Abstract
Heparan sulfate (HS) is a component of cell surface and matrix-associated proteoglycans (HSPGs) that collectively, play crucial roles in many physiologic processes including cell differentiation, organ morphogenesis and cancer. A key function of HS is to bind and interact with signaling proteins, growth factors, plasma proteins, immune-modulators and other factors. In so doing, the HS chains and HSPGs are able to regulate protein distribution, bio-availability and action on target cells and can also serve as cell surface co-receptors, facilitating ligand-receptor interactions. These proteins contain an HS/heparin-binding domain (HBD) that mediates their association and contacts with HS. HBDs are highly diverse in sequence and predicted structure, contain clusters of basic amino acids (Lys, Arg) and possess an overall net positive charge, most often within a consensus Cardin-Weintraub (CW) motif. Interestingly, other domains and residues are now known to influence protein-HS interactions, as well as interactions with other glycosaminoglycans, such as chondroitin sulfate. In this review we provide a description and analysis of HBDs in proteins including amphiregulin, fibroblast growth factor family members, heparanase, sclerostin and hedgehog protein family members. We discuss HBD structural and functional features and important roles carried out by other protein domains, and also provide novel conformational insights into the diversity of CW motifs present in Sonic, Indian and Desert hedgehogs. Finally, we review progress in understanding the pathogenesis of a rare pediatric skeletal disorder, Hereditary Multiple Exostoses (HME), characterized by HS deficiency and cartilage tumor formation. Advances in understanding protein-HS interactions will have broad implications for basic biology and translational medicine as well as for the development of HS-based therapeutics.
Keywords: Heparan sulfate proteoglycans, heparan sulfate/heparin-binding domains, signaling and growth factor proteins, extracellular matrix
Introduction
The heparan sulfate proteoglycans (HSPGs) constitute a large and important family of cell surface and extracellular matrix (ECM)-associated macromolecules. The HSPGs display distinct patterns of expression and regulate a variety of physiologic roles including cell differentiation, cell-cell interactions, tissue morphogenesis and organ function; when dys-regulated, they can also have roles in pathologies such as cancer or skeletal dysplasias (reviewed in 1,2,3,4). Each HSPG consists of a core protein to which one or more HS chains are covalently attached via hydroxyl groups on serine residues. The HS chains are polymerized sequentially, and the process initiates with the initial attachment of xylose to the serine residue followed by 2 galactose residues and glucuronic acid to form the linkage region. Polymerization continues with the sequential addition of glucuronic acid (GlcA) and N-acetyl- glucosamine (GlcNAc) to produce repetitive disaccharide units producing chains with an average size of 20–25 kDa (2,3). Individual saccharides along the HS chains are modified via epimerases and also by specific sulfotransferases. The latter catalyze the sulfation of carbohydrate carbons at position 2, 3 or 6 around the sugar rings, eliciting exceedingly complex sulfation patterns referred to as the “sulfation code” that have major biological significance (2). In toto, there are over 25 enzymes involved in HS chain polymerization and modification (reviewed by 2, 3). Additional complexity and subtleties are produced by selective removal of some of the sulfate groups by Sulf1 and Sulf2 sulfatases, eliciting segments with low/minimal sulfation along the HS chain intercalated by high sulfation segments (5). In addition, HS chains can be selectively removed from the cell surface or the ECM by the action of heparanase, the single entity in the human genome with the ability to do so (6). The family of mammalian HSPGs includes 4 syndecans whose core proteins span the cell surface bilayer and 6 glypicans whose core proteins are bound to the cell surface via a GPI anchor (Table 1) (2,3). It includes also a number of extracellular HSPGs such as perlecan, betaglycan and collagen XVIII and serglycan which resides in the secretory granules of mast cells (7, 8; see Table 1). As indicated above, HSPGs display selective patterns of expression in different tissues and organs and at different stages of development, adding to their functional complexity but also introducing significant specificity to their biological action and function (3).
Table 1.
Heparan Sulfate Proteoglycans Identified in Mammalian Cells*
| HSPG** | Location | Reference*** |
|---|---|---|
| Agrin | Transmembrane | 57 |
| Betaglycan | Transmembrane | 58 |
| Syndecans (4) | Transmembrane | 9,51,59 |
| Neuropilin-1 | Transmembrane | 60 |
| Glypicans (6) | Membrane, GPI anchored | 61 |
| Serglycin | Intracellular | 62 |
| Collagen XVIII | Extracellular | 57,63 |
| Perlecan | Extracellular | 64 |
| Testican (2) | Extracellular | 65 |
Adapted from 3.
Numbers in parenthesis- number of known gene family members.
Published Reviews
Because of their sulfation, the HS chains bear multiple anionic charges. One of the key functions of HSPGs stemming from this unique feature is their ability to interact with numerous proteins bearing a reciprocally charged HS-binding domain (HBD). The HS-binding proteins include plasma proteins, extracellular matrix components, cell surface proteins, and members of the major growth factor and signaling protein families including Wnt, hedgehog, bone morphogenetic protein, fibroblast growth factor and vascular endothelial growth factor families (9,10,11). The protein-HS interactions are very important and serve multiple functions including: modulating protein function and distribution; limiting protein range of action on target cells; stabilizing proteins and protect them from degradation; setting up morphogen protein gradients during development and growth; and presenting specific proteins to their cognate receptors for activation of signaling (2, 3). The nature and general traits of the HBD have been studied in many proteins and some overall features have become apparent (12,13). In general the HBDs contain basic residues (Arg and/or Lys), have an average pI > 11, and contain hydropathic amino acids spacing the basic residues apparently important for accommodating GAG chains in the binding pocket, but exhibit variable length and diverse amino acid sequence (Table 2). In general, the HBDs bind HS and heparin with high affinity when measured in standard biochemical assays, but can also bind chondroitin sulfate and hyaluronic acid, usually with lower affinity.
Table 2.
