Abstract
During collective migration of the Drosophila embryonic salivary gland, cells rearrange to form a tube of a distinct shape and size. Here, we report a novel role for the Drosophila KASH (Klarsicht-Anc-Syne Homology) domain protein Klarsicht (Klar) in the regulation of microtubule (MT) stability and integrin receptor localization during salivary gland migration. In wild-type salivary glands, MTs became progressively stabilized as gland migration progressed. In embryos specifically lacking the KASH domain containing isoforms of Klar, salivary gland cells failed to rearrange and migrate, and these defects were accompanied by decreased MT stability and altered integrin receptor localization. In muscles and photoreceptors, KASH isoforms of Klar work together with Klaroid (Koi), a SUN domain protein, to position nuclei; however, loss of Koi had no effect on salivary gland migration, suggesting that Klar controls gland migration through novel interactors. The disrupted cell rearrangement and integrin localization observed in klar mutants could be mimicked by overexpressing Spastin (Spas), a MT severing protein, in otherwise wild-type salivary glands. In turn, promoting MT stability by reducing spas gene dosage in klar mutant embryos rescued the integrin localization, cell rearrangement and gland migration defects. Klar genetically interacts with the Rho1 small GTPase in salivary gland migration and is required for the subcellular localization of Rho1. We also show that Klar binds tubulin directly in vitro. Our studies provide the first evidence that a KASH-domain protein regulates the MT cytoskeleton and integrin localization during collective cell migration.
Keywords: Klarsicht, KASH, microtubule, integrin, salivary gland, collective cell migration, Drosophila, morphogenesis, tubulin
INTRODUCTION
Collective cell migration is a fundamental process during embryogenesis as well as in pathological conditions, such as cancer. During morphogenesis, cells migrate collectively to form and shape tissues and organs. For cells to migrate collectively as an intact group, multiple cellular processes, such as cell shape changes, cell rearrangements and modulation of cellular adhesions, have to be coordinated spatially and temporally.
The mechanisms by which cells migrate collectively are still poorly understood. Studies in genetically tractable model organisms have made important contributions to dissecting these complex processes. In particular, the Drosophila embryonic salivary gland is an excellent system for studying how cells migrate collectively as an intact tubular structure. The salivary glands consist of a pair of elongated secretory tubes that are connected to the larval mouth by the duct tubes (Chung et al., 2014; Maruyama and Andrew, 2012; Pirraglia and Myat, 2010). After salivary gland cells invaginate from the ventral surface of the embryo, the tube is initially oriented dorsally. The entire salivary gland then turns posteriorly and migrates beginning with cells at the distal end. Collective migration of the salivary gland is dependent on distinct activities at the distal and proximal ends that are coordinated temporally and spatially. Proximal gland cells change shape from columnar to cuboidal, rearrange and migrate in a manner dependent on Rho and Rac GTPases (Pirraglia et al., 2013; Xu et al., 2011; Xu et al., 2008). In contrast, the distal gland cells elongate and extend basal membrane protrusions by forming integrin mediated contacts with surrounding mesoderm-derived tissues (Bradley et al., 2003; Pirraglia et al., 2013). During salivary gland migration, two classes of integrin adhesion receptors become enriched at gland-mesoderm contact sites: αPS1βPS (expressed in the salivary gland) and αPS2βPS (expressed in the surrounding mesoderm) (Jattani et al., 2009). This integrin accumulation is functionally important as the salivary gland fails to turn and migrate posteriorly in embryos mutant for integrin subunits, such as myospheroid (encoding the βPS subunit), multiple edamatous wings (encoding αPS1) or inflated (encoding αPS2) (Bradley et al., 2003). αPS1βPS integrin controls salivary gland migration by downregulating E-cadherin and promoting basal membrane protrusions through Rac1 in the distal gland cells (Pirraglia et al., 2013).
Nuclear envelope spectrin-repeat proteins (Nesprins) are well characterized as nuclear-cytoplasmic linker proteins. They localize to the outer nuclear membrane of the nuclear envelope (NE) via their KASH (Klarsicht/ANC-1/Syne Homology) domains. KASH proteins connect the nucleoskeleton and the cytoskeleton through their interactions with SUN domain proteins that reside in the inner nuclear membrane (Starr and Fischer, 2005; Yu et al., 2011). They interact either directly with F-actin or indirectly with intermediate filaments and microtubules (MT), the latter through associations with the MT motor proteins kinesin and dynein (Mellad et al., 2011; Starr and Fridolfsson, 2010). In Drosophila, the KASH-domain protein Klarsicht (Klar) mediates nuclear migration in photoreceptor cells (Fischer et al., 2004; Starr and Fischer, 2005; Welte et al., 1998) and regulates nuclear positioning in striated muscle (Elhanany-Tamir et al., 2012).
It is becoming increasingly clear that in addition to their established roles in linking the nucleoskeleton and cytoskeleton, nesprins and their relatives play critical functions in the cytoplasm. Numerous isoforms of the mammalian KASH proteins have been shown to localize to structures other than the nuclear envelope, such as the plasma membrane and Golgi (Raigor and Shanahan, 2013) and link, for example, P bodies to microtubules (Raigor et al., 2014). Drosophila Klar also performs cytoplasmic functions: we previously showed that Klar controls salivary gland lumen size, possibly through the targeted transport of the apical transmembrane protein Crumbs (Myat and Andrew, 2002). Klar also regulates the transport of lipid droplets in early embryos (Guo et al., 2005; Welte et al., 1998) and of oskar mRNA in the oocyte (Gaspar et al., 2014). For lipid-droplet and RNA transport, these cytoplasmic roles of Klar are mediated by distinct isoforms that lack the KASH domain and arise by alternative splicing (Gaspar et al., 2014; Guo et al., 2005; Kim et al., 2013). Here, we report a novel role for KASH-containing forms of Klar in salivary gland migration that is mediated by regulation of MT stabilization and integrin receptor localization.
RESULTS
Klarsicht is required for salivary gland migration
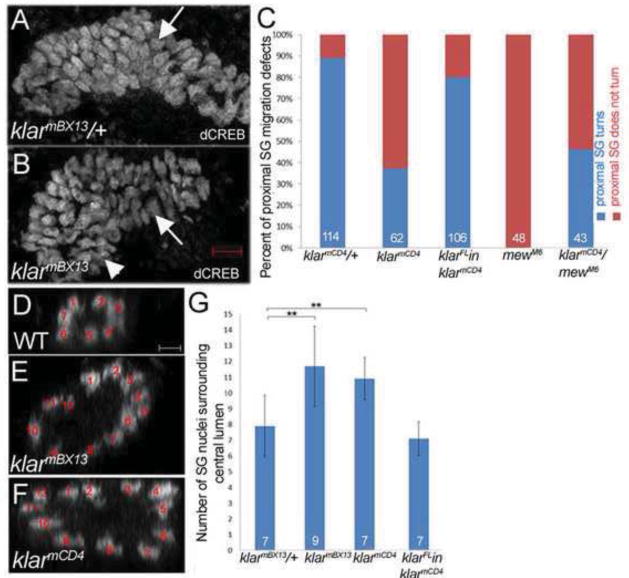
There are five isoforms of klar with klar α, γ and δ containing the conserved KASH domain (Supplementary Figure 1) (Kim et al., 2013). We previously showed that in Df(3L)emcE12 homozygous embryos that completely lack klar salivary gland morphology was grossly disrupted (Myat and Andrew, 2002). To determine a role for Klar in salivary gland migration, we analyzed embryos with lesions in klar. In particular, we focused on the function of KASH-domain containing isoforms and employed alleles that specifically lack the KASH domain. klarmCD4 contains a nonsense mutation just before the KASH domain whereas klarmBX13 has chromosomal breaks before the KASH domain (Fischer et al., 2004; Guo et al., 2005). Loss of klar resulted in glands that failed to migrate where the distal cells initiated the posterior turn but the proximal cells did not turn (Figure 1B). This is in contrast to glands of klar heterozygous embryos where both the distal and proximal gland cells turned and migrated posteriorly (Figure 1A). Quantification of the migration defect showed that 65% of klarmCD4 mutant glands failed to migrate completely by stage 14 as opposed to only 10% of glands of heterozygous siblings (Figure 1C). The gland migration defects of klar mutant embryos were accompanied by defects in the rearrangement of proximal gland cells as manifest by an increased number of cells surrounding the lumen of the gland (Figure 1D–G). In wild-type glands, eight cells on average surround the lumen whereas in klarmCD4 and klarmBX13 mutant glands 12 and 11 cells surround the lumen, respectively (Figure 1D–G). In wild-type salivary gland cells, endogenous Klar was enriched in the apical domain and also localized to discrete puncta in the basolateral and apical domains (Supplementary Figure 2A). Expression of wild-type Klar α, the longest KASH-domain containing isoform, specifically in the salivary glands of klar mutant embryos rescued the migration and cell rearrangement defects, demonstrating that klar is required cell-autonomously for gland migration (Figure 1C and G).
