Abstract
This report describes a novel, one-pot synthesis of hybrid nanoparticles formed by a nanostructured inorganic silica core and an organic pH-responsive hydrogel shell. This easy-to-perform, oil-in-water emulsion process synthesizes fluorescently-doped silica nanoparticles wrapped within a tunable coating of cationic poly(2-diethylaminoethyl methacrylate) hydrogel in one step. Transmission electron microscopy and dynamic light scattering analysis demonstrated that the hydrogel-coated nanoparticles are uniformly dispersed in the aqueous phase. The formation of covalent chemical bonds between the silica and the polymer increases the stability of the organic phase around the inorganic core as demonstrated by thermogravimetric analysis. The cationic nature of the hydrogel is responsible for the pH buffering properties of the nanostructured system and was evaluated by titration experiments. Zeta-potential analysis demonstrated that the charge of the system was reversed when transitioned from acidic to basic pH and vice versa. Consequently, small interfering RNA (siRNA) can be loaded and released in an acidic pH environment thereby enabling the hybrid particles and their payload to avoid endosomal sequestration and enzymatic degradation. These nanoparticles, loaded with specific siRNA molecules directed towards the transcript of the membrane receptor CXCR4, significantly decreased the expression of this protein in a human breast cancer cell line (i.e., MDA-MB-231). Moreover, intravenous administration of siRNA-loaded nanoparticles demonstrated a preferential accumulation at the tumor site that resulted in a reduction of CXCR4 expression.
Keywords: pH-responsive polymer, nanoparticles, lysosomal escape, siRNA delivery, cancer therapy
1. Introduction
New trends in polymer science have led to the development of materials responsive to the physical and chemical stimuli of the biological environment [1, 2]. Polymeric “smart” hydrogels have been designed that can swell [3], dissolve [4], and release [1, 5] a therapeutic payload in response to fluctuations in biological temperature and pH. Nanoscale, pH-responsive hydrogel particles are considered as promising drug delivery platforms due to their ability to release a payload in a timely fashion by responding to physiological changes of the microenvironment [1, 2, 6]. This approach has significant potential in improving the use of small interfering RNA (siRNA) as a therapeutic agent by protecting them from intracellular nucleases found within the acidic endolysosomal vesicles [2, 7] and favor their release in the cell cytoplasm after internalization. However, the use of a hydrogel matrix as a drug delivery vehicle is limited by its low structural integrity and poor mechanical stability [8]. Furthermore, although the mechanical properties of hydrogels can be strengthened by increasing its amount of crosslinking, this also can affect its environmental responsiveness and biocompatibility [9]. For example, pH-responsive hydrogels have been demonstrated to swell within a specific pH range, however, when such polymers are synthesized with an increased crosslinking density, they tend to swell less and particularly in more extreme pH conditions [10-15].
The immunogenic properties of hydrogels largely depend on their specific and qualitative composition rather than on their degree of crosslinking, thus the majority of polymeric smart hydrogels display little or no immunogenicity [16-21]. On the other hand, a high degree of crosslinking may affect the cytotoxicity of these polymers [3, 9, 22]. In most cases, the crosslinking process involves the free radical polymerization of short cross-linkers with a polymer chain. Unreacted free radicals that remain in the polymer network may adversely affect the cytocompatibility of the polymer system [9, 23]. These limitations can be overcome by developing a hybrid system based on the incorporation of a rigid inorganic core with a minimally crosslinked soft hydrogel matrix. The inorganic core stabilizes the hybrid system while the hydrogel matrix retains the environmental sensitivity [8, 24].
The synthesis of pH-responsive hybrid nanoparticle delivery platforms is typically based on coating the particle surface with poly(ethyleneimine) [4], poly(methacrylic acid) [1], or poly(2-diethylaminoethyl methacrylate) (PDEAEM) [25-27]. Synthesis usually entails a lengthy, multistep process consisting of: particle synthesis, surface modification with a chemical linker, hydrogel coating, and labeling with a fluorescent tag [25, 28]. Herein, we describe a one-pot, oil-in-water emulsion synthesis process for hybrid, monodispersed, pH-responsive, fluorescent nanoparticles capable of: (i) protecting a siRNA payload from the extracellular and intracellular microenvironment; (ii) escaping together with their payload from endosomal vesicles, and (iii) releasing the siRNA cargo into the cytoplasm where it can efficiently exert its biological action in vitro and in vivo.
2. Results and Discussion
2.1. Synthesis of hybrid nanoparticles
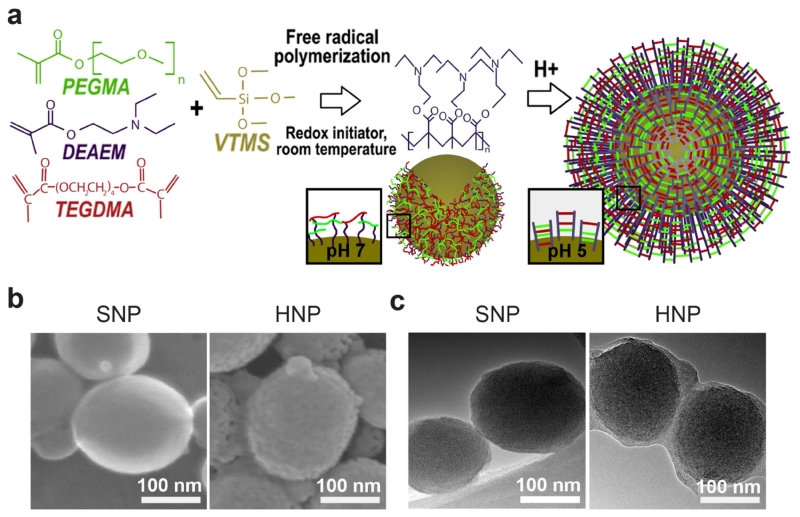
We have developed a novel one-pot synthesis approach to produce hybrid nanoparticles (HNP). We applied a self-assembly covalent stabilization strategy to produce a nanoscale single particle as a sphere with a core-shell morphology. This one-pot procedure is based on the free radical co-polymerization of the cationic monomer 2-(diethylamino)ethyl methacrylate (DEAEM) and the monomeric silica precursor vinyltrimethoxysilane (VTMS) in the presence of polyethylene glycol methacrylate (PEGMA) and triethylene glycol dimethacrylate (TEGDMA) linkers. This procedure yielded the in-situ formation of a cross-linked PDEAEM hydrogel shell around the silica nanoparticle (SNP) core (Fig. 1a). The resulting HNP consisted of a system able to buffer and swell in acidic pH. Scanning electron micrographs revealed that the HNP surface was rougher than the SNP surface (Fig. 1b), while transmission electron micrographs confirmed the presence of a distinctive 10 nm polymer layer surrounding the silica core (Fig. 1c).
Fig. 1.
(a) Schematic representation of hybrid nanoparticle (HNP) synthesis and pH-responsiveness; (b) SEM images of silica nanoparticles (SNP) and HNP, respectively; (c) TEM images of SNP and HNP, respectively.
2.2. Physicochemical characterization of hybrid nanoparticles
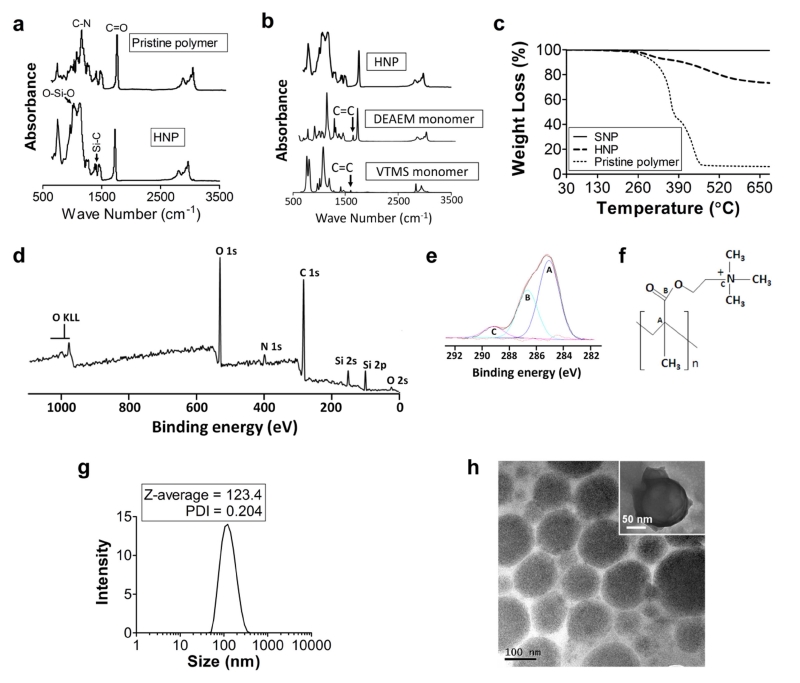
The Fourier transform infrared spectroscopy (FTIR) analysis of HNP confirmed the formation of chemical bonds between PDEAEM and silica (Fig. 2a). The infrared bands at 1135 cm−1 and 1702 cm−1 on the spectrum of the pristine hydrogel correspond to the C-N stretch of the tertiary amine and the C=O stretch of the PDEAEM carbonyl group, respectively [29, 30]. The HNP spectrum showed a band at 1080 cm−1 (Fig. 2a) that represents an oxygen-silicon-oxygen (O-Si-O) bond, as well as a band at 1451 cm−1 that represents the asymmetric distortion vibration of the Si-C bond [26, 28]. Moreover, a comparison between the FTIR spectra of the DEAEM monomer, the VTMS monomer and the HNP (Fig. 2b) demonstrated polymerization between the two monomers. The disappearance of the peaks that correspond to the unsaturated bond (C=C) of the VTMS and PDEAEM in the infrared spectrum of the HNP suggests that in this hybrid system, the silica was bound to the polymeric hydrogel through a C-C covalent linker that resulted from the polymerization of vinyl-terminated VTMS (C=C) with PDEAEM [31, 32].