Primary Sequences of HS/Heparin Binding Protein Domains
| Name* | Accession** | Position | Sequence | Reference*** |
|---|---|---|---|---|
| AAMP | AAA68889 | 14–25 | RRLRRMESESES | 66 |
| AREG | AAI46968 | 126–144 | RKKKGGKNGKNRRNRKKKN | 16 |
| Antistatin | AAB29420 | 93–120 | PNGLKRDKLGCEYCECRPKRKLIPRLS | 14 |
| AT III | BAC21176 | 64–71 | RRVWELSK | 12 |
| AT III | XP_005245255 | 105–118 | FFFAKLNCRLYRKA | 67 |
| AT III | AAA51794 | 156–177 | AKLNCRLYRKANKSSKLVSANR | 13 |
| AT III | AAB40025 | 319–332 | KPEKSLAKVEKELT | 68 |
| Apo B100 | CAA28420 | 3168–3182 | LSVKAQYKKNKHRHSI | 69 |
| Apo B100 | 1211338A | 3352–3371 | YKLEGTTRLTRKRGLKLATA | 69 |
| Apo E | 1NFN_A | 144–151 | LRKRLLRD | 70 |
| Apo E | AAB59518 | 229–236 | GERLRARM | 70 |
| Dhh | O43323 | 32–38 | RRRYARK (by homology) | |
| FGF 1 | CAA41788 | 34–54 | GLKKNGSCKRGPRTHYGQKAI | 71 |
| FGF 2 | AAK52309 | 84–101 | LKRTGQYKLGSKTGPGQK | 13 |
| FGF 2 | NP_001997 | 261–280 | KRTGQYKLGSKTGPGQKAIL | 24,22 |
| FGF 4 | NP_001998 | 181–200 | LSKNGKTKKGNRVSPTMKVT | 72 |
| FGF 7 | C46289 | 71–91 | LNQKGIPVRGKKTKKEQKTAH | 24 |
| FGF 8 | NP_001193318 | 78–98 | FTRKGRPRKGSKTRQHQREVH | 24 |
| FGF 10 | AAM46926 | 142–162 | LNGKGAPRRGQKTRRKNTSAH | 24 |
| FGF 18 | NP_003853 | 153–173 | FTKKGRPRKGPKTRENQQDVH | 24 |
| FBN | CAC86916 | 31–66 | R31-G66 | 73 |
| FBN | AAI17177 | 1847–1865 | YEKPGSPPREVVPRPRPGV | 74 |
| FBN | AAI17177 | 1887–1901 | KNNQKSEPLIGRKKT | 75 |
| Glia Nexin | NP_033281 | 82–105 | RYNVNGVGKVLKKINKAIVSKKNK | 13 |
| HGF | ACX45438 | 67–99 | A67-F99 | 76 |
| HB-EGF | NP_001936 | 93–113 | KRKKKGKGLGKKRDPCLRKYK | 77 |
| Hep Cofac 2 | NP_032249 | 181–198 | FRKLTHRLFRRNFGYTLR | 12 |
| Heparinase | NP_001159970 | 158–171 | KKFKNSTYSRSSVDC | 17 |
| IFN-γ | NP_000610 | 147–166 | AKTGKRKRSQMLFRGRRASQ | 30 |
| IGFBP-3 | CAA46087 | 242–259 | KKGFYKKKQCRPSKGRKR | 78 |
| IGFBP4 | NP_034647 | 202–216 | RNGNFHPKQCHPALDQ | 78 |
| IGFBP-5 | NP_000590 | 221–238 | RKGFYKRKQCKPSRGRKR | 79 |
| IGFBP-6 | NP_002169 | 192–209 | HRGFYRKRQCRSSQGQRR | 78 |
| IL10 | CAG46790 | 116–138 | LKTLRLRLRRCHRFLPCENKSKA (putative) | 80 |
| Ihh | Q14623 | 37–43 | RRRPPRK (by homology) | |
| Laminin | XP_006240058 | 662–681 | RYVVLPRPVCFEKGMNYTVR | 81 |
| Laminin | EAW83423 | 247–263 | RIQNLLKITNLRIKFVK | 82 |
| Laminin | EDL38338 | 3030–3051 | KQNCLSSRASFRGCVRNLRLSR | 82 |
| LPL | AAC61679 | 163–181 | RKNRCNNLGYEINKVRAKR | 83 |
| NCAM | AAH29119 | 150–167 | IWKHKGRDVILKKDVRFI | 84 |
| PDGF-A | EAW87161 | 198–215 | GRPRESGKKRKRKRLKPT | 85 |
| PRG4 | XP_009438019 | 94–107 | RSPKPPNKKKTKKV | 86 |
| Prot C Inhib | AAB60386 | 283–302 | SEKTLRKWLKMFKKRQLELY | 87 |
| Sclerostin | NP_079513 | 70–172 | F70-R170 | 31 |
| Serglycin | NP_035287 | 26–40 | YPARRARYQWVRCKP | 15 |
| Shh | NP_000184 | 32–38 | KRRHPKK | 39,40 |
| EC-SOD | AAA66000 | 223–240 | REHSERKKRRRESECKAA | 88 |
| Sulf1 | NP_001121678 | 417–735 | K417-K735 | 47 |
| TFPI | NP_006278 | 240–272 | G240-F272 | 89 |
| TGF-β1 | AAQ18642 | 14–32 | DFRKDLGWKWIHEPKGYHA | 90 |
| TSP | AAA61178 | 41–50 | RKGSGRRLVK | 91 |
| TSP | AAA61178 | 95–101 | RQMKKTR | 91 |
| VEGFA | NP_001165100 | 137–185 | A137-R185 | 26 |
| VTN | AAH05046 | 366–380 | AKKQRFRHRNRKGYR | 92 |
| vWF | CCQ25771 | 1328–1350 | YIGLKDRKRPSELRRIASQVKYA | 93 |
| XO | XP_003262893 | 781–795 | LGVPANRIVVRVKRM | 94 |
| XDH | AAA75287 | 1106–1122 | KKKNPSGSWEDWVTAAY | 13 |
Abbreviations: AAMP, angio-associated, migratory cell protein, AREG, amphiregulin, ATIII, Antithrombin III, FBN, fibronectin, HGF, hepatocyte growth factor, Hep Cof, Heparin cofactor II, LPL, Lipoprotein Lipase, TFP1, thrombospondin, VTN, vitronectin.
Genbank accession number.
Reference
Nature and primary structure of the HBDs
In pioneering work, Cardin and Weintraub set out to identify the domain(s) of proteins responsible for interaction with HS and other glycosaminoglycans and focused on four proteins: apolipoprotein B, apolipoprotein E, vitronectin, and platelet factor-4 (see 12). This comparative analysis allowed them to identify two binding motifs -XBBXBX and XBBBXXBX- in which B represents a basic residue and X represents any other residue. Analyses of additional proteins have since confirmed these findings and identified analogous motifs frequently containing clusters of basic amino acids -XBX, XBBX and XBBBX)- again separated by non-charged residues (14). The sequences first identified by Cardin and Weintraub have entered the vernacular and are referred to as the consensus Cardin-Weintraub (CW) motif. Indeed, Verrecchio and coworkers utilized consensus sequence templates to design peptides with high affinities for heparin and endothelial cell proteoglycans, and found that peptides with the highest affinity were tandem repeats of the sequence XBBBXXBX 15.
Amphiregulin (AREG) is an HS-binding growth factor that associates with and activates the epidermal growth factor (EGF) receptor tyrosine kinase. The mitogenic activity of AREG on cultured cells is blocked by addition of exogenous heparin and prior treatment of the cells with heparitinase (16). A peptide comprising residues 26–44 of fully processed AREG (Table 2), and comprising the putative HBD, blocks AREG binding to immobilized heparin, indicating that much of the HS-binding activity of AREG resides in that domain 16. Using solid phase binding assays, we have indeed found that the peptide does bind to heparin and HS with high affinity, but binds also to CS or HA albeit with lower affinity (Fig. 1). We carried out similar binding assays with human heparanase fragments. Not surprisingly, this enzyme also contains a HBD targeting it to its natural substrate. The HBD in human heparanase spans amino acids Lys158-Asp171 and can by itself block binding of full length enzyme to heparin (17). We synthesized the Lys158-Asp171 peptide and tested its binding capacities using solid phase assays with different GAGs as above. As one would expect, we did observe very high affinity binding to heparin and HS, but there was appreciable binding to CS and hyaluronic acid (HA) (not shown). These and similar experiments indicate that the HBD has a primary role in protein-HS interaction but other regions of the proteins are likely to be involved in order to enhance specificity of interaction and action.
Figure 1.
