. D., Kerr, W. E. & Paulino, Z. L. (1977) Genetics 86, 538–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yokoyama, S. & Nei, M. (1979) Genetics 91, 609–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cook, J. M. (1993) Heredity 71, 130–137. [Google Scholar]
- 27.Cook, J. M. (1993) Heredity 71, 421–435. [Google Scholar]
- 28.Dobson, S. L. & Tanouye, M. A. (1998) Genetics 149, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Butcher, R. D. J., Whitfield, W. G. F. & Hubbard, S. F. (2000) J. Evol. Biol. 13, 593–606. [Google Scholar]
- 30.Sawa, M., Fukunaga, A., Naito, T. & Oishi, K. (1989) Zool. Sci. 6, 541–547. [Google Scholar]
- 31.Becker, G. C. & Benjamin, D. M. (1967) Can. Entomol. 99, 146–159. [Google Scholar]
- 32.Koeniger, G. (1986) in Bee Genetics and Breeding, ed. Rinderer, T. E. (Academic, Orlando, FL), pp. 255–280.
- 33.Antolin, M. F. & Strand, M. R. (1992) Ecol. Entomol. 17, 1–7. [Google Scholar]
- 34.Foster, R. L. (1992) J. Kans. Entomol. Soc. 65 (3), 238–243. [Google Scholar]
- 35.Ode, P. J., Antolin, M. F. & Strand, M. R. (1995) Anim. Behav. 49, 1239–1248. [Google Scholar]
- 36.Cowan, D. P. (1979) Science 205, 1403–1405. [DOI] [PubMed] [Google Scholar]
- 37.Stahlhut, J. K. & Cowan, D. P. (2004) Mol. Ecol. 13, 631–638. [DOI] [PubMed] [Google Scholar]
- 38.Stahlhut, J. K. & Cowan, D. P. (2004) Heredity 92, 189–196. [DOI] [PubMed] [Google Scholar]
- 39.Cowan, D. P. (1981) Behav. Ecol. Sociobiol. 9, 95–102. [Google Scholar]
- 40.Krombein, K. V. (1967) Trap-Nesting Wasps and Bees: Life Histories, Nests, and Associates (Smithsonian Press, Washington, DC).
- 41.Chilcutt, C. F. & Cowan, D. P. (1993) Great Lakes Entomol. 26, 15–19. [Google Scholar]
- 42.Stahlhut, J. K., Cowan, D. P., Essani, K., Shoemaker, D. D. & Barkman, T. (2002) Mol. Ecol. Notes 2, 467–468. [Google Scholar]
- 43.Zar, J. H. (1999) Biostatistical Analysis (Prentice–Hall, Upper Saddle River, NJ).
- 44.Hogge, M. A. F. & King, P. E. (1975) J. Submicr. Cytol. 7, 81–96. [Google Scholar]
- 45.Hoage, T. R. & Kessel, R. G. (1968) J. Ultrastructure Res. 24, 6–32. [DOI] [PubMed] [Google Scholar]
- 46.Kukuk, P. & May, B. (1990) Evolution (Lawrence, Kans.) 44, 1522–1528. [DOI] [PubMed] [Google Scholar]
- 47.Roubik, D. W., Weight, L. A. & Bonilla, M. A. (1996) Evolution (Lawrence, Kans.) 50, 931–935. [DOI] [PubMed] [Google Scholar]
- 48.Chapman, T. W. & Stewart, S. C. (1996) Heredity 76, 65–69. [Google Scholar]
- 49.Rojas-Rousse, D., Eslami, J. & Periquet, G. (1988) J. Appl. Entomol. 106, 276–285. [Google Scholar]
- 50.Schmieder, R. G. & Whiting, P. W. (1947) Genetics 32, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skinner, S. W. & Werren, J. H. (1980) Genetics 94, s98. [Google Scholar]
- 52.Legner, E. F. (1979) Ann. Entomol. Soc. Am. 72, 114–118. [Google Scholar]
- 53.Beukeboom, L. W., Ellers, J. & Van Alphen, J. J. M. (2000) Heredity 84, 29–36. [DOI] [PubMed] [Google Scholar]
- 54.Hardy, I. C. W. (2002) Sex Ratios, Concepts and Research Methods (Cambridge Univ. Press, Cambridge, U.K.).
- 55.Brothers, D. J. & Carpenter, J. M. (1993) J. Hymenoptera Res. 2, 227–304. [Google Scholar]
- 56.Dowton, M. & Austin, A. D. (1994) Proc. Natl. Acad. Sci. USA 91, 9911–9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ross, K. G. & Fletcher, D. J. C. (1986) Behav. Ecol. Sociobiol. 19, 283–294. [Google Scholar]
- 58.Ratnieks, F. L. (1990) Behav. Ecol. Sociobiol. 26, 343–348. [Google Scholar]
- 59.Pamilo, P., Sundström, L., Fortelius, W. & Rosengren, R. (1994) Ethol. Ecol. Evol. 6, 221–235. [Google Scholar]
- 60.Page, R. E. (1980) Genetics 96, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crozier, R. H. & Page, R. E. (1985) Behav. Ecol. Sociobiol. 18, 105–115. [Google Scholar]
- 62.Hölldobler, B. & Wilson, E. O. (1990) The Ants (Belknap, Cambridge, MA).
- 63.Wilson, E. O. (1971) The Insect Societies (Belknap, Cambridge, MA).
- 64.Buschinger, A. (1989) J. Evol. Biol. 2, 265–283. [Google Scholar]