Fig. 2.
Chemico-physical characterization of the HNP. (a) Fourier transform infrared (FTIR) spectroscopy study of the HNP and pristine polymer. (b) FTIR study of the polymerization: HNP, DEAEM monomer and monomeric silica precursor VTMS. (c) thermogravimetric analysis of SNP, HNP and pristine polymer. XPS analysis: (d) XPS survey spectrum; (e) C1s core level spectrum of the HNP surface deconvoluted into the three component peaks (A, B and C) relative to the three types of carbon atoms. (f) Labeling of the different carbon moieties in the PDEAEM molecule. Dispersion study of the HNP: (g) Dynamic light scattering study; (h) TEM analysis (inset showing a single particle).
The hydrogel:silica ratio was evaluated by thermogravimetric analysis [33]. HNP, when heated to 700 °C, displayed an average weight loss of 25% (Fig. 2c). Conversely, pristine polymer heated to the same temperature completely decomposed, while the weight loss of the SNP was negligible. Also, the onset degradation temperature of HNP was higher than that of pristine polymer (ΔT=~40°C). This was likely due to the enhanced adhesion between the polymer chain and silica particles which acted as a barrier to the volatile materials and increased the HNP thermal stability [34, 35].
We also evaluated the HNP surface structure using X-ray photoelectron spectroscopy (XPS). The wide-scan XPS spectrum of the hybrid surface (Fig. 2d) shows the peaks corresponding to carbon, oxygen, nitrogen and silicon atoms at characteristic binding energies. Figure 2e illustrates the C1s core level spectra, deconvoluted into three component peaks (i.e., A, B, C). The intensity ratios of these deconvoluted peaks are in agreement with the stoichiometric carbon atoms in the chemical structure of PDEAEM as [A]:[B]:[C] = 5:3:1 (see Fig. 2f). These results confirm that the grafted polymer layer on the silica surface is composed of PDEAEM chains.
We next evaluated the polydispersity of HNP in aqueous media using dynamic light scattering (DLS) [36] analysis (polydispersity index 0.204, Fig. 2g) and transmission electron microscopy (TEM) (Fig. 2h). These experiments showed that the particles were uniformly dispersed when placed in an aqueous environment.
2.3. pH-responsiveness of the HNP
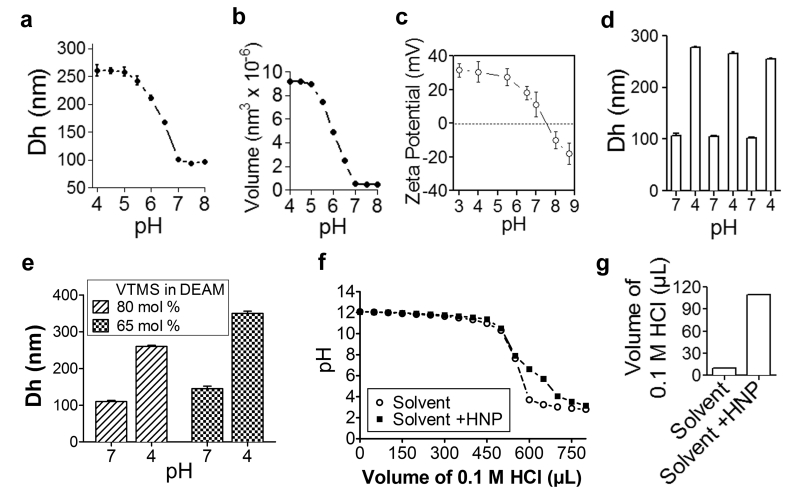
To determine pH-responsiveness, we studied a suspension of 0.1 mg mL−1 HNP at different pH values using DLS. This analysis demonstrated that the solvated hydrodynamic diameter (Dh) of HNP was inversely dependent on the environmental pH (i.e., the Dh increased as pH decreased) (Fig. 3a). In an acidic environment, the tertiary amine group of the PDEAEM shell is protonated, causing the shell to acquire a positive charge as previously reported [37]. The strong chain-solvent interactions and inter-molecule electrostatic repulsion induced polymer chain stretching and increased Dh (as schematically shown in Fig. 1a). This caused the hydrodynamic volume of the HNP to increase from 0.5 × 106 nm3 to 8.6 × 106 nm3 as the environmental pH changed from 7 to 5, resulting in a 17-fold increase in hydrodynamic volume (Fig. 3a,b). However, in neutral and alkaline conditions, the PDEAEM chains were deprotonated, thereby becoming hydrophobic and contracting against the surface of the silica core [38, 39]. This contraction favored the creation of polymer chain-chain interactions, which being stronger than chain-solvent interactions, induced chain aggregation and decreased the HNP Dh at physiological and alkaline pH (Fig. 3a). As expected, the analysis of the Dh as a function of the pH revealed a sigmoidal pattern during the HNP transition from a collapsed state to a swollen state [26, 37]. Zeta potential analysis confirmed the protonation of the nanogel coating as witnessed by a progressive shift towards a positive charge, shifting the pH towards acidic (Fig. 3c). This event was likely a result of the adsorption of the solvated ion (H+) at pH values below 7 [26]. Moreover, the HNP underwent a reversible swelling-shrinking cycle as the pH decreased from 7 to 4. The swelling ratio of the HNP remained almost constant with a 10% loss of polymer at the end of the third cycle, likely as a consequence of the increased solubility of PDEAEM in the acidic environment (Fig. 3d).
Fig. 3.
pH responsiveness study of HNP: (a) hydrodynamic diameter (Dh) and (b) volume of the nanogel particles in varying pH environments; (c) surface zeta-potential of the HNP in varying pH environments. (d) Reversible response of the HNP with varying pH. (e) Dh of HNP synthesized by using two different monomer-to-precursor ratios. (f) Titration study of the HNP: decreasing pH profiles of the experimental and control suspensions; (g) volume of HCl required to decrease the pH from 7.1 to 4.1 in the experimental and control suspensions.
To evaluate the tunability of our system, we synthesized HNP with varying thicknesses of polymer coating. HNP pH-dependent swelling was directly correlated with the amount of PDEAEM bound on the surface of the silica core (Fig. 3e). This observation indicates that the hydrogel coating functioned as an on-off switch, linked to the transition from neutral to acidic environments, and an indication that the swelling, as a result of pH, can be tailored by regulating the copolymer ratio and coating thickness.
Additionally, protonation of HNP has been shown to produce a buffering effect on the surrounding environment [40, 41]. We assessed this activity by titrating a basic aqueous solution of 0.1 M NaCl (solvent) with 0.1 M HCl, with and without HNP. Compared to the solvent, the presence of the HNP resulted in a slower decrease of the pH below 7 (Fig. 3f) and required a larger volume of HCl to decrease the pH of the solution from 7.1 to 4.1 (Fig. 3g). The HNP net buffering capacity was 11.0 which represents the μmol of H+ required to decrease the pH of 1 mg mL−1 of suspension from 7.1 to 4.1 [42].
These results are in agreement with those reported by Peppas and colleagues [26, 43], who demonstrated that PDEAEM has a pKa of about 7.2. Therefore the buffering properties of this polymer can exploit the endosomal acidic environment (pH < 7.2). In addition, these results strongly suggest that the hybrid nanogel prevented acidification of the suspension by chelating H+ ions. This process, known as the “proton sponge effect”, has been shown to favor the escape of nanoparticles from endosomal vesicles [39, 44, 45].