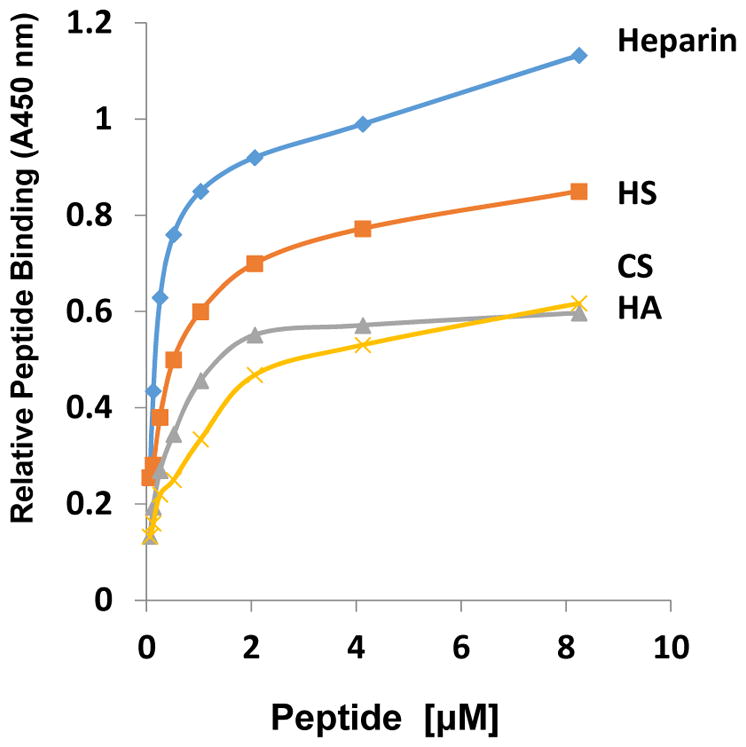
Differential binding of AREG peptide to different GAGs. The indicated type of GAG was immobilized on 96 well plates and the binding of Flag- tagged AREG (DYKDDDDKGG RKKKGGKNGKNRRNRKKKN; AREG sequence underlined) peptide was determined using an anti-Flag Ab and secondary antibody-HPR conjugate. Results included are from a representative experiment and were highly reproducible.
Some of the most detailed analyses of protein interactions with HS/heparin have been carried out with members of the FGF growth factor family. The family comprises 22 proteins with essential functions in cell growth, morphogenesis, tissue repair and angiogenesis (18). The FGFs interact with cognate cell surface tyrosine kinase receptors FGFR1, FGFR2, FGFR3 and FGFR4 and have an obligatory requirement for HS to exert their biological activity (Reviewed in18,19, 20). The most efficient signaling structure for FGF2 was recently shown to consist of a symmetrical complex [FGF2]2-[HS block]2-[FGFR]2 (21). Interestingly, a comparison of the HBDs in FGF-1, FGF-2 and FGF-7 with those in other FGFs shows that the HBDs are quite diverse in their primary sequences (22). Heparin/HS binding was found to be mediated in part by a “glycine box” with the consensus sequence XBXGXXBBG in which the location of the basic residues varies from FGF to FGF (23,24). Using a library of HS octasaccharides with defined 2-O and 6-O sulfation patterns, Ashikari-Hada et al. were able to define and classify the interaction of FGFs and other growth factors with specific patterns along the HS chains. It was determined that growth factor-HS interactions can be categorized into five groups: Group 1 includes FGFs with affinity for 2-O-sulfated octasaccharides (ex., FGF-2); Group 2 factors have affinity for 6-O-octasaccharides (ex., FGF-10); Group 3 factors have affinity for both 2-O-sulfated and 6-O-sulfated octasaccharides but prefer the 2-O-sulfated ones (FGF-18 and HGF); Group 4 requires both 2-O-sulfate and 6-O-sulfate within the octasaccharides for binding (FGF-4, FGF-7); and Group 5 includes FGFs and other proteins with weak binding to any octasaccharide tested (FGF-8, BMP6 and VEGF) (24). These results indicate that FGF binding is at least partially mediated by the glycine box sequence and differences in the primary sequence of this domain and sulfation patterns can both influence protein-GAG recognition in different FGFs and other proteins.
Secondary structure analyses of HBDs
In addition to amino acid sequence analysis, nuclear magnetic resonance (NMR) has been utilized to gain further structural insights into protein-HS interactions using approaches such as short and defined peptides. These studies have also yielded information about amino acid residues involved in binding that are distant from the primary HBD as the following examples demonstrate. Vascular endothelial growth factor (VEGF) is an important regulator of angiogenesis and its association and action on its cognate high affinity receptors involve HS and HSPGs (25). The HBD of VEGF comprises residues 116–165 at the C-terminus whose solution structure has been solved by NMR. The structure consists of a disordered N-terminal region followed by 2 disulfide-bonded subdomains containing two short stranded antiparallel β sheets followed by an α-helical region at its C-terminus. Most of the basic residues mediating HS/heparin binding turned out to reside on the N and C-terminal domains and are thus brought together along one face (26). Interestingly, cell surface-associated Syndecan 1 is frequently up-regulated in multiple myeloma27. The HS chains on Syndecan 1 are believed to play an important role in VEGF signaling and are actually implicated in the pathogenesis of multiple myeloma (28).
Mammalian heparanase (an endo-β-glucuronidase) is the only enzyme encoded in the genome that specifically cleaves HS at intra-chain sites. Its ability to recognize -and act upon HS and heparin- appears to reside in a specific HBD at position Lys158–Asp171 (31). NMR analysis of Lys158–Asp171 peptide mixed with a synthetic oligosaccharide mimicking heparin showed chemical shifts of a sub-domain from Lys158 to Asn162, suggesting that this sub-domain is critical for HS/heparin binding.
Similar experimental approaches were used to study IFNγ and its interactions with HS. This factor is an important immunomodulator and as such, needs to be strictly controlled within the extracellular milieu (29). Its interaction with HS/heparin involves a HBD residing at the C-terminus and spanning amino acids Arg 124 to Gln 143. NMR has revealed that two subdomains within this peptide, D1 (Lys 125- Arg 131) and D2 (Arg 137- Arg 140) exhibit the most pronounced chemical shifts following HS/heparin binding (30).
Sclerostin is a secreted cysteine-knot HS-binding protein that negatively regulates Wnt signaling and has critical roles in the regulation of bone formation and homeostasis (31). The interaction of Sclerostin with HS/heparin involves 3 loops within a structured HBD core region spanning residues 52-147: loop 1 spans Arg-57 to Val-80; loop 2 spans Gly-86 to Arg-109; and loop 3 spans Ile-111 to Ser-140. The loops are stabilized and cross-linked by a network of disulfide bonds, and are arranged together to form a linear and positively charged area covering one entire side of the protein. Following HS/heparin binding, chemical shifts were observed in amide nitrogens within loops 2 and 3 (31).
The above examples highlight the fact that domains and subdomains involved in HS/heparin binding in a given protein are structured to produce a congruent and often linear surface displaying all the negatively charged sugars, likely augmenting binding effectiveness, specificity and strength.