Figure 1. Klar is required for cell rearrangements in salivary gland migration.
In klarmBX13 heterozygous embryos (A), the salivary gland turns posteriorly (A, arrow) whereas in glands of homozygous siblings (B), the distal gland turns (B, arrow) but the proximal gland does not (B, arrowhead). Quantification of salivary gland migration defect (C). Salivary gland cells rearrange to form a narrow tube in wild-type embryos (D) whereas in klarmBX13 (E) and klarmCD4 (F) homozygous embryos, cells fail to rearrange and widened tubes are formed. Graph depicting number of nuclei surrounding the central lumen (G). In panels C and G, KlarFL in klarmCD4 is the fkh-GAL4 driven expression of UAS full-length klar α cDNA specifically in the salivary glands. Panels D–F show 1 μm orthogonal sections of the proximal region of salivary glands. Embryos shown are at stage 14 and were stained for dCREB to mark the salivary gland nuclei and for β-galactosidase (β-gal; not shown) to distinguish the heterozygous embryos with the marked balancer chromosome from the homozygous embryos. Numbers shown indicate number of embryos or glands scored. Scale bars represent 5 μm. **=p<0.01.
Integrin localization is altered in klar mutant salivary gland cells
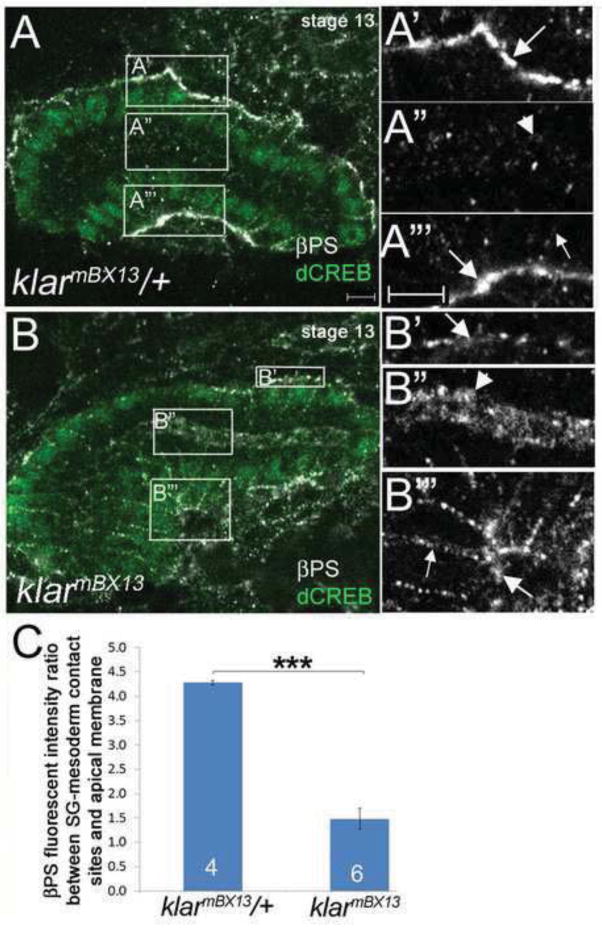
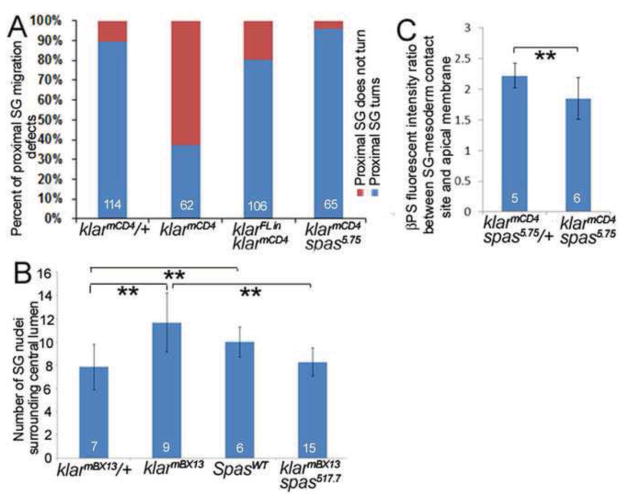
The salivary gland migration defect of klar mutant embryos resembled that of embryos with mutations in integrin subunits where distal gland cells turn posteriorly but proximal gland cells do not (Bradley et al., 2003; Pirraglia et al., 2013). For example, in embryos homozygous for mewM6, a mutation in the αPS1 integrin subunit, 100% of salivary glands fail to complete posterior turning (see also Figure 1C). To test if klar and integrins act in the same pathway, we analyzed embryos trans-heterozygous for klarmBX13and mewM6: 55% of glands failed to complete migration (Figure 1C). Because of this genetic interaction, we tested if klar mutations affect the subcellular localization of the βPS integrin receptor in migrating salivary glands. We previously showed that the βPS and αPS2 integrin subunits become enriched at sites of contact between the migrating salivary gland and the overlying circular visceral mesoderm and underlying fat body (Jattani et al., 2009). In salivary glands of klarmBX13 heterozygous embryos, as in wild-type embryos, βPS integrin was localized predominantly at the basal membranes in contact with surrounding tissues and only at low levels at the apical membrane (Figure 2A and data and shown). By contrast, in glands of klarmBX13 homozygous embryos, βPS integrin was not enriched at gland-mesoderm contact sites and instead was enriched in the apical domain (Figure 2B). Quantification of βPS integrin levels based on measurements of fluorescent intensity ratio between the basal gland-mesoderm contact site and the apical membrane revealed significant enrichment in the apical membrane and reduction at the gland-mesoderm contact site in klarmBX13 homozygous embryos compared to heterozygous siblings (Figure 2C). In contrast, apical membrane markers, such as aPKC and Crumbs and the adherens junction protein E-cadherin, which marks apical-lateral membranes, showed no difference in salivary gland cells of klar homozygous and heterozygous embryos (Supplementary Figure 3 and data not shown). Thus, the altered localization of βPS integrin in klar mutant salivary glands does not reflect loss of apical-basal polarity and, rather, represents a specific defect. Klar regulation of βPS integrin likely occurs post-transcriptionally since RNA in situ hybridization for myospheroid encoding βPS integrin showed no differences between klar heterozygous and homozygous embryos (data not shown).