2.4. siRNA loading and release
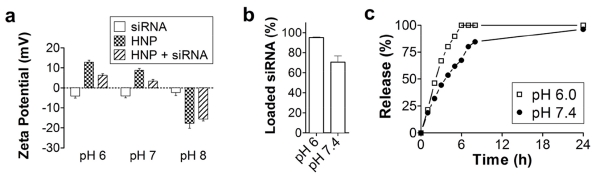
Loading of siRNA was driven via the electrostatic interaction between the negatively charged molecules of nucleic acid and the positive charge of the HNP polymer shell. The measurement of the surface charge of the HNP revealed that the particles maintained a positive surface charge at pH 6 and 7, whereas a negative charge was recorded at pH 8 (Fig. 4a). Conversely, free siRNA displayed a negative surface charge throughout all pH conditions (Fig. 4a) and resulted in a slight pH shift when loaded within HNP. We next evaluated the loading efficiency of the siRNA molecules in acidic and physiological pH conditions, and obtained a loading efficiency of 95% at pH 6 compared to 70% at pH 7.4 (Fig. 4b). This difference was likely due to the higher affinity the particles had for siRNA as a direct result of polymer protonation in acidic conditions. Therefore, in the subsequent experiments, the siRNA was loaded into the HNP only at pH 6.
Fig. 4.
siRNA loading and release. (a) Surface z-potential analysis of siRNA and the HNP at different pH values before and after loading. (b) Loading of siRNA at pH 6 and 7.4. (c) Release of siRNA from the HNP at pH 6 and 7.4.
The release of siRNA was investigated in physiologic and acidic conditions. After 4 h of incubation, the cumulative siRNA release from the siRNA-HNP reached ~80% at pH 6 with a 53% release at pH 7.4 (Fig. 4c). Despite the higher affinity of the HNP for the payload, siRNA release was accelerated in acidic conditions likely as a result of hydrogel swelling (Fig. 3a), thereby increasing the solubility of the coating and favoring the diffusion of the payload into the surrounding environment. Conversely, the release of siRNA was reduced in physiological pH even though the affinity of the HNP for siRNA was lower in this environmental condition. We assume that the entrapment of siRNA in the collapsed hydrogel structure delayed the release of siRNA molecules in the presence of physiologic pH.
2.5. Biological effects of the HNP
2.5.1. Cytotoxicity and endolysosomal effect of the HNP
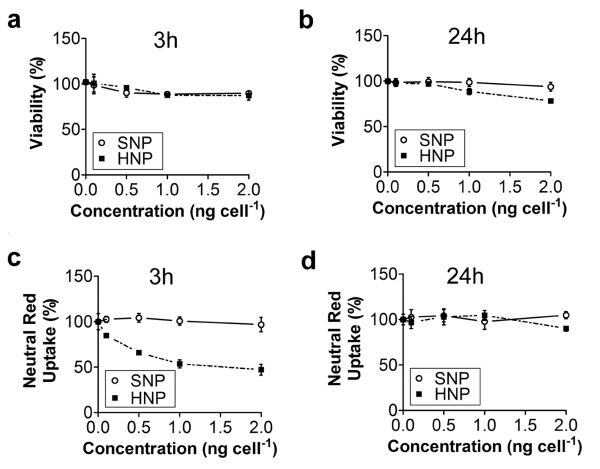
To evaluate the ability of HNP to deliver a bioactive payload, we used a human breast cancer cell line, MDA-MB-231. The effect of particles on cell viability was assessed using a (3-(4,5-dimethylthiazol-2-yl)2,5-diphenyltetrazolium bromide) (MTT) assay which showed that both SNP and HNP had a negligible impact at concentrations of 0-2 ng cell−1 concentrations (Fig. 5a,b).
Fig. 5.
Evaluation of the impact of SNP and HNP on cell viability measured by MTT assay at 3 h (a) and 24 h (b). The data are normalized against untreated control cells. In the concentration range and time points tested, the effect of SNP and HNP on cell viability did not exceed 20%. Evaluation of neutral red dye uptake in endolysosomal vesicles at 3 h (c) and 24 h (d) after treatment with SNP and HNP. The data are normalized against untreated control cells. HNP greatly impaired the ability of lysosomal vesicles to retain this viable dye 3 h after treatment. Error bars represent standard deviation (n=6)
The endolysosomal function of MDA-MB-231 cells exposed to HNP, at doses ranging from 0.5 ng cell−1 to 2 ng cell−1, was evaluated by measuring the ability of cells to incorporate neutral red dye into the endolysosomal vesicles [46, 47]. As expected, HNP impaired the ability of the endosomal compartment to retain the dye. In particular, 3 h after treatment, the HNP reduced endosomal function, probably as a consequence of the “proton sponge effect” (Fig. 5c). The ability to incorporate the dye into the intracellular acidic vesicles was restored 24 h after treatment with the lowest concentrations of HNP (0.5 - 1.5 ng cell−1), whereas this ability was still impaired 24 h after treatment with 2 ng cell−1 of HNP (Fig. 5d). Taken together, these results suggest that high doses of HNP do not affect MDA-MB-231 cell viability, associated with brief obstruction of endosomal cell functions likely attributed to the disruption of endosomal vesicles.
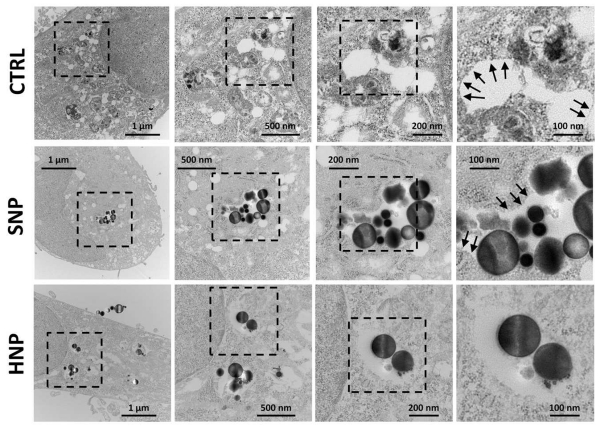
To further evaluate the potential of HNP to escape endolysosomal compartments, we analyzed cells treated with HNP, SNP, and untreated cells by TEM (Fig. 6). Cells were analyzed 3 h after treatment with 0.3 ng cell−1 of particles, a dose that did not cause any cytotoxic effect (Fig. 5a), while the same dose caused the loss of neutral red uptake (Fig. 5c). TEM images showed that untreated and SNP-treated cells maintained distinguishable vesicles as demonstrated by a regular, round shape delimited by a compact border. Conversely, cells treated with HNP contained subcellular structures that resembled vesicular or vacuolar organelles, although these structures were irregular in shape and had a smooth, non-continuous border. Additionally, SNP tended to remain grouped and were clearly confined to vesicular structures that had relatively compact borders (Fig. 6) while HNP were scattered throughout the cytoplasm. These observations suggest that the HNP broke out from endolysosomal vesicles and, at least partially, compromised the normal intracellular vesicle trafficking.
Fig. 6.
TEM images of MDA-MB-231 cells 3 h after treatment with HNP. SNP-treated and untreated (CTRL) cells served as a control.
2.5.2. siRNA intracellular delivery
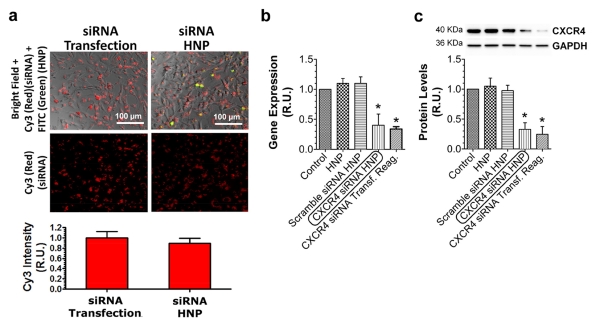
To evaluate the efficiency of the HNP in mediating siRNA cellular internalization, we performed fluorescent microscopy analysis on MDA-MB-231 cells after internalization of fluorescently labeled siRNA either delivered by HNP or transfected using a commercial liposomal transfection reagent. HNP were loaded with the same amount of siRNA (0.166 pmol siRNA g−1 HNP; 0.3 ng particle cell−1; 5 × 10−2 fmol siRNA cell−1) that was used for transfection with reagent (5 × 10−2 fmol siRNA cell−1). This analysis revealed that the efficiency of the HNP in mediating siRNA cellular internalization was similar to that of the transfection reagent (Fig. 7a).
Fig. 7.
(a) Fluorescent microscopy analysis of MDA-MB-231 cells shows that the intracellular delivery of Cy3-labeled siRNA (red) mediated by FITC-labeled HNP (green) has an efficiency comparable to that of transfection. Error bars represent standard deviations (n=16). Evaluation of CXCR4 silencing in MDA-MB-231 cells 48 h after treatment: (b) Real-time RT-qPCR analysis of gene expression and (c) Western blot analysis of protein levels. siRNA-loaded HNP effectively decreased the expression of CXCR4 mRNA and protein. Error bars represent standard deviation (n=3). *=p<0.01 (compared to Control).