More complex interactions
Recent studies on certain signaling factors have shown that interactions with HS are more complex both structurally and functionally than previously realized and can actually involve concurrent interactions of other protein domains with other GAG types. A prominent example of this phenomenon can be found in members of the hedgehog family that include Sonic (Shh), Indian (Ihh) and Desert (Dhh) hedgehogs (11, 32,33, 34). The Hh proteins are very potent signaling factors and have critical functions in embryonic development as well as tissue function and homeostasis and skeletal and non-skeletal pathologies including cancer (4,35, 36, 37). Interactions with HS and HSPGs have been shown to regulate the distribution and action of Hh proteins on target cells and tissues and also their ability to form morphogen gradients within/amongst tissues during embryonic development (38). The Shh signaling pathway in particular is frequently up-regulated in pancreatic cancer and interestingly, the HS/heparin binding activity of Shh is required for its action on the proliferation and metastatic spread of pancreatic ductal adenocarcinoma cells (PDAC) (see 39).
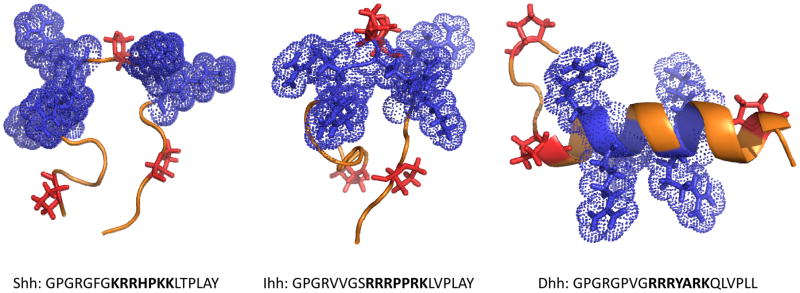
The HBD resides within the amino-terminal and biologically active portion of Hh proteins and contains a CW motifs with the consensus BBBXXBB (Table 3; Reviewed in 11, 35, 40). Site- specific mutagenesis has shown that the Lys residues at position 37 and 38 within the CW are important for Shh function, including its ability to induce osteoblast differentiation in C3H/10T1/2 cells (39). Somewhat unexpectedly, these studies have also shown that a Lys residue at position 178 and thus far away from the CW is equally important for Shh function. A recent important crystallographic study on murine Shh combining site-directed mutagenesis, biological analysis and protein-GAG and protein-protein interactions has confirmed that the above 3 residues are involved in HS/heparin binding and function, but has also uncovered new aspects of Hh biology (40). The study has revealed that additional basic amino acids at position K88, R124, R154 and R156 (corresponding to K87, R123, R153, R155 in human SHH) influence Shh interactions with HS, and this region was termed the “Hh core GAG-binding site” (40). The CW at the N-terminus and the core site were shown to both be involved in Shh interactions with HS, but also mediated interactions of Shh with CS. Because the Hh proteins oligomerize to diffuse, to form morphogen gradients and to interact with cells, the authors examined these aspects of Hh biology and found that HS favors oligomerization much more than CS. Equally interesting was the finding that the GAG binding site partially overlaps with the domain of Shh interactions with other protein partners including cell surface receptor Patched and interference hedgehog proteins, suggesting unique mechanisms of Hh signaling modulation by HS, CS and other interacting proteins.
Table 3.
Sequence of CW motif and flanking amino acids in Sonic, Indian and Desert Hedgehogs.
| Hh* | Organism | Sequence**, *** | Accession**** |
|---|---|---|---|
| SHH | Human | GPGRGFGKRRHPKKLTPLAY | Q15465 |
| SHH | Mouse | GPGRGFGKRRHPKKLTPLAY | NP_033196 |
| SHH | Sheep | GPGRGFGKRRNPKKLTPLAY | XP_004008418 |
| SHH | Aardvark | GPGRGFGKRRHPKKLTPLAY | XP_007951664 |
| SHH | Python | GPGRGFGKRRHPKKLTPLAY | XP_007433256 |
| SHH | Chicken | GPGRGIGKRRHPKKLTPLAY | AAA72428 |
| IHH | Human | GPGRVVGSRRRPPRKLVPLAY | Q14623 |
| IHH | Mouse | GPGRVVGSRRRPPRKLVPLAY | AAH46984 |
| IHH | Aardvark | GPGRVVGSRRRPPRKLVPLAY | XP_007957594 |
| IHH | Python | GPGRVVGSRRRPPRKLIPLAY | XP_007419884 |
| IHH | Chicken | GPGRVVGSRRRPPRKLIPLAY | NP_990288 |
| DHH | Human | GPGRGPVGRRRYARKQLVPLL | NP_066382 |
| DHH | Mouse | GPGRGPVGRRRYVRKQLVPLL | EDL04156 |
| DHH | Aardvark | GPGRGPVGRRRYVRKQLVPLL | XP_007936046 |
| DHH | Dolphin | GPGRGPVGRRRYVRKQLVPLL | XP_004326854 |
| DHH | Python | GPGRGPVGRKQSSRKSLAPLQ | XP_007437346 |
Hedgehog protein
Sequence of CW motif (Bold type) and flanking amino acids; sequence variations underlined.
CW consensus sequence BBBXXBB.
GenBank accession number.
We have uncovered additional interesting aspects of Hh biology by examining further and comparing the sequences of the CW motifs of Shh, Ihh and Dhh and surrounding sequences. As summarized in Table 3, uniformly spaced Gly residues are present on the N-terminal side of the CW in all three Hh proteins. It is well established that glycines provide conformational flexibility to polypeptide chains (41) and likely increase plasticity that may enable the CWs to adjust and optimize their interactions with GAG chains. Intriguingly, a highly conserved and invariant proline is present in the 5th position of Shh’s CW domain, while two prolines occupy positions 4 and 5 in Ihh’s CW and none are present in Dhh’s CW. Proline residues induce bends or kinks in polypeptide chains (42) and presumably, are expected to have a major impact on overall protein conformation. Thus, we used a secondary structure prediction program in the PSIPRED Protein Sequence Analysis Workbench (Link: bioinf.cs.ucl.ac.uk/psipred) and scanned the PDB using the I-TASSER server (43,44,45) to predict and analyze the CW conformation in Shh, Ihh and Dhh. The data obtained point to substantial conformational differences (Figure 2). Specifically, the CW of Shh is predicted to have a random coil-like conformation with a central kink generated by its single proline, while the CW of Ihh would have a similar configuration but with a more pronounced and complex kink generated by its 2 prolines. Because Dhh’s CW lacks prolines, its configuration is predicted to be a continuous and uniform helix as one would expect (Figure 2). Though these predictions need to be verified by more stringent methods, they raise the interesting possibility that the CWs of the 3 Hh proteins may be intrinsically different, could differentially affect the interactions of each Hh member with HS chains, different sites with distinct sulfation patterns and/or different HSPGs, and could in turn influence protein distribution, metabolism, overall conformation and perhaps even function (see 34). In view of the identification of the “Hh core GAG-binding site” (40), we also wonder whether the different CWs could have repercussions on that sites function as well in each Hh member.
Figure 2.
Structure of CW motifs in Shh, Ihh and Dhh. Secondary structure predications were carried out using the I-TASSER server (45) and resulting structures were visualized using PyMol. The peptides are oriented with the N-terminus on the left; side chains of basic residues within the CW motifs are in blue while the side groups of proline are in red. Note that CWs of Shh and Ihh have a largely random coil configuration with central kinks due to the proline residue(s) while the CW of Dhh exhibits a more helical configuration. The sequences examined are presented below each structure; the CW motif is in bold type and flanking residues are in plain typeset.