Figure 2. βPS integrin localization is altered in klar mutant salivary gland cells.
In klarmBX13 heterozygous embryos at stage 13 (A), βPS integrin accumulates at gland-mesoderm contact sites (A′ and A‴, large arrows) and is also found at low levels in the apical (A″, arrowhead) and basolateral (A‴, small arrow) domains. In stage 13 klarmBX13 homozygous embryos (B), βPS is present at low levels at gland-mesoderm contact sites (B′ and B‴, large arrows) but is enriched in the apical domain (B″, arrowhead) and basolateral membrane (B‴, small arrow). A′-A‴ and B′-B‴ correspond to boxed regions in A and B, respectively. (C) Graph depicting βPS fluorescent intensity ratio between the salivary gland (SG)-mesoderm contact site and apical membrane in klarmBX13 heterozygous and homozygous embryos. **=p<0.001. Numbers shown indicate number of glands scored. Scale bars represent 5μm.
Klaroid is not required for salivary gland migration
In photoreceptors and muscle cells, KASH-domain isoforms of Klar have critical roles in positioning of nuclei (Elhanany-Tamir et al., 2012; Fischer et al., 2004). Here, Klar works together with Klaroid (Koi), the only Drosophila SUN-domain protein expressed outside the testis (Kracklauer et al., 2007). In wild-type salivary gland cells, endogenous Koi localized to the nuclear envelope (Supplementary Figure 2B). This is in contrast to endogenous Klar, which localized predominantly to the apical domain. To test if Koi is required for salivary gland migration, we analyzed embryos mutant for the koi null allele koi84. We did not observe a significant defect in salivary gland cell rearrangement in koi84 homozygous embryos (Supplementary Figure 4C). koi84 mutant embryos also did not show defects in βPS integrin localization compared to wild-type embryos (data not shown). The absence of salivary gland defects in koi84 mutant embryos could be due to the strong maternal contribution of koi. These studies suggest that Klar likely controls gland migration by associating with as yet unidentified interactors.
Klar regulates microtubule stability
During lipid droplet movement in the early embryo, Klar associates indirectly with the MT cytoskeleton through the MT motors cytoplasmic dynein and kinesin-1 (Gross et al., 2000; Shubeita et al., 2008), and in oocytes Klar physically interacts with kinesin-1 and modulates kinesin-1-dependent motility of oskar mRNPs (Gaspar et al., 2014). Thus, we tested the hypothesis that klar regulates salivary gland migration by regulating the MT cytoskeleton and/or its associated motor proteins. We first analyzed embryos mutant for the null allele of the kinesin heavy chain (khc8), the null allele of dynein heavy chain (dhc64C4-19), or dynein light chain 90F (dlc90F05089) for salivary gland migration defects. We observed no defects in gland migration or cell rearrangement in khc8, dhc64C4-19 or dlc90F05089 homozygous embryos (data not shown). khc8 mutant embryos also showed no defect in integrin localization whereas dhc64C4-19 and dlc90F05089 mutant embryos showed enhanced enrichment of βPS integrin at the gland-mesoderm contact site, a phenotype distinct from that of klar mutant embryos (data not shown). Although we cannot rule out that maternally provided motors compensate for the lack of zygotic expression in these embryos, our data suggest that klar likely regulates gland migration by interacting with regulators other than kinesin-1 or dynein.
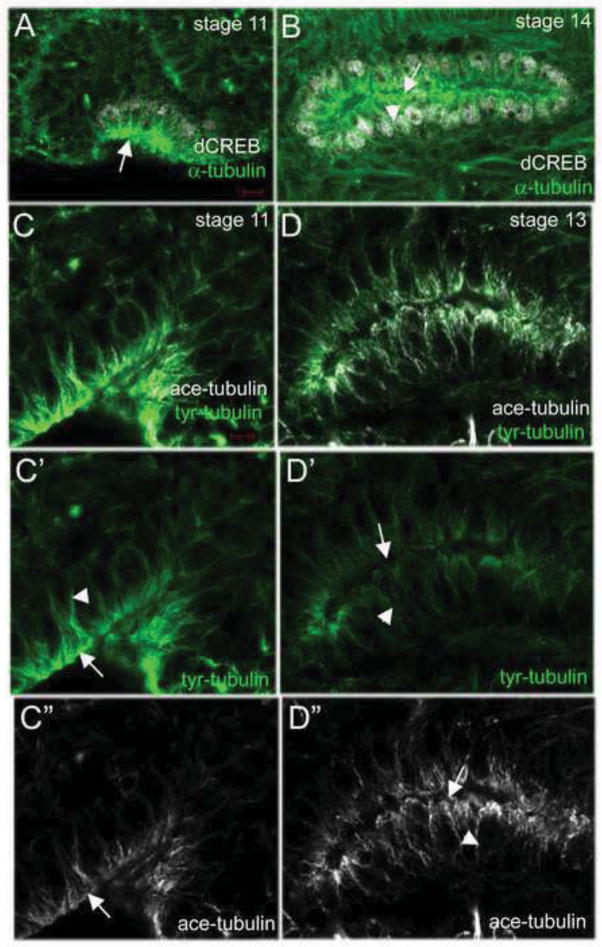
We next determined if klar controls the MT cytoskeleton during salivary gland migration by analyzing MT organization and structure in migrating gland cells. Staining for α-tubulin revealed that in invaginating and migrating salivary gland cells, MTs were enriched in the apical domain and extended as long fibers into the basolateral membrane (Figure 3A and B), as previously reported for Drosophila tracheal cells (Brodu et al., 2010). We also examined tubulin tyrosination, which is associated with newly formed MTs, as well as tubulin acetylation, which is associated with stable MTs (Hammond et al., 2008). We observed that during salivary gland invagination tyrosinated MTs were enriched in the apical and basolateral domains (Figure 3C). As salivary gland migration progressed, tyrosinated MTs decreased throughout the cell (Figure 3D). In contrast to tyrosinated MTs, acetylated (stable) MTs were less abundant in invaginating gland cells (Figure 3C). However, as salivary gland migration progressed, stable MTs became enriched in the apical domain and extended to the basolateral domain (Figure 3D).
Figure 3. MT organization during salivary gland migration.
During salivary gland invagination (A) and migration (B), MTs, as detected by α-tubulin staining (A and B, green), are enriched in the apical domain (A and B, arrows) and extend to the basolateral domain (A and B, arrowhead). In invaginating salivary glands (C), tyrosinated-tubulin (tyr-tubulin) is enriched in the apical and basolateral domains (C and C′, arrow and arrowhead, respectively) whereas acetylated tubulin (ace-tubulin) is predominantly found in the apical domain (C and C″, arrow). As the salivary gland migrates (D), tyr-tubulin is reduced in the apical and basolateral domains (D and D′, arrow and arrowhead, respectively) whereas ace-tubulin is enriched in the apical and basolateral domains (D and D″, arrow and arrowhead, respectively). Embryos in A and B were stained for dCREB (white) and α-tubulin (green) whereas those in C and D were stained for tyr-tubulin (green) and ace-tubulin (white). Scale bars represent 5 μm.