We evaluated the ability of the HNP to deliver active siRNA by treating the cells with siRNA-loaded HNP (3.3 pmol siRNA μg−1 HNP; 0.3 ng HNP cell−1) directed against CXCR4 mRNA (which encodes a chemokine receptor) [48] compared to HNP alone and scrambled siRNA-loaded HNP as negative controls. The expression levels of the CXCR4 gene and protein were analyzed 48 h after treatment using real-time qPCR and Western blot (Fig. 7b and c, respectively). CXCR4 siRNA-loaded HNP significantly reduced the expression of both the CXCR4 transcript and protein. Moreover, the silencing efficacy of siRNA-loaded HNP was similar to that of the liposomal transfection reagent at the same dose of siRNA (1 fmol cell−1). Indeed, the dose of siRNA we used was relatively high compared to other reports [49-54]; however, preliminary results indicated that lower concentrations of siRNA (0.05-0.2 fmol cell−1) did not cause a significant reduction of CXCR4 gene expression, after transfection with the liposomal reagent (data not shown).
2.5.3. siRNA in vivo delivery
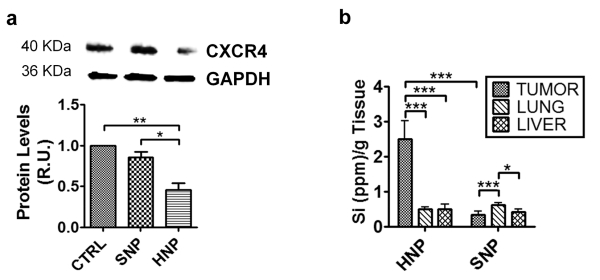
In order to evaluate the efficacy of HNP to deliver siRNA directed against CXCR4 in vivo, we employed an orthotopic human breast cancer mouse model. To this aim, MDA-MB-231 cells were inoculated into the mammary fat pad of female athymic nude mice. When mouse tumors reached approximately 5 mm diameter, they were randomly subdivided into three groups (n=10) and treated by tail vein injections. Mice were treated with: (i) CXCR4 siRNA-loaded HNP (3.3 pmol siRNA/μg NP, 92 μg NP/mouse, 0.3 nmol (5 μg) siRNA/mouse) resuspended in a sterile 0.9% saline solution; (ii) sterile 0.9% saline solution; (iii) or SNP and CXCR4 siRNA resuspended in sterile 0.9% saline solution, using the same amounts of NP and siRNA that was used for the first group (92 μg NP/mouse, 0.3 nmol [5 μg] siRNA/mouse). The treatments were repeated 3 and 5 days later and mice were sacrificed on day 7. Western blot analyses of extracts from homogenized tumors showed that siRNA-HNP effectively silenced CXCR4 expression, whereas CXCR4 expression was not significantly lower in tumors of mice treated with SNP and siRNA than in untreated controls (Fig. 8a). Moreover, the reduction of CXCR4 expression seemed to be comparable with that measured in the cell culture experiments (Fig. 7c). This suggest that, besides having a good transfection efficacy, HNP preferentially accumulated in the tumor. To verify this aspect, we analyzed the biodistribution of HNP and SNP in these mouse models. To this aim, we measured the silicon content in the tumor, heart, lungs, liver, kidney, spleen and peripheral blood of the three groups of mice using Inductively Coupled Plasma–Atomic Emission Spectroscopy. Readings for blood, kidney, spleen and heart were below the detection limits, while significant amounts of silicon were found in the tumor, lungs, and liver (Fig. 8b). As shown in Fig. 8b, the level of silicon was 10-fold higher in the tumor of HNP-treated animals than in those of SNP-treated animals. Moreover, in SNP-treated mice, the level of silicon was higher in lung than in the tumor. We can argue that HNP bearing polyethylene glycol (PEG) chains on their surface (Fig. 1a) remained longer in the circulation than did SNP and thus had a greater chance of penetrating the tumor microenvironment, as in the case of many PEG-based systems [55, 56]. Indeed, in HNP-treated animals the silicon content was 5-fold higher in the tumor than in the lung or liver. Taken together, these data suggest that the in vivo efficacy of gene silencing with siRNA-HNP was due not only to the latters’ endolysosomal escape but also to their propensity to accumulate at the tumor site.
Fig. 8.
siRNA-HNP in vivo delivery. (a) Western blot analysis of CXCR4 levels in the tumor of mice models of human breast cancer shows a high gene silencing efficacy of siRNA-HNP. The results represent the average of three independent experiments (n=3) and error bars represent standard deviations. *=p<0.05, **=p<0.01. (b) NP biodistribution analysis by ICP-AES shows that HNPs target the tumor while SNPs accumulate prevalently in the lung. The results represent the average of four independent experiments (n=4) and error bars represent standard deviations. *=p<0.05, **=p<0.01, ***=p<0.005.
3. Conclusions
The one-pot synthesis described in this paper is an easy-to-perform efficient means with which to produce a nanovector able to deliver a payload in the cytoplasm. The in situ incorporation of a fluorescent dye within the silica lattice during the synthesis procedure resulted in the production of inherently pH-responsive fluorescent nanoparticles that do not require further functionalization with a fluorescent tag. The basic nature of the tertiary amine-terminated monomer favored the silane hydrolysis [57] and the subsequent co-polymerization reaction between the polymer chain and the silica nanoparticles resulted in a stable and uniform polymeric nanolayer with a well dispersed and reproducible composition. Moreover, HNP exhibited an increase in thermal stability when compared to pristine polymer and displayed favorable buffering properties in an acidic environment associated with an increased hydrodynamic volume. In addition, the tuning of pH-response by varying the silica:hydrogel ratio enabled the pH-dependent loading and release of siRNA and facilitated the endosomal escape of the nanoparticles through the “proton sponge effect”.
As demonstrated by the group of Peppas [26, 43], PDEAEM has a pKa of ~7.2, therefore, its buffering properties can exploit the endosomal acidic environment (pH < 7.2). Thanks to this phenomenon, CXCR4 siRNA was efficiently delivered into the cytoplasm of MDA-MB-231 breast cancer cells and inhibited the protein expression of CXCR4 with an efficacy comparable to that of a commercial transfection reagent [58, 59]. Moreover, HNP demonstrated an excellent efficacy in delivering siRNA in vivo. In fact, HNP preferentially accumulated in the tumors of human breast cancer-bearing mice and mediated CXCR4 silencing in these tumors. In conclusion, we describe a new synthesis protocol with which to produce cost-effective and quickly synthesizable nanovectors potentially useful as non-viral carriers for siRNA therapeutics; this protocol bypasses all the intermediate steps of surface functionalization that are usually necessary to produce hybrid particles.
4. Material and Methods
Chemicals
DEAEM (99%), tetraethylene glycol dimethacrylate (TEGDMA, 90%), N,N,N’,N’-tetraethylmethanediamine (TEMED, 97%), ammonium persulfate (APS, 98% ACS reagent), sodium metabisulfite (NaMBS), Triton X-100, myristyltrimethylammonium bromide (Mytab), vinyltrimethoxysilane (VTMS, 98%), and (3-aminopropyl)triethoxysilane (APTES, 99%) were purchased from Sigma-Aldrich (St. Louis, MO). Poly(ethylene glycol)monomethyl ether monomethacrylate (PEGMA, Mn=1000) and fluorescein isothiocyanate (FITC) were obtained from Polyscience Inc. (Warrington, PA) and Thermo Scientific (Thermo Fisher Scientific Inc., Waltham, MA) respectively.