Related and very interesting insights have been gained in a recent study that analyzed the secreted sulfatase Sulf1. As pointed out above, this enzyme is involved in remodeling the 6O-sulfation state of cell surface HSPGs and must thus possess the ability to recognize and act selectively upon its natural substrate (5,46). However, full-length Sulf 1 was found to bind not only to HS/heparin, but also to CS and dermatan sulfate (DS) in affinity chromatography assays. Further structural and biochemical analyses using different Sulf1 deletion mutants showed that the interactions of Sulf1 with HS encompass a very large protein region called the hydrophilic domain (HD), spanning 319 amino acids Lys417-Lys735 (47). This large region includes a consensus HBD and when assayed in solution and solid phase binding assays, was found to bind HS with high affinity and specificity but not to CS, DS or 6-O-desulfated HS (48). HS/heparin binding was found to be mediated by 2 sites located in the inner and C-terminal regions of HD (48). The authors concluded that the substrate specificity of Sulf1 is mediated by the HD, involves at least two separate HS-binding sites, and clearly depends on presence of 6-O sulfation, that is its own substrate.
HS- and HSPG-associated pathologies
Given the important roles that HS and HSPGs play in normal cell and tissue physiology, it is not surprising that genetic and metabolic defects in these macromolecules are linked to a number of pathological changes and disorders, in addition to those pointed out above. For instance, loss-of-function mutations in Glypican-3 cause the human X-linked disorder Simpson-Golabi-Behmel syndrome that is characterized by embryonic and postnatal overgrowth, cardiac malformations and predisposition to certain tumors (49,50). Abnormally high shedding of syndecan-2 from the cell surface has been linked to progression and malignancy of various human tumors (51,52). For sometime now, we have been studying the pediatric skeletal disorder Hereditary Multiple Exostoses (HME). This disease is characterized by benign cartilaginous outgrowths –termed exostoses- that form next to, but never within, the growth plates in developing and growing skeletal elements including long bones, ribs, vertebrae and pelvis (4,53). The majority of HME cases are linked to heterozygous loss-of-function mutations in EXT1 or EXT2 and as a result, the patients have a systemic deficiency in HS levels amounting to about half of that seen in healthy people (54). It has remained unclear and controversial as to whether and how the HS deficiency causes exostosis formation, why the exostoses form near but not inside the growth plates, whether the exostoses are formed by aberrant function of growth plate chondrocytes themselves or involve perichondrial progenitors, and what growth factors trigger and sustain exostosis outgrowth (4,53). In studies we and others carried out previously, we found that mutant mice lacking one allele of Ext1 or Ext2 did not reproduce the human HME skeletal phenotype, while double heterozygous Ext1+/−; Ext2+/− mice or conditionally ablated Ext1−/− mice did (55, 56). The data strongly indicated that a partial decrease in HS levels such as that elicited by a simple heterozygous EXT mutation is insufficient for exostosis formation and HME progression, but a deeper decrease is required. The mechanisms for the latter are still unclear, but several possibilities have been suggested including loss of heterozygosity or a second hit to an unrelated gene (4). In order to identify growth factors involved in exostosis formation, we monitored BMP signaling -a key chondrogenic pathway- in Ext1 conditionally-ablated mice (4). Specifically, we ablated both alleles of Ext1 along the perichondrium of growing long bones in mice and monitored BMP signaling and exostosis formation over time. We found that BMP signaling levels revealed by phosphorylated SMAD immunohistochemistry were very low in perichondrium in control mice, but were significantly higher in the mutant mice. This was followed by initiation and growth of exostoses at later time points, indicating that ectopic BMP signaling precedes exostosis formation and may be a major driver in their outgrowth (4). In good agreement, studies by others have shown that the distribution and signaling of Ihh also become abnormally wide and off-target within the growth plate of Ext1 hypomorphic mice that produce about 10–20% of normal HS levels, leading to abnormal growth plate organization, chondrocyte function and skeletal growth retardation (38). Together, the above studies have indicated that a major role of HS and HSPGs in the growth plate is in fact to regulate growth factor distribution and restrict and delimit signaling action of growth factors on target cells. Thus, a wider and uncontrolled distribution of growth factors, resulting from severe decrease of Ext gene expression and HS levels, would derange normal growth plate and/or perichondrial function and cause pathologies including exostosis formation.
Discussion and Prospective
The ability of many signaling proteins, growth factors and other proteins to interact with HS and HSPGs has long been known, but the present review and analysis of current literature show that while there is been considerable progress, significant gaps in our understanding remain. There is little doubt that because of its primary structure and concentration of positively-charged amino acid residues, the CW motif plays a very important role in establishing and favoring HS-protein interactions and may even initiate them. However, the specific sequence, organization and predicted configuration of the HBDs vary considerably from protein to protein (see Table 2), and the significance and consequences of this variability remain incompletely understood. The diversity of these domains does suggest however, that they may have evolved to have multiple and subtle functions, to selectively interact with defined regions present in HS and possibly other GAGs, and to contribute to define and regulate the roles and activities of the interacting proteins. Since different HSPGs are expressed in different tissues and at different developmental stages and because the sulfation patterns along their HS chains differ significantly as well (2,3,4), the diversity of HBDs could thus introduce elements of specificity and selectivity of HS-protein interactions and permit differential retention, accumulation and/or activity of given signaling and growth factor proteins on cells. This feature could be particularly important to insure that distinct factors, present within the same tissue and organ, would be able to coordinately interact with their specific targets. We provide above clear example of these paradigms regarding Ihh, a powerful signaling protein that becomes abnormally and broadly distributed and active at ectopic sites when HS and/or HSPGs are deficient or deficiently expressed (4,38).
As we describe above, it has become clear that in addition to CW motifs, other protein domains and regions are involved in regulating protein-GAG interactions and in particular those with HS and CS. The recent models proposed for Sulf1 and Shh are particularly attractive and revealing. In the case of Sulf1, portions of the protein at the N- and C-termini would provide surfaces for interactions with CS-rich proteoglycans within the matrix, serving as a support and guiding system to allow the more centrally-located catalytic hydrophilic domain (HD) to act on the 6-O-sulfate substrate groups on neighboring HS chains and carry out sequential desulfation (48). In the case of Shh, the CW motif at the N-terminus would have a major role in binding to HS and signaling function, but the centrally-located Hh core GAG-binding site would be equally important for interactions with HS and CS and overall hedgehog protein distribution, structural features and function (11,40). As summarized above, interactions of Shh with HS would favor oligomerization and thus function and signaling, whereas interactions with CS would provide a lower degree of oligomerization, possibly promoting storage of Shh for subsequent function. Our own analysis of the CW in Shh indicates that its configuration is quite different from the CW of Ihh and Dhh, suggesting that proteins as powerful as hedgehogs may have acquired additional structural features during evolution, introducing additional elements of regulation. It is thus clear that the interactions of proteins with GAG chains and their respective proteoglycans introduce several levels of regulation and as pointed out in recent reviews, can in fact fine-tune cell and tissue physiology and developmental processes and create pathologies when abnormal (2,3,4). All the above findings, insights and speculations are attractive, intriguing and interesting, but much remains to be learned about their basis, true relevance and roles and implications. Advances and new insights in these complex research fields hopefully in the near future will have broad repercussions and significance for basic biology and translational medicine as well as for the conception and creation of HS-based therapeutics.