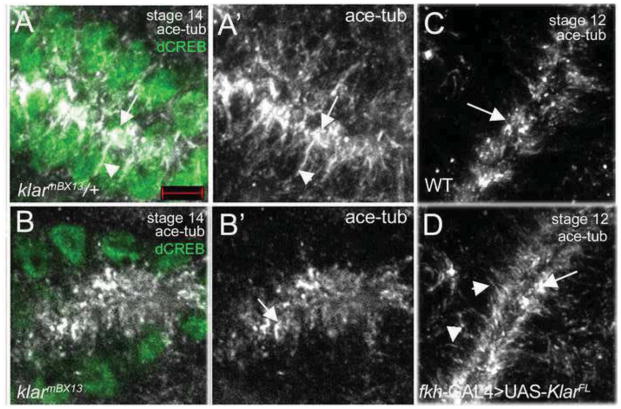
In salivary glands of stage 14 klarmBX13 or klarmCD4 heterozygous embryos, acetylated MTs were found as short fibers in the apical domain and as long fibers that extended basally around nuclei as in the wild type (Figure 4A and data not shown). By contrast, in glands of klarmBX13 or klarmCD4 homozygous embryos acetylated tubulin staining was found in the apical domain in a diffused pattern and not as fibers that projected basally (Figure 4B and data not shown). Staining for α-tubulin to label the MT cytoskeleton or tyrosinated tubulin to specifically label newly polymerized MTs revealed no difference between klar hetero- and homozygous embryos (Supplementary Figures 5 and 6). Thus, the effect of loss of Klar appears to be specific to stable MTs. These findings are consistent with those reported in the muscle of klar mutant embryos where the MT cytoskeleton is altered (Elhanany-Tamir et al., 2012).
Figure 4. Klar is required for microtubule stability.
In salivary glands of klarmBX13 heterozygous embryos (A), stable MTs marked by acetylated tubulin (ace-tub) are enriched in the apical domain (A and A′, arrows) and extend basally (A and A′, arrowheads) whereas in glands of homozygous siblings (B), ace tub-stained MTs appear as puncta in the apical domain (B and B′, arrow). In wild-type salivary glands at stage 12 (C), stable MTs are detected only in the apical domain (C, arrow) whereas in wild-type glands overexpressing Klar α (D), stable MTs are enriched apically (D, arrow) and also extend basally (D, arrowheads). Embryos were stained for acetylated tubulin (ace-tub, white), dCREB (green) and β-gal (not shown). Scale bars represent 5 μm.
Overexpression of Klar α was even sufficient to promote increased MT stability in a wild-type background. In stage 12 wild-type glands, stable MTs were only found in the apical domain (Figure 4C). By contrast, in klar α-overexpressing glands stable MTs accumulated prematurely in the apical domain and extended into the basolateral domain (Figure 4D). In contrast to klar, loss of koi had no effect on MT stability (data not shown). We conclude that Klar controls the stability of MTs during salivary gland migration.
A role of Klar in MT stability is not completely unprecedented. A genetic modifier screen in the Drosophila retina identified CLASP (cytoplasmic linker protein [CLIP]-associated protein), a well-conserved MT plus-end protein, as an interactor of Klar (Lowery et al., 2010). Subsequent MT plus-end imaging studies in Drosophila S2 cells suggested Klar to stabilize MTs by acting as an inhibitor of rescue and pause events (Long et al., 2013), though the effects we observed on the MT network in the salivary gland are much more dramatic. Because of this link between CLASP and Klar, we analyzed salivary gland migration in embryos lacking CLASP, Df(3L)chb4. We found that loss of CLASP disrupted salivary gland cell rearrangement during migration, similar to loss of Klar (Supplementary Figure 4A–C). Klar and CLASP also interacted genetically; 10 nuclei surrounded the lumen of salivary glands in klarmCD4 Df(3L)chb4 trans-heterozygous embryos (n=10 salivary glands). Moreover, in Df(3L)chb4 homozygous embryos staining for acetylated tubulin showed that stable MTs did not extend to the basolateral domain as in heterozygous siblings, similar to klar mutant embryos (Supplementary Figure 4D and E). Thus, Klar may regulate MT stability in the salivary gland in a manner dependent on CLASP.
Klar interacts genetically with spastin
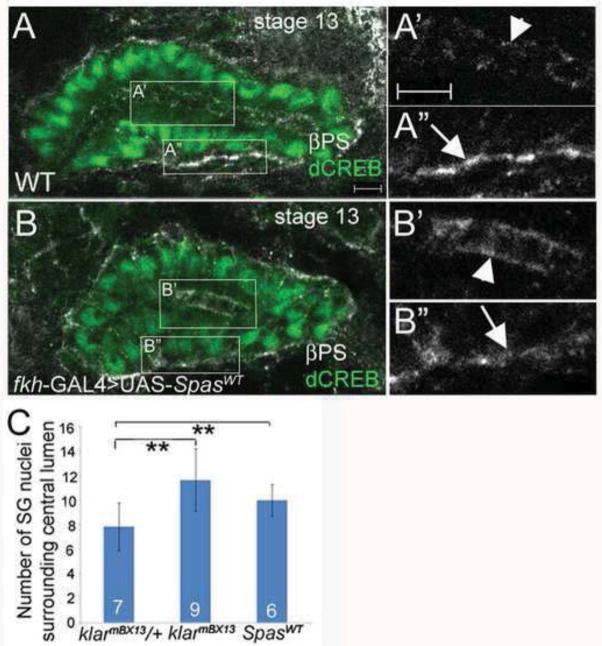
Loss of Klar causes both a failure in salivary gland migration and a loss of stable MTs. To determine if the effects on MT stability contribute to failed gland migration, we sought to disrupt MTs by independent means. When we overexpressed the AAA ATPase Spastin (Spas), a MT severing protein (Lumb et al., 2012; Roll-Mecak and Vale, 2005) specifically in the salivary glands of wild-type embryos, it resulted in a gradual loss of stable MTs, such that by stage 13 stable MTs were barely detectable (data not shown). Spas overexpression disrupted proximal salivary gland cell rearrangement and resulted in loss of βPS integrin subunits at the gland-mesoderm contact sites and their accumulation in the apical membrane (Figure 5A–C). Thus, Spas overexpression phenocopied loss of klar and disrupted salivary gland cell rearrangement and integrin localization during gland migration.
Figure 5. Spastin overexpression disrupts salivary gland cell rearrangement and integrin localization.
In wild-type salivary glands at stage 13 (A), βPS integrin accumulates at gland-mesoderm contact sites (A″, arrow) and is also present at the apical domain (A′, arrowhead). In salivary glands overexpressing wild-type Spastin (B), βPS integrin is enriched in the apical domain (B′, arrowhead) and is reduced from the gland-mesoderm contact site (B″, arrow). Graph depicting number of salivary gland (SG) nuclei surrounding the central lumen in klarmBX13 heterozygous and homozygous embryos, and wild-type embryos expressing SpasWT in the gland (C). Embryos shown were stained for βPS (white) and dCREB (green). Numbers shown indicate number of glands scored. Scale bars represent 5μm. **=p<0.001.
To test to what extent the phenotypes of klar mutant salivary glands are due to loss of MT stability, we reduced spas function in klar homozygous embryos using a hypomorphic (spas17-7) or a null (spas5.75) allele of spas (Sherwood et al., 2004). In klarmCD4spas5.75 and klarmBX13spas17-7 double mutant embryos, salivary gland migration, proximal gland rearrangement and integrin localization were significantly rescued compared to klar homozygous embryos alone (Figure 6A–C and data not shown). We conclude that the primary defect in klar mutant salivary glands is loss of MT stability.
Figure 6. Reduction of Spastin rescues the salivary gland defects of klar mutant embryos.
Graphs depicting the frequency of salivary gland migration defects (A), number of nuclei surrounding the lumen (B) and distribution of βPS integrin (C) in klarmCD4 and klarmBX13 heterozygous and homozygous embryos, klarmCD4 embryos expressing klar α, wild-type embryos expressing SpasWT, and klarmCD4 spas5.75 and klarmBX13 spas17-7 double mutant embryos. Numbers shown indicate number of embryos or glands scored. **=p<0.01.