Synthesis of hybrid nanogel
In-situ polymerization of DEAEM occurs in the presence of TEGDMA and PEGMA that act as crosslinkers with VTMS monomer (silica precursor). Synthesis occurred by free radical polymerization through an oil-in-water emulsion technique. All monomers were passed through a column of basic alumina powder to remove inhibitors before use. 4.5 mL of DEAEM was mixed with 80 mol% of VTMS and 1 mol% of TEGDMA in a glass flask. The mixture also contained the polymerizing activator TEMED. To incorporate FITC into the silica matrix, 1.3 mL of APTES and 16 mg of FITC were introduced into the same flask. In a separate flask, 5% (w/v) solution of PEGMA in 100 mL of Millipore (Millipore, Billerica, MA)water was stirred until it formed a homogeneous solution. 0.4 g of Mytab and 100 μL of triton X-100 were introduced in the flask while stirring. The aqueous solution of PEGMA was then mixed with the monomer mixture into an oil-in-water emulsion. The mixture was then emulsified for 5 minutes using a Misonix probe sonicator (Misonix S-4000, Qsonica, LLC, Newtown, CT) at 80% amplitude and 60 W with 15 sec pulse on and 15 sec pulse off in a pyrex tube which was partially submerged in a stirred ice water bath. The emulsion was then purged with nitrogen gas for 20 minutes to eliminate dissolved oxygen. Two mol% (with respect to the PDEAEM) of initiator mixer (APS and NaMBS) in distilled water was added to the reaction mixture under stirring. The reaction was carried out for 24 h under continuous agitation in a flask equipped with a mechanical stirrer (IKA, Staufen, Germany) at 400 rpm at room temperature. The products were separated from the bulk mixture by centrifugation at 14,000 rpm for 10 minutes. The particles were then allowed to swell by introducing 0.5 M HCl solution followed by the addition of acetone for 10 minutes, and then centrifuged at 14,000 rpm for 10 minutes. To eliminate the surfactants and unreacted monomers, the particles were washed several times in repeated cycles of resuspension in acetone and water, and then centrifuged. The particles were then lyophilized on a Freezone freeze dryer and stored in a desiccator until use. To obtain particles that differed in the thickness of the polymer layer, similar experiments were done in parallel with 65 mol% VTMS with respect to the DEAEM monomer.
SEM and TEM analyses of nanoparticles
Scanning electron microscopy (SEM) images were acquired under high vacuum, at 20.00 kV, spot size 3.0–5.0, using a FEI Quanta 400 FEG ESEM equipped with an ETD (SE) detector (FEI Co., Hillsboro, OR). Samples were prepared by introducing 1 drop (about 2 μL) of aqueous suspension (0.1mg/mL) of the particles on the SEM stub (Ted Pella Inc., Redding, CA) and allowing the solvent to dry in air. Samples were sputter-coated with a 10 nm layer of gold using a Plasma Sciences CrC-150 sputtering system (Torr International Inc., New Windsor, NY) before the analysis. High-resolution transmission electron microscopy was performed using a FEI Technai electron microscope (FEI Co.) at a tension of 200 kV. Samples were prepared by placing 1 drop of 0.1 mg mL−1 aqueous suspension of the hybrid nanogel particles on the 300 mesh carbon-coated copper grids (Ted Pella) and allowing the solvent to evaporate in air.
FTIR analyses
Fourier transform infrared (FT-IR) spectra were obtained on a Nicolet 6700 spectrometer (Thermo Scientific) with a deuterated triglycine sulfate (DTGS) detector and potassium bromide (KBr) beam splitter. Typically, 64 scans were performed in the wave number range of 4000–600 cm−1, both forward and backward at 4 kHz, with a manual gain of one. Pellets of about 10 mg of sample and 200 mg KBr (spectroscopy grade) (Thermo Scientific) were pressed using a Carver laboratory press. Spectra were collected by transmitting IR light through the pellets.
XPS analyses
We examined the surface of the hybrid nanogel particles also using X-ray photoelectron spectroscopy (XPS). XPS measurements can probe a maximum sampling depth of approximately 3-5 nm and therefore provide information about surface properties. XPS measurements were made with a PHI Quantera XPS (Physical Electronics, Inc, Chanhassen, MN), which uses a focused monochromatic Al Kα X-ray (1486.7 eV) source for excitation. The 40W, 15kV and 200 μm diameter X-rays were used on the sample. The vacuum pressure was maintained below 5.0 × 10−8 Tor during the analysis. The XPS survey scan spectra in the 1100.0 eV binding energy range were recorded in 0.5 eV steps with a pass energy of 140eV. High resolution scan spectra of C1s were recorded in 0.1 eV steps with a pass energy of 26 eV. Low energy electrons and Ar+ ions were conducted for specimen neutralization in each measurement. The spectral lines were referenced against the C1s signal at 284.50 eV. In spectral deconvolution, the widths of Gaussian peaks were maintained constant for all components in each spectrum.
DLS, Zeta-potential and pH titration
The hydrodynamic diameters of the hybrid nanoparticles were characterized using a ZetaPALS Zeta Potential Analyzer with a Multi Angle Particle Sizing Option installed (Brookhaven Instruments Corporation, Holtsville, NY), operating with a 25 mW laser at a wavelength of 660 nm. Scattered light was detected at 90° to the incident beam. Each sample was subjected to 3 three-minute measurements. Typically, 2.5 mL suspensions of the hybrid nanogel particles in phosphate buffer with physiological ionic strength (100 mM) at a concentration of 0.1 mg mL−1 were used for the characterization at a temperature of 25 °C. Phosphate buffer with a different pH was prepared and measured with an ISFET probe using a SevenEasy Mettler Toledo pH meter (Mettler Toledo International, Greifensee, Switzerland). In a separate experiment, the hybrid particles (using the same concentration as above) were dispersed in PB first at pH 7 and then were centrifuged and redispersed at pH 4; this cycle was carried out three times and the Dh was measured after each pH change (see Fig. 3c). The hydrodynamic diameter of the dispersed particles was measured by density light scattering to evaluate the reversible pH responsiveness in two different pH environments. The Zeta-potential was measured by electrophoretic light scattering using the ZetaPALS Zeta Potential Analyzer (Brookhaven Instruments Corporation, Holtsville, NY). 1.4 mL suspension of the hybrid nanogel particles in PB with an ionic strength of 100 mM with varying pH at a concentration of 0.1 mg mL−1 was used for Zeta-potential characterization. Scattered light was detected at an angle of 15° and at a temperature of 25°C. Each sample was subjected to three measurements.
To determine the buffering capacity of the hybrid nanogel particles in aqueous environment, we examined protonation of the particles by pH titration. In a typical experiment, 1 mg mL−1 suspension of the particles in 20 mL NaCl solution with an ionic strength of 0.1 M was initially rendered basic by introducing 500 μL of 0.1 M NaOH. While the HCl (0.1M) was added stepwise to each suspension the pH was measured using a pH meter (AB15, Accumet Basic, Fisher Scientific). The same experiment was done with a blank solution lacking any particle as a control.
TGA
Thermogravimetric analysis of hybrid nanogels, pristine polymer and bare silica particles was performed after these samples (~16.0 mg) were dried under vacuum at 40°C for 24 h. TGA was performed using a Q600 SDT (TA Instruments Inc., New Castle, DE) by heating the particles from 30°C to 700°C in nitrogen atmosphere at a rate of 20°C min−1.
Loading & release of siRNA
A Cy3-labeled reporter siRNA (Ambion, Thermo Fisher Scientific Inc.) was used to determine the pH-mediated loading and release kinetics of the HNP system. Polyplexes were formed by mixing 5 nmol of Cy3-labeled siRNA with the HNP at an amino/phosphate (N/P) ratio of 40 in PB solutions at a pH of 6 or 7.4. In a typical procedure, a 100 μL aliquot of 300 μg HNP were mixed with 100 μL aliquot of 0.3325 μg siRNA in PB solution. The mix was gently shaked at room temperature for 15 minutes. The resulting mixture was left at room temperature for another 15 minutes. The solution was then spun down at 21,000 g for 5 minutes. The loading was evaluated by analyzing the fluorescence intensity of the supernatant with a spectrofluorimeter. Standard procedures were followed to prepare the PB solution with certain pH before the complex formation. To investigate the pH-mediated release of siRNA, the siRNA-HNP complexes loaded at pH 7 were incubated in PB solution at pH 6 or 7.4. The amount of siRNA released was monitored by evaluating Cy3 fluorescence intensity with a spectrofluorimeter.
Cell cultures
The cells used in this study were from the human “triple-negative” breast cancer cell line MDA-MB-231 (ATCC# HTB-26, LGC Standards, Teddington, Middlesex, UK ). They were cultured at 37°C and 5% CO2 in DMEM (Thermo Scientific) culture medium supplemented with 10% (v/v) FBS and 1% (v/v) penicillin-streptomycin solution (Thermo Scientific).
MTT assay
Cells were seeded in 96-well microplates (Costar, Corning Inc., Corning, NY) at a density of 25 × 103. Twenty-four hours after cell attachment, plates were washed with PBS, and the cells were treated with increasing concentrations, from 0.1 to 2 ng cell−1, of both SNP and HNP for 3 or 24 hours. Six replicate wells were used for each control and tested concentration. The tetrazolium salt (MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide]) was dissolved in PBS (5 mg/mL) and added to MDA-MB-231 cells (100 μL mL−1 DMEM without serum or phenol red) according to the method of Mosmann.[60] After incubation for 3 hours at 37°C, a solution of 1 N hydrogen chloride-isopropanol (1:24, v:v) was pipetted to each well, and mixed to dissolve the dark-blue formazan crystals formed. After a few minutes at room temperature, the plates were read at 570 nm in a BioTek Microplate reader.