Acknowledgments
We thank Dr. Sriram Krishnaswamy in the Division of Thrombosis and Hemostasis here at the Children’s Hospital of Philadelphia for helpful suggestions and discussion and peptide modeling. The original studies described here were supported by NIAMS RO1 grant AR061758.
Abbreviations
- AREG
Amphiregulin
- CS
chondroitin sulfate
- CW
Cardin-Weintraub motif
- ECM
extracellular matrix
- FGF
fibroblast growth factor
- G-box
glycine box
- GAG
glycosaminoglycan
- GlcA
glucuronic acid
- GlcNAc
N-Acetyl-D-glucosamine
- HA
hyaluronic acid
- HBD
heparin/heparan binding domain
- HME
Hereditary Multiple Exostoses
- HS
heparan sulfate
- HSPGs
heparan sulfate proteoglycans
References
- 1.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–7. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 2.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couchman JR, Pataki Ca. An introduction to proteoglycans and their localization. J Histochem Cytochem. 2012;60:885–97. doi: 10.1369/0022155412464638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huegel J, et al. Heparan sulfate in skeletal development, growth, and pathology: the case of hereditary multiple exostoses. Dev Dyn. 2013;242:1021–32. doi: 10.1002/dvdy.24010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond E, Khurana A, Shridhar V, Dredge K. The Role of Heparanase and Sulfatases in the Modification of Heparan Sulfate Proteoglycans within the Tumor Microenvironment and Opportunities for Novel Cancer Therapeutics. Front Oncol. 2014;4:195. doi: 10.3389/fonc.2014.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vlodavsky I, Iozzo RV, Sanderson RD. Heparanase: multiple functions in inflammation, diabetes and atherosclerosis. Matrix Biol. 2013;32:220–2. doi: 10.1016/j.matbio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Rönnberg E, Pejler G. Serglycin: the master of the mast cell. Methods Mol Biol. 2012;836:201–17. doi: 10.1007/978-1-61779-498-8_14. [DOI] [PubMed] [Google Scholar]
- 8.Wernersson S, Pejler G. Mast cell secretory granules: armed for battle. Nat Rev Immunol. 2014;14:478–94. doi: 10.1038/nri3690. [DOI] [PubMed] [Google Scholar]
- 9.Couchman JR, Gopal S, Lim HC, Nørgaard S, Multhaupt HAB. Syndecans: from peripheral coreceptors to mainstream regulators of cell behaviour. Int J Exp Pathol. 2014 doi: 10.1111/iep.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Couchman JR. Transmembrane signaling proteoglycans. Annu Rev Cell Dev Biol. 2010;26:89–114. doi: 10.1146/annurev-cellbio-100109-104126. [DOI] [PubMed] [Google Scholar]
- 11.Ryan KE, Chiang C. Hedgehog secretion and signal transduction in vertebrates. J Biol Chem. 2012;287:17905–13. doi: 10.1074/jbc.R112.356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cardin aD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arterioscler Thromb Vasc Biol. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 13.Hileman RE, Fromm JR, Weiler JM, Linhardt RJ. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays. 1998;20:156–67. doi: 10.1002/(SICI)1521-1878(199802)20:2<156::AID-BIES8>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 14.Fromm JR, Hileman RE, Caldwell EE, Weiler JM, Linhardt RJ. Pattern and spacing of basic amino acids in heparin binding sites. Arch Biochem Biophys. 1997;343:92–100. doi: 10.1006/abbi.1997.0147. [DOI] [PubMed] [Google Scholar]
- 15.Verrecchio A, et al. Design of peptides with high affinities for heparin and endothelial cell proteoglycans. J Biol Chem. 2000;275:7701–7. doi: 10.1074/jbc.275.11.7701. [DOI] [PubMed] [Google Scholar]
- 16.Johnson GR, Wong L. Heparan sulfate is essential to amphiregulin-induced mitogenic signaling by the epidermal growth factor receptor. J Biol Chem. 1994;269:27149–54. [PubMed] [Google Scholar]
- 17.Levy-Adam F, et al. Identification and characterization of heparin/heparan sulfate binding domains of the endoglycosidase heparanase. J Biol Chem. 2005;280:20457–66. doi: 10.1074/jbc.M414546200. [DOI] [PubMed] [Google Scholar]
- 18.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–53. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10:116–29. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 20.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8:235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sterner E, et al. Fibroblast growth factor-based signaling through synthetic heparan sulfate blocks copolymers studied using high cell density three-dimensional cell printing. J Biol Chem. 2014;289:9754–65. doi: 10.1074/jbc.M113.546937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye S, et al. Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs. Biochemistry. 2001;40:14429–39. doi: 10.1021/bi011000u. [DOI] [PubMed] [Google Scholar]
- 23.Luo Y, et al. The glycine box: a determinant of specificity for fibroblast growth factor. Biochemistry. 1998;37:16506–15. doi: 10.1021/bi9816599. [DOI] [PubMed] [Google Scholar]
- 24.Ashikari-Hada S, et al. Characterization of growth factor-binding structures in heparin/heparan sulfate using an octasaccharide library. J Biol Chem. 2004;279:12346–54. doi: 10.1074/jbc.M313523200. [DOI] [PubMed] [Google Scholar]
- 25.Chiodelli P, et al. Heparan sulfate proteoglycans mediate the angiogenic activity of the vascular endothelial growth factor receptor-2 agonist gremlin. Arterioscler Thromb Vasc Biol. 2011;31:e116–27. doi: 10.1161/ATVBAHA.111.235184. [DOI] [PubMed] [Google Scholar]
- 26.Fairbrother WJ, Champe MA, Christinger HW, Keyt BA, Starovasnik MA. Solution structure of the heparin-binding domain of vascular endothelial growth factor. Structure. 1998;6:637–48. doi: 10.1016/s0969-2126(98)00065-3. [DOI] [PubMed] [Google Scholar]
- 27.Seidel C, et al. Serum syndecan-1: a new independent prognostic marker in multiple myeloma. Blood. 2000;95:388–92. [PubMed] [Google Scholar]
- 28.Khotskaya YB, et al. Syndecan-1 is required for robust growth, vascularization, and metastasis of myeloma tumors in vivo. J Biol Chem. 2009;284:26085–95. doi: 10.1074/jbc.M109.018473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larkin J, Ahmed CM, Wilson TD, Johnson HM. Regulation of interferon gamma signaling by suppressors of cytokine signaling and regulatory T cells. Front Immunol. 2013;4:469. doi: 10.3389/fimmu.2013.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saesen E, et al. Insights into the mechanism by which interferon-γ basic amino acid clusters mediate protein binding to heparan sulfate. J Am Chem Soc. 2013;135:9384–90. doi: 10.1021/ja4000867. [DOI] [PubMed] [Google Scholar]