Klar genetically interacts with Rho1 GTPase and controls its subcellular localization
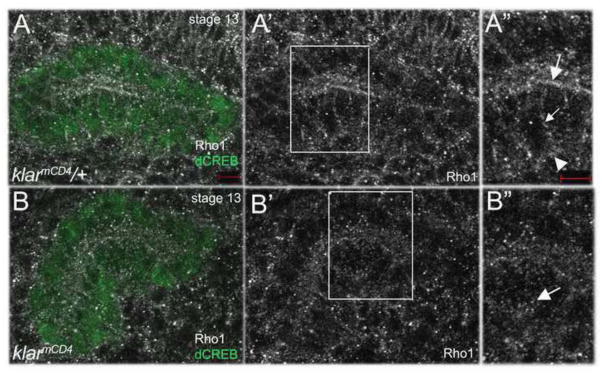
To understand how Klar might affect MT stability, we took a candidate approach. Like Klar, Rho1 GTPase is required for salivary gland migration and cell rearrangement in Drosophila (Xu et al., 2011; Xu et al., 2008). In mammalian cells, Rho1 mediates the selective stabilization of MTs in response to lysophosphatidic acid (Cook et al., 1998). Thus, we tested if klar promotes MT stability in migrating salivary glands in a Rho1-dependent manner. We first confirmed that loss of Rho1 decreased MT stability like loss of klar (Supplementary Figure 7). In embryos trans-heterozygous for Rho11B and klarmBX13, salivary glands failed to migrate and gland cells failed to rearrange, similar to that of Rho11B or klarmBX13 homozygous embryos (Supplementary Figure 8). In klarmCD4 heterozygous embryos, endogenous Rho1 localized to the apical and basolateral membranes in addition to being present as cytoplasmic puncta (Figure 7A). By contrast, in klarmCD4 homozygous embryos, Rho1 failed to localize to the apical and basolateral membranes and instead was found predominantly as cytoplasmic puncta (Figure 7B). From these data we conclude that Klar may regulate MT stability by affecting the subcellular localization of Rho1.
Figure 7. Klar controls Rho1 localization.
In klarmCD4 heterozygous embryos (A), endogenous Rho1 localizes to the apical (A-A″, large arrow), basolateral (A-A″, small arrow) and basal (A-A″, arrowhead) membranes, whereas in homozygous siblings (B), Rho1 is found predominantly in cytoplasmic puncta (B-B″, arrow). Panels A″ and B″ represent magnified views of boxed regions in panels A′ and B′. Embryos shown were stained for dCREB (green), Rho1 (white) and β-gal (not shown). Scale bars represent 5 μm.
Klar binds tubulin in vitro
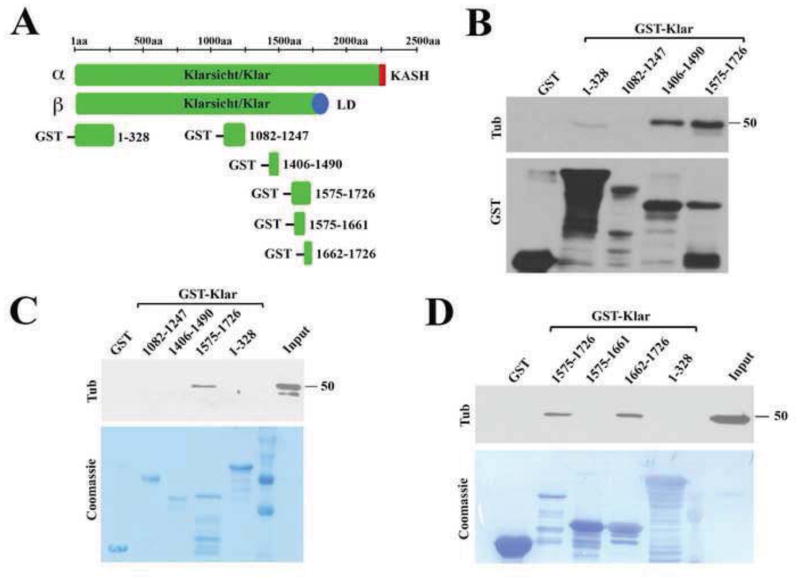
Because of the effects of loss of Klar on MT stability, we tested if Klar associates with tubulin directly or indirectly, by generating GST-tagged fragments of Klar. In oocytes where the klar β isoform is the predominant type, Klar also affects MT stability (Gaspar et al., 2014). As the α and β isoforms share the N-terminal portion of Klar (amino acids 1-1726) (Kim et al., 2013), we focused our attention on this shared region and generated constructs containing the following fragments of Klar: 1-328, 1082-1247, 1406-1490 and 1575-1726 (Figure 8A). We found that Klar[1575-1726] could precipitate endogenous tubulin from whole embryo lysates (Figure 8B). This interaction may be direct as the same construct was also able to precipitate purified tubulin (Figure 8C). Using smaller fragments of Klar[1575-1726], we mapped the tubulin-binding activity to its C-terminal half (Figure 8D). We propose that a region of Klar encompassing amino acids 1662 through 1726 mediates Klar binding to tubulin in vitro and thus might contribute to MT stabilization in vivo by recruiting Klar directly to MTs. Alignment of sequences from fourteen arthropod species showed significant conservation of the putative microtubule binding domain among the six Diptera analyzed (Supplementary Figure 9).
Figure 8. Klarsicht binds tubulin.
(A) Schematic diagram showing various fragments of Klar expressed as GST-fusions. GST pull down assays were performed with either embryo lysate (B, D) or with purified tubulin (C); tubulin was detected with anti-tubulin antibodies, and GST-fusions either by anti-GST antibodies or by Coomassie staining. (B) Both Klar[1406-1490] and Klar[1575-1726] are able to precipitate tubulin from whole embryo lysates; the tubulin binding activity of Klar[1575-1726] was further mapped to its C-terminal half, Klar[1662-1726] (D). Klar[1575-1726] was also able to precipitate purified tubulin (C), suggesting a direct interaction.
DISCUSSION
Although it is widely accepted that MTs are essential for the migration of cultured cells, it is only in recent years that a role for MTs in cell migration during embryogenesis is recognized. Thus, we know little of how MTs and their associated proteins regulate such complex processes as the collective cell movements that occur during embryogenesis. Mammalian nesprin proteins have been shown to regulate the migration of a number of cell types, such as keratinocytes and endothelial cells (King et al., 2014; Rashmi et al., 2012); however, their control of cell migration is thought to occur through regulation of the actin cytoskeleton. Our studies provide the first evidence for a non-nuclear role of KASH-containing Klar in controlling cell migration through regulation of the MT cytoskeleton. We showed that Klar controls MT stability and integrin receptor localization during the collective migration of the Drosophila embryonic salivary gland. It is unlikely that Klar control of integrin localization indirectly affects MT stability since stable MTs accumulated in the same temporal and spatial manner in mewM6 mutant embryos as in WT embryos (data not shown). Instead, Klar may regulate integrin localization through its effects on the MT cytoskeleton. For example, in migrating cells integrins are known to be trafficked to the leading edge and MTs are implicated in this process (Gu et al., 2011; Pellinen and Ivaska, 2006). Alternatively, or in addition, Klar may control the membrane trafficking machinery that localizes integrins to the basal membrane of salivary gland cells independent of Klar’s function in MT stability. Further studies are necessary to elucidate how stable MTs regulate the localization of integrin receptors during cell migration in embryogenesis.