Neutral red uptake assay
Human breast cancer MDA-MB-231cells were seeded as described above and treated with SNP or HNP and exposed to a medium (without serum or phenol red) containing 50 μg mL−1 neutral red dye and processed according to the method of Borenfreund and Puerner [61]. After incubation for 3 hours at 37°C, cells were washed twice in PBS and fixed according to the procedure described by Riddell et al [62]. T he plates were then left at room temperature for 10 minutes, and the absorbance of the released neutral red dye was read at 540 nm in a BioTek Microplate reader (BioTek Instruments, Winooski, VT).
TEM analyses of cells
For TEM analyses N × 105 MDA-MB-231 cells were seeded in each well of a 6-well plate. The next day, the cells were treated with 0.3 ng cell−1 of HNP or SNP. Untreated cells were used as a control. Three hours after the treatment, the cells were washed and fixed in a solution of 2% (w/v) paraformaldehyde (Electron Microscopy Sciences; Hatfield, PA) and 3% (w/v) glutaraldehyde (Sigma-Aldrich) in 0.01 M phosphate buffered saline pH 7.4 (2 mL well−1; Sigma-Aldrich) for one hour. Fixed samples were washed in 0.1 M cacodylate buffer and treated with 0.1% Millipore-filtered buffered tannic acid, postfixed with 1% buffered osmium tetroxide for 30 min, and stained en bloc with aqueous 1% Millipore-filtered uranyl acetate. The samples were washed several times in water, then dehydrated in increasing concentrations of ethanol, infiltrated, and embedded in LX-112 medium. The samples were polymerized in a 60°C oven for about 3 days. Ultrathin sections were cut in a Leica Ultracut microtome (Leica, Deerfield, IL), stained with uranyl acetate and lead citrate in a Leica EM Stainer, and examined in a JEM 1010 transmission electron microscope (JEOL, USA, Inc., Peabody, MA) at an accelerating voltage of 80 kV at High Resolution Electron Microscopy Facility at the University of Texas MD Anderson Cancer Center. Digital images were obtained using AMT Imaging System (Advanced Microscopy Techniques Corp, Danvers, MA).
Evaluation of siRNA delivery through fluorescent microscopy
2 × 104 MDA-MB-231 cells were seeded in each well of a 96-well plate. The next day, cells were either transfected with siRNA using the HiPerFect transfection reagent (Qiagen, Hilden, Germany) or treated with siRNA-loaded hybrid nanoparticles. The siRNA was “Cy3-Labeled Negative Control siRNA” (Life Technologies, Thermo Fisher Scientific Inc.). For transfection, we used 1 pmol siRNA and 1 μL HiPerfect reagent in a final volume of 200 μL culture medium per well. Hybrid nanoparticles were loaded by using 0.16 pmol siRNA μg−1 NPs in PB buffer at pH 6, then they were washed twice in PB buffer at pH 7.4 and resuspended in the cell medium. 6 μg NPs per well were used in a final volume of 200 μL culture medium. After 3 hours cells treated with NPs were washed twice with PBS and the fresh culture medium was added. Twenty-four hours later cells were washed twice with PBS and supplemented with fresh culture medium. All experiments were performed in four replicates. The cells were observed with a Nikon fluorescence microscope using a 10× lens, and the images were acquired. Images were analyzed by using “NIS elements” (Nikon) and “ImageJ” (NCBI) softwares. To evaluate fluorescence intensity, four different fields of view were acquired from each well.
CXCR4 silencing
For the evaluation of gene silencing through real-time RT-PCR, 1.2 × 105 MDA-MB-231 cells were seeded in each well of a 12-well plate. The next day, cells were either transfected with siRNA by using the HiPerFect transfection reagent (Qiagen) or treated with siRNA-loaded hybrid nanoparticles. We used a cocktail (1:1 ratio) of two types of siRNA molecules (Qiagen, SI02664235, SI02664242) directed against two different regions of the CXCR4 transcript. For the transfection with the reagent we used 120 pmol siRNA and 6 μL HiPerfect reagent in a final volume of 1.2 mL culture medium per well. Hybrid nanoparticles were loaded by using 3.3 pmol siRNA μg−1 NPs in PB buffer at pH 6, then they were washed twice in PB buffer at pH 7.4 and resuspended in the cell medium. 36 μg NPs per well were used in a final volume of 1.2 mL culture medium. Every experiment was performed in 3 independent replicates.
To measure the reduction of the CXCR4 protein, 2.4 × 105 MDA-MB-231 cells were seeded in each well of a 6-well plate. The next day, cells were either transfected with siRNA by using the HiPerFect transfection reagent (Qiagen) or treated with siRNA-loaded hybrid nanoparticles. For the transfection we used 240 pmol siRNA and 12 μL HiPerfect reagent in a final volume of 2.4 mL culture medium per well. Hybrid nanoparticles were loaded by using 3.3 pmol siRNA μg−1 NPs and 72 μg NPs per well in a final volume of 2.4 mL culture medium. Each experiment was performed in 3 independent replicates.
Real time RT-qPCR analyses
Forty-eight hours after transfection and treatments, total RNA was extracted from cells with the RNeasy Mini Kit (Qiagen). cDNA was obtained through retrotranscription of 0.8 μg RNA by using Superscript II (Life Technologies) as reverse transcriptase and Oligo(dT) (Life Technologies) as primers. Real-time qPCR analysis was performed by using the StepOnePlus System (Life Technologies) as real-time thermal cycler, TaqMan Gene Expression Assays and TaqMan Gene Expression Master Mix. In particular, CXCR4 transcript levels were evaluated using, the TaqMan Gene Expression Assay CXCR4 Hs00607978_s1 (Life Technologies) and normalized using TaqMan Gene Expression Assay GAPDH Hs02758991_g1 (Life Technologies). The analysis was performed in triplicate for each biological replicate. Relative quantification of CXCR4 gene was obtained from averages of threshold cycle (Ct) using the 2−ΔΔCt method [63].Statistical significance was assessed by one-way two-tail paired T-test.
Cell lysis, protein extraction and Western blot analysis
Forty-eight hours after transfection, whole cell protein extracts were obtained by cell lysis using RIPA buffer (Thermo Scientific) with 1 μL Halt protease inhibitor cocktail (Thermo Scientific) per 100 μL of buffer. 40 μg of whole cell protein extracts were loaded onto SDS/PAGE and then transferred on to a PVDF membrane for Western blot analysis, as previously described [64]. Blots were incubated with rabbit anti-CXCR4 (Millipore) and rabbit anti-GAPDH (Abcam, Cambridge, UK) primary antibodies and then with horseradish peroxidase-conjugated anti-Rabbit IgG (Sigma–Aldrich) as secondary antibody. The bands were revealed with the SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific) and images were visualized and acquired with the ChemiDoc XRS+ System and Image Lab software (Bio-Rad Laboratories, Hercules, CA). Quantitative analysis of the bands was performed by using ImageJ software (NIH) and CXCR4 signals were normalized with GAPDH. The reported results are the average of three (n=3) independent experiments. Statistical significance was assessed by one-way two-tail paired T-test.
Mice models of human breast cancer
Female athymic nude mice (NCr-Fox1nu; 6-8 week old) were purchased from Charles Rivers Laboratories (Wilmington, MA, USA) and maintained as previously described) [65]. Orthotopic breast cancer models were established by implanting 2×106 MDA-MB231 cells in the mouse mammary fat pad. Experiments were performed in conformity with protocols approved by the veterinary department of the Italian Ministry of Health and in accordance to the ethical and safety rules and guidelines for the use of animals in biomedical research provided by the relevant Italian laws and European Union’s directives (no. 2010/63/EU). All efforts were made to minimize the animal’s suffering.
In vivo delivery of siRNA-HNP
Tumor-bearing mice were selected for treatment when the tumors reached approximately ~5 mm in diameter as measured with a digital caliper (14–21 days post-inoculation). These mice were randomly distributed in 3 groups (n = 10 per group). Prior to treatments, each mouse was anesthetized, and 100 μL of sterile 0.9% saline solution containing CXCR4 siRNA-loaded HNP were then injected intravenously via the tail vein. In particular, for each dose, 92 μg of NP were loaded with 0.3 nmol (5 μg) siRNA (3.3 pmol siRNA/ μg−1 NP). In the sham experiments, mice were injected with 100 μL of sterile 0.9% saline solution. Finally, treatment of the third group consisted in the injection of 100 uL of sterile 0.9% saline solution containing 92 μg of SNP incubated with 0.3 nmol (5 μg) siRNA. Treatments were repeated 3 and 5 days later. Mice were sacrificed on day 7 and tumor samples were collected for protein extraction and expression analysis.