- 31.Veverka V, et al. Characterization of the structural features and interactions of sclerostin: molecular insight into a key regulator of Wnt-mediated bone formation. J Biol Chem. 2009;284:10890–900. doi: 10.1074/jbc.M807994200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bürglin TR. The Hedgehog protein family. Genome Biol. 2008;9:241. doi: 10.1186/gb-2008-9-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan JA, et al. Proteoglycan interactions with Sonic Hedgehog specify mitogenic responses. Nat Neurosci. 2009;12:409–17. doi: 10.1038/nn.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pathi S, et al. Comparative biological responses to human Sonic, Indian, and Desert hedgehog. Mech Dev. 2001;106:107–17. doi: 10.1016/s0925-4773(01)00427-0. [DOI] [PubMed] [Google Scholar]
- 35.Beachy PA, Hymowitz SG, Lazarus RA, Leahy DJ, Siebold C. Interactions between Hedgehog proteins and their binding partners come into view. Genes Dev. 2010;24:2001–12. doi: 10.1101/gad.1951710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jochmann K, Bachvarova V, Vortkamp A. Heparan sulfate as a regulator of endochondral ossification and osteochondroma development. Matrix Biol. 2014;34:55–63. doi: 10.1016/j.matbio.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Koyama E, et al. Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis. Development. 2007;134:2159–69. doi: 10.1242/dev.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev Cell. 2004;6:801–13. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Chang SC, Mulloy B, Magee AI, Couchman JR. Two distinct sites in sonic Hedgehog combine for heparan sulfate interactions and cell signaling functions. J Biol Chem. 2011;286:44391–402. doi: 10.1074/jbc.M111.285361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whalen DM, Malinauskas T, Gilbert RJC, Siebold C. Structural insights into proteoglycan-shaped Hedgehog signaling. Proc Natl Acad Sci U S A. 2013;110:16420–5. doi: 10.1073/pnas.1310097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okoniewska M, Tanaka T, Yada RY. The pepsin residue glycine-76 contributes to active-site loop flexibility and participates in catalysis. Biochem J. 2000;349:169–77. doi: 10.1042/0264-6021:3490169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morgan AA, Rubenstein E. Proline: the distribution, frequency, positioning, and common functional roles of proline and polyproline sequences in the human proteome. PLoS One. 2013;8:e53785. doi: 10.1371/journal.pone.0053785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang J, et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods. 2014;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayano S, et al. Roles of heparan sulfate sulfation in dentinogenesis. J Biol Chem. 2012;287:12217–29. doi: 10.1074/jbc.M111.332924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frese MA, Milz F, Dick M, Lamanna WC, Dierks T. Characterization of the human sulfatase Sulf1 and its high affinity heparin/heparan sulfate interaction domain. J Biol Chem. 2009;284:28033–44. doi: 10.1074/jbc.M109.035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milz F, et al. Cooperation of binding sites at the hydrophilic domain of cell-surface sulfatase Sulf1 allows for dynamic interaction of the enzyme with its substrate heparan sulfate. Biochim Biophys Acta. 2013;1830:5287–98. doi: 10.1016/j.bbagen.2013.07.014. [DOI] [PubMed] [Google Scholar]
- 49.Ng A, et al. Loss of glypican-3 function causes growth factor-dependent defects in cardiac and coronary vascular development. Dev Biol. 2009;335:208–15. doi: 10.1016/j.ydbio.2009.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tenorio J, et al. Simpson-golabi-behmel syndrome types I and II. Orphanet J Rare Dis. 2014;9:138. doi: 10.1186/s13023-014-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fears CY, Woods A. The role of syndecans in disease and wound healing. Matrix Biol. 2006;25:443–56. doi: 10.1016/j.matbio.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 52.Iozzo RV, Sanderson RD. Proteoglycans in cancer biology, tumour microenvironment and angiogenesis. J Cell Mol Med. 2011;15:1013–31. doi: 10.1111/j.1582-4934.2010.01236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jones KB, Pacifici M, Hilton MJ. Multiple hereditary exostoses (MHE): elucidating the pathogenesis of a rare skeletal disorder through interdisciplinary research. Connect Tissue Res. 2014;55:80–8. doi: 10.3109/03008207.2013.867957. [DOI] [PubMed] [Google Scholar]
- 54.Anower-E-Khuda MF, et al. Glycosaminoglycans in the blood of hereditary multiple exostoses patients: Half reduction of heparan sulfate to chondroitin sulfate ratio and the possible diagnostic application. Glycobiology. 2013;23:865–76. doi: 10.1093/glycob/cwt024. [DOI] [PubMed] [Google Scholar]
- 55.Sgariglia F, et al. Epiphyseal abnormalities, trabecular bone loss and articular chondrocyte hypertrophy develop in the long bones of postnatal Ext1-deficient mice. Bone. 2013;57:220–31. doi: 10.1016/j.bone.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zak BM, et al. Compound heterozygous loss of Ext1 and Ext2 is sufficient for formation of multiple exostoses in mouse ribs and long bones. Bone. 2011;48:979–87. doi: 10.1016/j.bone.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uechi G, Sun Z, Schreiber EM, Halfter W, Balasubramani M. Proteomic View of Basement Membranes from Human Retinal Blood Vessels, Inner Limiting Membranes, and Lens Capsules. J Proteome Res. 2014;13:3693–3705. doi: 10.1021/pr5002065. [DOI] [PubMed] [Google Scholar]
- 58.Bilandzic M, Stenvers KL. Betaglycan: a multifunctional accessory. Mol Cell Endocrinol. 2011;339:180–9. doi: 10.1016/j.mce.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 59.Choi Y, Chung H, Jung H, Couchman JR, Oh ES. Syndecans as cell surface receptors: Unique structure equates with functional diversity. Matrix Biol. 2011;30:93–9. doi: 10.1016/j.matbio.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 60.Nasarre P, Gemmill RM, Drabkin HA. The emerging role of class-3 semaphorins and their neuropilin receptors in oncology. Onco Targets Ther. 2014;7:1663–87. doi: 10.2147/OTT.S37744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fransson L-åke. Glypicans. Int J Biochem Cell Biol. 2003;35:125–129. doi: 10.1016/s1357-2725(02)00095-x. [DOI] [PubMed] [Google Scholar]
- 62.Kolset SO, Pejler G. Serglycin: a structural and functional chameleon with wide impact on immune cells. J Immunol. 2011;187:4927–33. doi: 10.4049/jimmunol.1100806. [DOI] [PubMed] [Google Scholar]
- 63.Marneros AG, Olsen BR. Physiological role of collagen XVIII and endostatin. FASEB J. 2005;19:716–28. doi: 10.1096/fj.04-2134rev. [DOI] [PubMed] [Google Scholar]
- 64.Farach-Carson MC, Warren CR, Harrington DA, Carson DD. Border patrol: insights into the unique role of perlecan/heparan sulfate proteoglycan 2 at cell and tissue borders. Matrix Biol. 2014;34:64–79. doi: 10.1016/j.matbio.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgell CJS, BaSalamah MA, Marr HS. Testican-1: a differentially expressed proteoglycan with protease inhibiting activities. Int Rev Cytol. 2004;236:101–22. doi: 10.1016/S0074-7696(04)36003-1. [DOI] [PubMed] [Google Scholar]