Acetylation of α-tubulin is one of the earliest post-translational modifications of MTs to occur (L’Hernault and Rosenbaum, 1983; L’Hernault and Rosenbaum, 1985); however it is not well understood what effect tubulin acetylation has on cellular function. In klar mutant salivary gland cells, acetylated tubulin was detected as puncta dispersed in the apical domain instead of a tubular network that extended basally, as found in wild-type cells. Because acetylated tubulin signal was still detected in klar mutant salivary gland cells, our findings suggest that loss of klar did not necessarily affect the acetylation of tubulin and instead affected the structure of stable MTs. Reported studies suggest that intracellular organelles, such as the ER and mitochondria, preferentially move along acetylated MTs (Friedman et al., 2010; Lee and Chen, 1988; Waterman-Storer and Salmon, 1998). Thus, one mechanism by which Klar-mediated stabilization of MTs affects salivary gland migration could be through organelle movement during migration. The Drosophila salivary gland provides a unique experimental system to further address the role of Klar and stable MTs in collective cell migration during morphogenesis.
Our studies suggest that Klar might affect MT stability in several different ways. First, the disruption of Rho1 localization in klar mutant salivary gland cells suggests that Klar controls MT stability by regulating Rho1 activity. In cultured mammalian cells, Rho1 acts downstream of lysophosphatidic acid (LPA) to selectively stabilize stable MTs oriented toward the leading edge of migrating cells (Cook et al., 1998). Second, Klar may promote MT stability through interactions with CLASP. Third, the direct binding we observed between Klar and tubulin also suggests that Klar may directly influence the MT cytoskeleton. This is supported by recent studies in Drosophila S2 cells which suggest Klar to affect MT stability by inhibiting MT rescue and pause events (Long et al., 2013). Thus, in migrating salivary gland cells, Klar may associate directly with MTs and influence the activities of CLASP and/or other MT plus-tip proteins, such as Rho1, to promote MT stabilization. The KASH domain of Klar may be required for correct intracellular localization of Klar to ensure interaction with MTs through the MT binding domain. Alternatively, Klar may act as an adaptor protein where the MT binding domain interacts with MTs and the KASH domain interacts with effector proteins, thus bringing the effector protein to the MTs. It is also possible that the absence of a functional KASH domain in the klarmBX13 and klarmCD4 alleles analyzed here could affect the correct folding of the MT binding domain.
Our analysis of kinesin-1 and dynein mutant embryos did not show any defects in salivary gland cell rearrangement or migration. This was somewhat surprising considering that kinesin and dynein have both been shown to be required for nuclear movement during myogenesis (Folker et al., 2012 ; Folker et al., 2014; Metzger et al., 2012). It is possible that other kinesins are involved in salivary gland migration. Alternatively, the maternal supply of kinesin could be sufficient for salivary gland migration which occurs earlier in embryogenesis than myogenesis.
In support of a role for KASH domain proteins in cell migration, mammalian Nesprin-2 controls keratinocyte migration during wound healing, and Nesprin-1 and Nesprin-2 regulate endothelial cell shape and migration (King et al., 2014; Rashmi et al., 2012). This role of Nesprins in cell migration is attributed to their effects on actin polymerization and stress fiber assembly. Thus, our demonstration that Klar-regulated MT stability is important for cell migration during embryogenesis demonstrates that KASH proteins are required for cell migration by regulating not only the actin cytoskeleton but the MT cytoskeleton as well. MTs are thought to contribute to directed cell migration in a number of ways, including their role in polarized trafficking of essential signaling and adhesion proteins and their mechanical properties. Interestingly, in migrating cultured cells selectively stabilized MTs are oriented towards the direction of migration (Gundersen and Bretscher, 2003; Gundersen and Bulinski, 1988). While MT stabilization is observed to correlate with tracheal morphogenesis in the Drosophila embryo (Brodu et al., 2010), it is not known what role MT stabilization plays in morphogenesis. Our studies shed novel insight into MT stability and collective cell migration during salivary gland morphogenesis and identify Klar as an important regulator of MT stabilization. It will be of interest to determine whether mammalian Nesprins control cell migration by regulating the MT cytoskeleton as well.
MATERIALS AND METHODS
Drosophila Strains and Genetics
Canton-S flies were used as wild-type controls. klarmCD4, klarmBX13, khc8, dhc64C4-19, dlc90F05089, mewM6, Rho11B, Rho1E3.10, Df(3L)chb4 and UAS-klarα, described in (Fischer et al., 2004) as Klar full-length (klarFL), were obtained from the Bloomington Stock Center and are described in FlyBase (http://flybase.bio.indiana.edu/). spas5.75, spas17-7 and UAS-Spastin (full-length wild-type spastin (Sherwood et al., 2004) were kind gifts from Nina Sherwood (Duke University, Durham, NC). koi84 was a kind gift of Janice Fischer (University of Texas, Austin). For salivary gland-specific expression of the UAS constructs, we used fork head (fkh)-GAL4.
Immunocytochemistry and in situ hybridization
Embryo fixation and antibody staining were performed as previously described (Xu et al., 2011). Immunostaining for Klar was performed as described (Guo et al., 2005). Embryos were prepared for MT staining as previously described (Lee et al., 2003). Briefly, embryos were fixed with a methanol solution of 90% methanol, 5 mM sodium bicarbonate (pH 9), and 3% formaldehyde chilled to −20°C for 10 minutes. The following antisera were used at the indicated dilutions: rat Koi antisera at 1:10 (Kracklauer et al., 2007); mouse Klar-C antisera at 1:5; mouse acetylated tubulin antisera (SIGMA, St. Louis, MO)and mouse tyrosinated tubulin antisera (Abcam, Cambridge, MA) at 1:250 and 1:200, respectively; FITC conjugated mouse antisera to α-tubulin (SIGMA) at 1:100; mouse antisera to βPS integrin, Crumbs and Rho1 at 1:500, 1:10 and 1:10, respectively, and rat antisera to DE-cadherin at 1:20 (Developmental Studies Hybridoma Bank, DSHB; Iowa City, IA); rabbit aPKC antiserum (Santa Cruz Biotechnology, Dallas, TX) at 1:500; mouse β-galactosidase (β-gal) antiserum (Promega, Madison, WI) at 1:500. Appropriate biotinylated- (Jackson Immunoresearch Laboratories, Westgrove, PA), AlexaFluor 488-, 647- or Rhodamine- (Molecular Probes, Eugene, OR) conjugated secondary antibodies were used at a dilution of 1:500. Stained embryos were mounted in Aqua Polymount (Polysciences, Inc., Warrington, PA) and thick (1 μm) fluorescence images were acquired on a Zeiss Axioplan microscope (Carl Zeiss) equipped with LSM 510 (Weill Cornell Medical College optical core facility, New York, NY) or LSM 710 (Medgar Evers College – CUNY, New York, NY). In situ hybridization was performed using myospheroid cDNA (Open Biosystems, GE Healthcare) as a template for generating antisense digoxygenin-labeled RNA probes as previously described (Patel et al., 2012).
Quantification of βPS fluorescence intensity
Fluorescent intensity ratio between the basolateral and apical domains was obtained using an established method (Pines et al., 2011). Fluorescent intensity within an area of approximately 1.7 μm2 (3.08 μm × 0.56 μm) in the apical region of the cell and a region of the exact same size in the basolateral region was measured using the Image J software (National Institute of Health, Bethesda, MD) and its ratio (Apical/Baso-lateral) calculated. Statistical analysis was done using Microsoft Excel.