Western blot analysis of CXCR4 levels in tumor mice models
An appropriate volume (1ml of buffer for 10 mg of tissue) of ice-cold homogenizer buffer (T-PER Tissue Protein Extraction Reagent, Thermo Scientific, Rockford, IL, USA) containing protease inhibitors (Thermo Scientific) was pipetted into pre-chilled homogenization tubes. The tumors were rinsed in cold PBS to remove blood, cut in small pieces and transferred into the tubes. Then, they were homogenized using a tissue homogenizer (Bio-Gen PRO200, PRO scientific) following the protocol supplied by the manufacturer. The homogenate was transferred to a 2 mL centrifuge tube, incubated on ice for 15 min and then centrifuged at maximum speed (16,100 g) for 30 min. The supernatant was collected in a clean tube and protein concentration was checked by Bradford assay using albumin as standard. 50 μg of whole cell protein extracts were loaded onto 12% (w/v) polyacrylamide gel and then transferred on to a PVDFmembrane for Western blot analysis, as previously described. Blots were incubated with rabbit anti-CXCR4 (Millipore) and rabbit anti-GAPDH (Abcam) primary antibodies and then with horseradish peroxidase-conjugated anti-Rabbit IgG (Sigma–Aldrich) as secondary antibody. The bands were revealed with the SuperSignal West Dura Chemiluminescent Substrate (Thermo Scientific) and images were visualized and acquired with the ChemiDoc XRS+ System and Image Lab software (Bio-Rad Laboratories, Hercules, CA). Quantitative analysis of the bands was perfomed using ImageJ software. The reported results are the average of tumor analyses from three (n=3) different mice for each treatment group. Statistical significance was assessed by one-way two-tail paired T-test.
Analysis of HNP biodistribution by ICP-AES
For Inductively Coupled Plasma–Atomic Emission Spectroscopy (ICP-AES) analysis, organs (tumor, heart, lungs, liver, kidneys, and spleen) were harvested, washed in PBS, weighed and homogenized. They were then centrifuged at 5000 rpm for 20 min and supernatant was collected, filtered using 0.45 μm nylon centrifugal filter microfuge tubes (VWR), and diluted with DI water for elemental analysis by ICP-OES (Varian 720 ES, Varian Inc., Walnut Creek, CA, USA) as previously described [65, 66]. The reported results are the average from analysis of tumors of four (n=4) different mice for each treatment group. Statistical significance was assessed by one-way two-tail unpaired T-test.
Statistical analyses
All the numeric data are the result of a minimum of three independent experiments. Statistical computation was performed with GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA) or Microsoft Excel software (Microsoft, Redmond, WA). The statistical significance was calculated using paired or unpaired one-way two-tail Student’s T-test.
Acknowledgements
The authors acknowledge support from the Alliance for NanoHealth Department of Defense Telemedicine & Advanced Technology Research Center (09-W81XWH-10-2-0125 and 08-W81XWH-09-02-0139), the National Institutes of Health (1R21CA173579-01A1), the Department of Defense/Breast Cancer Research Program (W81XWH-12-10414 ). The High Resolution Electron Microscopy Facility at the University of Texas MD Anderson Cancer Center is supported by the National Cancer Institute (P30CA16672). This study was partially supported by the Italian Ministry of Health RF-2010-2318372 (to AP and FS) and by the Bianca Garavaglia Association, Busto Arsizio, Va, Italy. We thank Matthew Landry for graphical assistance, Mr. Kenneth Dunner for assistance with transmission electron microscopy, and Jean Ann Gilder (Scientific Communication srl., Naples, Italy) and Megan Livingston for editing the text.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Qiu Y, Park K. Environment-sensitive hydrogels for drug delivery. Advanced Drug Delivery Reviews. 2012 Dec;64:49–60. doi: 10.1016/s0169-409x(01)00203-4. [DOI] [PubMed] [Google Scholar]
- [2].Parodi A, Corbo C, Cevenini A, Molinaro R, Palomba R, Pandolfi L, Agostini M, Salvatore F, Tasciotti E. Enabling cytoplasmic delivery and organelle targeting by surface modification of nanocarriers. Nanomedicine (Lond) 2015 Jul;10(12):1923–1940. doi: 10.2217/nnm.15.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hennink WE, van Nostrum CF. Novel crosslinking methods to design hydrogels. Advanced Drug Delivery Reviews. 2012 Dec 64;:223–236. doi: 10.1016/s0169-409x(01)00240-x. [DOI] [PubMed] [Google Scholar]
- [4].Hoffman AS. Hydrogels for biomedical applications. Advanced Drug Delivery Reviews. 2012 Dec;64:18–23. doi: 10.1016/s0169-409x(01)00239-3. [DOI] [PubMed] [Google Scholar]
- [5].Hamner KL, Alexander CM, Coopersmith K, Reishofer D, Provenza C, Maye MM. Using Temperature-Sensitive Smart Polymers to Regulate DNA-Mediated Nanoassembly and Encoded Nanocarrier Drug Release. Acs Nano. 2013 Aug;7(8):7011–7020. doi: 10.1021/nn402214e. [DOI] [PubMed] [Google Scholar]
- [6].Hoffman AS. Stimuli-responsive polymers: Biomedical applications and challenges for clinical translation. Advanced Drug Delivery Reviews. 2013 Jan;65(1):10–16. doi: 10.1016/j.addr.2012.11.004. [DOI] [PubMed] [Google Scholar]
- [7].Wang J, Lu Z, Wientjes MG, Au JLS. Delivery of siRNA Therapeutics: Barriers and Carriers. Aaps Journal. 2010 Dec;12(4):492–503. doi: 10.1208/s12248-010-9210-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yang J, Deng LH, Han CR, Duan JF, Ma MG, Zhang XM, Xu F, Sun RC. Synthetic and viscoelastic behaviors of silica nanoparticle reinforced poly(acrylamide) core-shell nanocomposite hydrogels. Soft Matter. 2013;9(4):1220–1230. [Google Scholar]
- [9].Buwalda SJ, Boere KW, Dijkstra PJ, Feijen J, Vermonden T, Hennink WE. Hydrogels in a historical perspective: From simple networks to smart materials. J Control Release. 2014 Sep 28;190C:254–273. doi: 10.1016/j.jconrel.2014.03.052. [DOI] [PubMed] [Google Scholar]
- [10].Robinson DN, Peppas NA. Preparation and characterization of pH-responsive poly(methacrylic acid-g-ethylene glycol) nanospheres. Macromolecules. 2002 Apr 23;35(9):3668–3674. [Google Scholar]
- [11].Thomas JB, Creecy CM, McGinity JW, Peppas NA. Synthesis and properties of lightly crosslinked poly((meth)acrylic acid) microparticles prepared by free radical precipitation polymerization. Polymer Bulletin. 2006 May;57(1):11–20. [Google Scholar]
- [12].Tamura G, Shinohara Y, Tamura A, Sanada Y, Oishi M, Akiba I, Nagasaki Y, Sakurai K, Amemiya Y. Dependence of the swelling behavior of a pH-responsive PEG-modified nanogel on the cross-link density. Polymer Journal. 2012 Mar;44(3):240–244. [Google Scholar]
- [13].Kharlampieva E, Kozlovskaya V, Zavgorodnya O, Lilly GD, Kotov NA, Tsukruk VV. pH-responsive photoluminescent LbL hydrogels with confined quantum dots. Soft Matter. 2010;6(4):800–807. [Google Scholar]
- [14].Bennour S, Louzri F. Study of Swelling Properties and Thermal Behavior of Poly (N, N Dimethylacrylamide-co-Maleic Acid) Based Hydrogels. Advances in Chemistry. 2014:2014. [Google Scholar]
- [15].Bian QQ, Xiao Y, Zhou C, Lang MD. Synthesis, self-assembly, and pH-responsive behavior of (photo-crosslinked) star amphiphilic triblock copolymer. Journal of Colloid and Interface Science. 2013 Feb 15;392:141–150. doi: 10.1016/j.jcis.2012.08.076. [DOI] [PubMed] [Google Scholar]
- [16].Park CW, Yang HM, Lee HJ, Kim JD. Core-shell nanogel of PEG-poly(aspartic acid) and its pH-responsive release of rh-insulin. Soft Matter. 2013;9(6):1781–1788. [Google Scholar]
- [17].Mukhopadhyay P, Sarkar K, Soam S, Kundu PP. Formulation of pH-responsive carboxymethyl chitosan and alginate beads for the oral delivery of insulin. Journal of Applied Polymer Science. 2013 Jul 15;129(2):835–845. [Google Scholar]