- 66.Beckner ME, et al. Identification of a new immunoglobulin superfamily protein expressed in blood vessels with a heparin-binding consensus sequence. Cancer Res. 1995;55:2140–9. [PubMed] [Google Scholar]
- 67.Tyler-Cross R, Sobel M, Marques D, Harris RB. Heparin binding domain peptides of antithrombin III: analysis by isothermal titration calorimetry and circular dichroism spectroscopy. Protein Sci. 1994;3:620–7. doi: 10.1002/pro.5560030410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Villanueva GB. Predictions of the secondary structure of antithrombin III and the location of the heparin-binding site. J Biol Chem. 1984;259:2531–6. [PubMed] [Google Scholar]
- 69.Weisgraber KH, Rall SC. Human apolipoprotein B-100 heparin-binding sites. J Biol Chem. 1987;262:11097–103. [PubMed] [Google Scholar]
- 70.Cardin AD, et al. Binding of a high reactive heparin to human apolipoprotein E: identification of two heparin-binding domains. Biochem Biophys Res Commun. 1986;134:783–9. doi: 10.1016/s0006-291x(86)80489-2. [DOI] [PubMed] [Google Scholar]
- 71.Wong P, et al. Analysis of putative heparin-binding domains of fibroblast growth factor-1. Using site-directed mutagenesis and peptide analogues. J Biol Chem. 1995;270:25805–11. doi: 10.1074/jbc.270.43.25805. [DOI] [PubMed] [Google Scholar]
- 72.Bellosta P, et al. Identification of receptor and heparin binding sites in fibroblast growth factor 4 by structure-based mutagenesis. Mol Cell Biol. 2001;21:5946–57. doi: 10.1128/MCB.21.17.5946-5957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barkalow FJ, Schwarzbauer JE. Localization of the major heparin-binding site in fibronectin. J Biol Chem. 1991;266:7812–8. [PubMed] [Google Scholar]
- 74.McCarthy JB, et al. RGD-independent cell adhesion to the carboxy-terminal heparin-binding fragment of fibronectin involves heparin-dependent and -independent activities. J Cell Biol. 1990;110:777–87. doi: 10.1083/jcb.110.3.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haugen PK, McCarthy JB, Skubitz AP, Furcht LT, Letourneau PC. Recognition of the A chain carboxy-terminal heparin binding region of fibronectin involves multiple sites: two contiguous sequences act independently to promote neural cell adhesion. J Cell Biol. 1990;111:2733–45. doi: 10.1083/jcb.111.6.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou H, et al. The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site. Structure. 1998;6:109–16. doi: 10.1016/s0969-2126(98)00012-4. [DOI] [PubMed] [Google Scholar]
- 77.Thompson SA, et al. Characterization of sequences within heparin-binding EGF-like growth factor that mediate interaction with heparin. J Biol Chem. 1994;269:2541–9. [PubMed] [Google Scholar]
- 78.Fowlkes JL, Thrailkill KM, George-Nascimento C, Rosenberg CK, Serra DM. Heparin-binding, highly basic regions within the thyroglobulin type-1 repeat of insulin-like growth factor (IGF)-binding proteins (IGFBPs) -3, -5, and -6 inhibit IGFBP-4 degradation. Endocrinology. 1997;138:2280–5. doi: 10.1210/endo.138.6.5182. [DOI] [PubMed] [Google Scholar]
- 79.Arai T, Parker A, Busby W, Clemmons DR. Heparin, heparan sulfate, and dermatan sulfate regulate formation of the insulin-like growth factor-I and insulin-like growth factor-binding protein complexes. J Biol Chem. 1994;269:20388–93. [PubMed] [Google Scholar]
- 80.Salek-Ardakani S, Arrand JR, Shaw D, Mackett M. Heparin and heparan sulfate bind interleukin-10 and modulate its activity. Blood. 2000;96:1879–88. [PubMed] [Google Scholar]
- 81.Charonis AS, et al. A novel synthetic peptide from the B1 chain of laminin with heparin-binding and cell adhesion-promoting activities. J Cell Biol. 1988;107:1253–60. doi: 10.1083/jcb.107.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kouzi-Koliakos K, Koliakos GG, Tsilibary EC, Furcht LT, Charonis AS. Mapping of three major heparin-binding sites on laminin and identification of a novel heparin-binding site on the B1 chain. J Biol Chem. 1989;264:17971–8. [PubMed] [Google Scholar]
- 83.Hata A, et al. Binding of lipoprotein lipase to heparin. Identification of five critical residues in two distinct segments of the amino-terminal domain. J Biol Chem. 1993;268:8447–57. [PubMed] [Google Scholar]
- 84.Kallapur SG, Akeson RA. The neural cell adhesion molecule (NCAM) heparin binding domain binds to cell surface heparan sulfate proteoglycans. J Neurosci Res. 1992;33:538–48. doi: 10.1002/jnr.490330406. [DOI] [PubMed] [Google Scholar]
- 85.Khachigian LM, Owensby DA, Chesterman CN. A tyrosinated peptide representing the alternatively spliced exon of the platelet-derived growth factor A-chain binds specifically to cultured cells and interferes with binding of several growth factors. J Biol Chem. 1992;267:1660–6. [PubMed] [Google Scholar]
- 86.Jin C, et al. Human synovial lubricin expresses sialyl Lewis x determinant and has L-selectin ligand activity. J Biol Chem. 2012;287:35922–33. doi: 10.1074/jbc.M112.363119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shirk RA, Elisen MG, Meijers JC, Church FC. Role of the H helix in heparin binding to protein C inhibitor. J Biol Chem. 1994;269:28690–5. [PubMed] [Google Scholar]
- 88.Sandström J, Carlsson L, Marklund SL, Edlund T. The Heparin-binding domain of extracellular superoxide dismutase C and formation of variants with reduced heparin affinity. J Biol Chem. 1992;267:18205–18209. [PubMed] [Google Scholar]
- 89.Enjyoji K, Miyata T, Kamikubo Y, Kato H. Effect of heparin on the inhibition of factor Xa by tissue factor pathway inhibitor: a segment, Gly212-Phe243, of the third Kunitz domain is a heparin-binding site. Biochemistry. 1995;34:5725–35. doi: 10.1021/bi00017a004. [DOI] [PubMed] [Google Scholar]
- 90.McCaffrey TA, Falcone DJ, Du B. Transforming growth factor-beta 1 is a heparin-binding protein: identification of putative heparin-binding regions and isolation of heparins with varying affinity for TGF-beta 1. J Cell Physiol. 1992;152:430–40. doi: 10.1002/jcp.1041520226. [DOI] [PubMed] [Google Scholar]
- 91.Murphy-Ullrich JE, Gurusiddappa S, Frazier WA, Höök M. Heparin-binding peptides from thrombospondins 1 and 2 contain focal adhesion-labilizing activity. J Biol Chem. 1993;268:26784–9. [PubMed] [Google Scholar]
- 92.De Boer HC, Preissner KT, Bouma BN, de Groot PG. Binding of vitronectin-thrombin-antithrombin III complex to human endothelial cells is mediated by the heparin binding site of vitronectin. J Biol Chem. 1992;267:2264–8. [PubMed] [Google Scholar]
- 93.Sobel M, Solern DF, Kerrnodes C, Harrisn B. Localization and Characterization of a Heparin Binding Domain Peptide of Human von Willebrand Factor. 1992;267:8857–8862. [PubMed] [Google Scholar]
- 94.Fukushima T, Adachi T, Hirano K. The heparin-binding site of human xanthine oxidase. Biol Pharm Bull. 1995;18:156–8. doi: 10.1248/bpb.18.156. [DOI] [PubMed] [Google Scholar]