Generation of Klar constructs
We generated different GST constructs of Klar consisting of both the N-terminal and C-terminal regions of Klar in the pGEX-6P-1 vector (GE Healthcare). These are 1-328, 1082-1247, 1406-1490, 1575-1726, 1575-1661 and 1662-1726.
| Primers- | |
| 1-328 sense: | 5′ GGGGAATTCATGGAAATGCAACAGGAAAAC 3′ |
| 1-328 anti-sense: | 5′ GGGCTCGAGCGTGGATGCAATTTCATGG 3′ |
| 1082-1247 sense: | 5′ GGGGGATCCATGCCCCATCCCATCAAGC 3′ |
| 1082-1247anti-sense: | 5′ GGGGTCGACACCGGACGGAGAGTTTTCC 3′ |
| 1406-1490 sense: | 5′ GGGGAATTCGATGGCGAGGGAGGGG 3′ |
| 1406-1490 anti-sense: | 5′ GGGGTCGACGTCCATGCGCGGCGGC 3′ |
| 1575-1726 sense: | 5′ GGGGGATCCAAGAATCAGTCGACCAGC 3′ |
| 1575-1726 anti-sense: | 5′ GGGCTCGAGTTTTGTCTGGCCAGCGC 3′ |
| 1575-1661 sense: | 5′ GGGGGATCCAAGAATCAGTCGACCAGC 3′ |
| 1575-1661 anti-sense: | 5′ GGGCTCGAGCTCCTGCTCTCCGTCC 3′ |
| 1662-1726 sense: | 5′ GGGGGATCCATGCGATCCCTGCTGCAG 3′ |
| 1662-1726 anti-sense: | 5′ GGGCTCGAGTTTTGTCTGGCCAGCGC 3′ |
GST pull-down assays
GST and GST-klar proteins were produced in BL21 E. coli and purified by using glutathione beads (GE healthcare). The beads were washed with high salt solution [500 mM NaCl, 50 mM Tris-HCl (pH 7.5), 100 mM PMSF]. Both GST and GST fusion proteins bound to beads were equilibrated with homogenizing buffer [20mM HEPES (pH 7.4), 10% Glycerol, 0.2 mM EDTA. 150 mM NaCl, 0.1% NP40, 0.5 mM DTT, 1mM PMSF, 1X Protease inhibitor cocktail from Sigma] and incubated with either embryo lysate or pure bovine tubulin (Cytoskeleton, Denver, CO, Catalog # TL238-B) overnight at 4°C. Bound beads were washed five times with homogenizing buffer. The bound proteins were subjected to SDS-PAGE followed by western blotting using a mouse monoclonal anti-alpha-tubulin antibody (Sigma-Aldrich) at 1:80,000.
Alignment of klar α sequences
The alignment shown in Supplementary Figure 9 employed Klar α sequences predicted from fourteen arthropod species. The species and GenBank accession numbers of the sequences are as follows: Drosophila melanogaster (NP_523873), Drosophila pseudobscura (XP_001354250), Drosophila willistoni (XP_002065747), Bactrocera cucurbitae (XP_011183765), Ceratitis capitata (XP_004526913), Musca domestica (XP_005189601), Aedes aegypti (XP_001653019), Anopheles sinensis (KFB38974), Pogonomyrmex barbatus (XP_011639645), Acromyrmex echinator (EGI67743), Apis mellifera (XP_001122432), Nasonia vitripennis (XP_008201871), and Daphnia pulex (EFX72496). A putative Klar α sequence from the centipede Strigamia maritime was retrieved from the Ensembl Genomes server (gene SMAR003016) (Kersey et al., 2014). Sequences were aligned with ClustalX 2.0.10. The conservation score for each position was calculated in CulstalX using Gonnet PAM 80 as the protein weight matrix.
Supplementary Material
Acknowledgments
We are grateful to Janice Fischer, Nina Sherwood, the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for providing fly lines and antisera. We are indebted to Geri Kreitzer and members of the Kreitzer lab for providing valuable insight and discussions during the course of these studies and for critical reading of the manuscript. We also thank the Weill Cornell Medical College optical core facility. This work was supported in part by NIH grants GM082996 to M.M.M and GM64687 to MAW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bradley PL, Myat MM, Comeaux CA, Andrew DJ. Posterior migration of the salivary gland requires an intact visceral mesoderm and integrin function. Developmental Biology. 2003;257:249–262. doi: 10.1016/s0012-1606(03)00103-9. [DOI] [PubMed] [Google Scholar]
- Brodu V, Baffet A, Le Droguen PM, Casanova J, Guichet A. A Developmentally Regulated Two-Step Process Generates a Noncentrosomal Microtubule Network in Drosophla Tracheal Cells. Dev Cell. 2010;18:790–801. doi: 10.1016/j.devcel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- Chung S, Hanlon C, Andrew D. Building and specializng epithelial tubular organs: the Drosophila salivary gland as a model system for revealing how epithelial organs are specified, form and specialize. Wiley Interdiscip Rev Dev Biol. 2014;3:281–300. doi: 10.1002/wdev.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook T, Nagasaki T, Gundersen G. Rho guanosine triphosphate mediates the selective stabilization of microtubules induced by lysophosphatidic acid. J Cell Biol. 1998;141:175–85. doi: 10.1083/jcb.141.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhanany-Tamir H, Yu Y, Shnayder M, Jain A, Welte M, Volk T. Organelle positioning in muscles requires coorperation between two KASH proteins and microtubules. J Cell Biol. 2012;198:833–846. doi: 10.1083/jcb.201204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer J, Acosta S, Kenny A, Cater C, Robinson C, Hook J. Drosophila klarsicht has distinct subcellular localization domains for nuclear envelope and microtubule localization in the eye. Genetics. 2004;168:1385–1393. doi: 10.1534/genetics.104.028662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker E, Schulman V, Baylies M. Muscle length and myonuclear position are independently regulated by distinct Dynein pathways. Development. 2012;139:3827–3837. doi: 10.1242/dev.079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker E, Schulman V, Baylies M. Translocating myonuclei have distinct leading and lagging edges taht require kinesin and dynein. Development. 2014;141:355–366. doi: 10.1242/dev.095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Webster B, Mastronarde D, Verhey K, Voeltz G. ER sliding dynamics and ER-mitochondrial contacts occur on acetylated microtubules. J Cell Biol. 2010;190:363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar I, Yu Y, Cotton S, Kim D, Ephrussi A, Welte M. Klar ensures thermal robustness of oskar localization by restraining RNP motility. J Cell Biol. 2014;206:199–215. doi: 10.1083/jcb.201310010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Welte M, Block S, Wieschaus E. Dynein-mediated cargo transport in vivo: a switch controls travel distance. J Cell Biol. 2000;148:945–956. doi: 10.1083/jcb.148.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Noss E, Hsu V, Brenner M. Integrins traffic rapidly via circular dorsal ruffles and macropinocytosis during stimulated cell migration. J Cell Biol. 2011;193:61–70. doi: 10.1083/jcb.201007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen G, Bretscher A. Cell Biology. Microtubule assymetry. Science. 2003;300:2040–2041. doi: 10.1126/science.1084938. [DOI] [PubMed] [Google Scholar]
- Gundersen G, Bulinski J. Selective stabilization of microtubules oriented toward the direction of cell migration. Proc Natl Acad Sci USA. 1988;85:5946–5950. doi: 10.1073/pnas.85.16.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Jangi S, Welte M. Organelle-specific control of intracellular transport: distinctly targeted isoforms of the regulator Klar. Mol Biol Cell. 2005;16:1406–1416. doi: 10.1091/mbc.E04-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J, Cai D, Verhey K. Tubulin modifications and their cellular functions. Curr Opin Cell Biol. 2008;20:71–76. doi: 10.1016/j.ceb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jattani R, Patel U, Kerman B, Myat M. Deficiency screen identifies a novel role for beta 2 tubulin in salivary gland and myoblast migration in the Drosophila embryo. Developmental Dynamics. 2009;238:853–863. doi: 10.1002/dvdy.21899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersey P, Allen J, Christensen M, Davis P, Falin L, Grabmueller C, Hughes D, Humphrey J. Ensembl Genomes 2013: scaling up access to genome-wide data. Nucleic acids research. 2014;42:D546–D552. doi: 10.1093/nar/gkt979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Cotton S, Manna D, Welte M. Novel isoforms of the transport regulator Klar. PLoS One. 2013;8:e55070. doi: 10.1371/journal.pone.0055070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S, Nowak K, Suryavanshi N, Holt I, Shanahan C, Ridley A. Nesprin–1 and Nesprin-2 regulate endothelial cell shape and migration. Cytoskeleton. 2014 doi: 10.1002/cm.21182. [DOI] [PubMed] [Google Scholar]
- Kracklauer M, Banks S, Xie X, Wu Y, Fischer J. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly (Austin) 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- L’Hernault S, Rosenbaum J. Chlamydomonas alpha-tubulin is posttranslationally modified in the flagella during flagellar assembly. J Cell Biol. 1983;97:258–263. doi: 10.1083/jcb.97.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- L’Hernault S, Rosenbaum J. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- Lee C, Chen L. Dynamic behavior of endoplasmic reticulum in living cells. Cell. 1988;54:37–46. doi: 10.1016/0092-8674(88)90177-8. [DOI] [PubMed] [Google Scholar]
- Long J, Bagonis M, Lowery L, Lee H, Danuser G, Van Vactor D. Multiparametric Analysis of CLASP-Interacting Protein Functions during Interphase Microtubule Dynamics. Molecular and Cellular Biology. 2013;33:1528–1545. doi: 10.1128/MCB.01442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowery L, Lee H, Lu C, Murphy R, Obar R, Zhai B, Schedl M, Van Vactor D, Zhan Y. Parallel genetic and proteomic screens identify Msps as a CLASP-Abl pathway interactor in Drosophila. Genetics. 2010;185:1311–1325. doi: 10.1534/genetics.110.115626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumb J, Connell J, Allison R, Reid E. The AAA ATPase spastin links microtubule severnig to membrane modelling. Biochimica et Biophysica Acta. 2012;1823:192–197. doi: 10.1016/j.bbamcr.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Maruyama R, Andrew D. Drosophila as model for epithelial tube formation. Dev Dyn. 2012;241:119–135. doi: 10.1002/dvdy.22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellad J, Warren D, Shanahan C. Nesprins LINC the nucleus and cytoskeleton. Current Opinion in Cell Biology. 2011;23:47–54. doi: 10.1016/j.ceb.2010.11.006. [DOI] [PubMed] [Google Scholar]
- Metzger T, Gache V, Xu M, Cadot B, Folker E, Richardson B, Gomes E, Baylies M. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myat MM, Andrew DJ. Epithelial tube morphology is determined by the polarized growth and delivery of apical membrane. Cell. 2002;111:879–891. doi: 10.1016/s0092-8674(02)01140-6. [DOI] [PubMed] [Google Scholar]
- Patel U, Davies S, Myat MM. Receptor-type guanylyl cyclase Gyc76C is required for development of the Drosophila embryonic somatic muscle. Biology Open. 2012;1:507–515. doi: 10.1242/bio.2012943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellinen T, Ivaska J. Integrin traffic. J Cell Sci. 2006;119:3723–3731. doi: 10.1242/jcs.03216. [DOI] [PubMed] [Google Scholar]
- Pines M, Fairchild M, Tanentzapf G. Distinct regulatory mechanisms control integrin adhesive processes during tissue morphogenesis. Developmental Dynamics. 2011;240:36–51. doi: 10.1002/dvdy.22488. [DOI] [PubMed] [Google Scholar]
- Pirraglia C, Myat MM. Genetic Regulation of Salivary Gland Development in Drosophila melanogaster. In: Tucker A, Miletich I, editors. Salivary Glands: Development, Adaptations and Disease. 14. S. Karger; 2010. pp. 32–47. [DOI] [PubMed] [Google Scholar]
- Pirraglia C, Walters J, Ahn N, Myat M. Rac1 GTPase acts downstream of αPS1βPS integrin to control collective migration and lumen size in the Drosophila salivary gland. Dev Biol. 2013;377:21–32. doi: 10.1016/j.ydbio.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raigor D, Mellad J, Soong D, Rattner J, Fritzler M, Shanahan C. Mammalian microtubule P-body dynamics are mediated by nesprin-1. J Cell Biol. 2014;205:457–475. doi: 10.1083/jcb.201306076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raigor D, Shanahan C. Nesprins: from the nuclear envelope and beyond. Expert Rev Mol Med. 2013;15:e5. doi: 10.1017/erm.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashmi R, Eckes B, Glockner G, Groth M, Neumann S, Gloy J, Sellin L, Walz G, Schneider M, Karakesisoglou I, et al. The nuclear envelope protein Nesprin-2 has roles in cell proliferation and differentiation during wound healing. Nucleus. 2012;3:172–186. doi: 10.4161/nucl.19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A, Vale R. The Drosophila Homologue of the Hereditary Spastic Paraplegia Protein, Spastin, Severs and Disassembles Microtubules. Curr Biol. 2005;15:650–655. doi: 10.1016/j.cub.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Sherwood N, Sun Q, Xue M, Zhang B, Zinn K. Drosophila Spastin Regulates Synaptic Microtubule Networks and Is Required for Normal Motor Function. PLoS Biology. 2004;2:e429. doi: 10.1371/journal.pbio.0020429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubeita G, Tran S, Xu J, Vershinin M, Cermelli S, Cotton S, Welte M, Gross S. Consequence of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 2008;135:1098–1107. doi: 10.1016/j.cell.2008.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D, Fridolfsson H. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fischer JA. KASH’n Karry: the KASH domain family of cargo-specific cytoskeletal adaptor proteins. Bioessays. 2005;27:1136–1146. doi: 10.1002/bies.20312. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer C, Salmon E. Endoplasmic reticulum membrane tubules are distributed by microtubules in living cells using three distinct mechanisms. Curr Biol. 1998;8:798–806. doi: 10.1016/s0960-9822(98)70321-5. [DOI] [PubMed] [Google Scholar]
- Welte MA, Gross SP, Postner M, Block SM, Wieschaus EF. Developmental Regulation of Vesicle Transport in Drosophila Embryos: Forces and Kinetics. Cell. 1998;92:547–557. doi: 10.1016/s0092-8674(00)80947-2. [DOI] [PubMed] [Google Scholar]
- Xu N, Bagumian G, Galiano M, Myat MM. Rho GTPase controls Drosophila salivary gland lumen size through regulation of the actin cytoskeleton and Moesin. Development. 2011;138:5415–5427. doi: 10.1242/dev.069831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Keung B, Myat M. Rho GTPase controls invagination and cohesive migration of the Drosophila salivary gland through Crumbs and Rho-kinase. Developmental Biology. 2008;321:88–100. doi: 10.1016/j.ydbio.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Yu J, Lei K, Zhou M, Craft C, Xu G, Zhuang Y, Xu R, Han M. KASH protein Syne-2/Nesprin-2 and SUN proteins SUN1/2 mediate nuclear migration during mammalian retinal development. Hum Mol Genet. 2011;20:1061–1073. doi: 10.1093/hmg/ddq549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.