- [18].Kirschner CM, Anseth KS. Hydrogels in healthcare: From static to dynamic material microenvironments. Acta Materialia. 2013 Feb;61(3):931–944. doi: 10.1016/j.actamat.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nature Reviews Genetics. 2014 Aug;15(8):541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- [20].Yang J, Chen J, Pan D, Wan Y, Wang Z. pH-sensitive interpenetrating network hydrogels based on chitosan derivatives and alginate for oral drug delivery. Carbohydrate Polymers. 2013 Jan 30;92(1):719–725. doi: 10.1016/j.carbpol.2012.09.036. [DOI] [PubMed] [Google Scholar]
- [21].Du H, Liu M, Yang X, Zhai G. The design of pH-sensitive chitosan-based formulations for gastrointestinal delivery. Drug Discov Today. 2015 Aug;20(8):1004–1011. doi: 10.1016/j.drudis.2015.03.002. [DOI] [PubMed] [Google Scholar]
- [22].Mironi-Harpaz I, Wang DY, Venkatraman S, Seliktar D. Photopolymerization of cell- encapsulating hydrogels: crosslinking efficiency versus cytotoxicity. Acta Biomater. May;8(5):1838–1848. doi: 10.1016/j.actbio.2011.12.034. [DOI] [PubMed] [Google Scholar]
- [23].Mazzoccoli JP, Feke DL, Baskaran H, Pintauro PN. Mechanical and cell viability properties of crosslinked low- and high-molecular weight poly(ethylene glycol) diacrylate blends. Journal of Biomedical Materials Research Part A. 2010 May;93A(2):558–566. doi: 10.1002/jbm.a.32563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ji J, Shu S, Wang F, Liu JJ, Yu ZZ. Revisit to the self-assembled hybrid acrylate/silica core-shell structured particles in the presence of unmodified silica particles. Colloids and Surfaces a-Physicochemical and Engineering Aspects. 2014 Apr 5;446:156–162. [Google Scholar]
- [25].Sun JT, Hong CY, Pan CY. Fabrication of PDEAEMA-Coated Mesoporous Silica Nanoparticles and pH-Responsive Controlled Release. Journal of Physical Chemistry C. 2010 Jul 29;114(29):12481–12486. [Google Scholar]
- [26].Marek SR, Conn CA, Peppas NA. Cationic nanogels based on diethylaminoethyl methacrylate. Polymer. 2010 Mar 11;51(6):1237–1243. doi: 10.1016/j.polymer.2010.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Liechty WB, Caldorera-Moore M, Phillips MA, Schoener C, Peppas NA. Advanced molecular design of biopolymers for transmucosal and intracellular delivery of chemotherapeutic agents and biological therapeutics. Journal of Controlled Release. 2011 Oct 30;155(2):119–127. doi: 10.1016/j.jconrel.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gao Q, Xu Y, Wu D, Sun YH, Li XA. pH-Responsive Drug Release from Polymer-Coated Mesoporous Silica Spheres. Journal of Physical Chemistry C. 2009 Jul 23;113(29):12753–12758. [Google Scholar]
- [29].Coates J. Interpretation of infrared spectra, a practical approach. Encyclopedia of analytical chemistry. 2000 [Google Scholar]
- [30].Kim DJ, Lee KB, Chi YS, Kim WJ, Paik HJ, Choi IS. Biomimetic formation of silica thin films by surface-initiated polymerization of 2-(dimethylamino) ethyl methacrylate and silicic acid. Langmuir. 2004 Sep 14;20(19):7904–7906. doi: 10.1021/la048657b. [DOI] [PubMed] [Google Scholar]
- [31].Gulari E, McKeigue K, Ng K. Raman and FTIR spectroscopy of polymerization: bulk polymerization of methyl methacrylate and styrene. Macromolecules. 1984;17(9):1822–1825. [Google Scholar]
- [32].Cokbaglan L, Arsu N, Yagci Y, Jockusch S, Turro NJ. 2-Mercaptothioxanthone as a novel photoinitiator for free radical polymerization. Macromolecules. 2003;36(8):2649–2653. [Google Scholar]
- [33].Khaled SM, Sui R, Charpentier PA, Rizkalla AS. Synthesis of TiO2-PMMA nanocomposite: Using methacrylic acid as a coupling agent. Langmuir. 2007 Mar 27;23(7):3988–3995. doi: 10.1021/la062879n. [DOI] [PubMed] [Google Scholar]
- [34].Huang XY, Brittain WJ. Synthesis and characterization of PMMA nanocomposites by suspension and emulsion polymerization. Macromolecules. 2001 May 8;34(10):3255–3260. [Google Scholar]
- [35].Hackley VA, Clogston JD. Measuring the hydrodynamic size of nanoparticles in aqueous media using batch-mode dynamic light scattering. Characterization of Nanoparticles Intended for Drug Delivery: Springer. 2007:35–52. doi: 10.1007/978-1-60327-198-1_4. [DOI] [PubMed] [Google Scholar]
- [36].Amalvy JI, Wanless EJ, Li Y, Michailidou V, Armes SP, Duccini Y. Synthesis and characterization of novel pH-responsive microgels based on tertiary amine methacrylates. Langmuir. 2004 Oct 12;20(21):8992–8999. doi: 10.1021/la049156t. [DOI] [PubMed] [Google Scholar]
- [37].De SK, Aluru NR, Johnson B, Crone WC, Beebe DJ, Moore J. Equilibrium swelling and kinetics of pH-responsive hydrogels: Models, experiments, and simulations. Journal of Microelectromechanical Systems. 2002 Oct;11(5):544–555. [Google Scholar]
- [38].Ostroha J, Pong M, Lowman A, Dan N. Controlling the collapse/swelling transition in charged hydrogels. Biomaterials. 2004 Aug;25(18):4345–4353. doi: 10.1016/j.biomaterials.2003.11.019. [DOI] [PubMed] [Google Scholar]
- [39].Pack DW, Hoffman AS, Pun S, Stayton PS. Design and development of polymers for gene delivery. Nature Reviews Drug Discovery. 2005 Jul;4(7):581–593. doi: 10.1038/nrd1775. [DOI] [PubMed] [Google Scholar]
- [40].Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL. The Possible “Proton Sponge” Effect of Polyethylenimine (PEI) Does Not Include Change in Lysosomal pH. Molecular Therapy. 2013 Jan;21(1):149–157. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Nel AE, Madler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nanobio interface. Nature Materials. 2009 Jul;8(7):543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- [42].You YZ, Manickam DS, Zhou QH, Oupicky D. Reducible poly(2-dimethylaminoethyl methaerylate): Synthesis, cytotoxicity, and gene delivery activity. Journal of Controlled Release. 2007 Oct 8;122(3):217–225. doi: 10.1016/j.jconrel.2007.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Fisher OZ, Peppas NA. Polybasic Nanomatrices Prepared by UV-Initiated Photopolymerization. Macromolecules. 2009 May 12;42(9):3391–3398. doi: 10.1021/ma801966r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Miller DK, Griffiths E, Lenard J, Firestone RA. Cell killing by lysosomotropic detergents. J Cell Biol. 1983 Dec;97(6):1841–1851. doi: 10.1083/jcb.97.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Moreira C, Oliveira H, Pires LR, Simoes S, Barbosa MA, Pego AP. Improving chitosan-mediated gene transfer by the introduction of intracellular buffering moieties into the chitosan backbone. Acta Biomater. 2009 Oct;5(8):2995–3006. doi: 10.1016/j.actbio.2009.04.021. [DOI] [PubMed] [Google Scholar]
- [46].Albini A, Mussi V, Parodi A, Ventura A, Principi E, Tegami S, Rocchia M, Francheschi E, Sogno I, Cammarota R, Finzi G, Sessa F, Noonan DM, Valbusa U. Interactions of single-wall carbon nanotubes with endothelial cells. Nanomedicine. 2009 Apr;6(2):277–288. doi: 10.1016/j.nano.2009.08.001. [DOI] [PubMed] [Google Scholar]
- [47].Parodi A, Quattrocchi N, van de Ven AL, Chiappini C, Evangelopoulos M, Martinez JO, Brown BS, Khaled SZ, Yazdi IK, Enzo MV, Isenhart L, Ferrari M, Tasciotti E. Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat Nanotechnol. Jan;8(1):61–68. doi: 10.1038/nnano.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Liang ZX, Yoon YH, Votaw J, Goodman MM, Williams L, Shim H. Silencing of CXCR4 blocks breast cancer metastasis. Cancer Research. 2005 Feb 1;65(3):967–971. [PMC free article] [PubMed] [Google Scholar]
- [49].Lee H, Lytton-Jean AK, Chen Y, Love KT, Park AI, Karagiannis ED, Sehgal A, Querbes W, Zurenko CS, Jayaraman M, Peng CG, Charisse K, Borodovsky A, Manoharan M, Donahoe JS, Truelove J, Nahrendorf M, Langer R, Anderson DG. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat Nanotechnol. 2012 Jun;7(6):389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Arima H, Tsutsumi T, Yoshimatsu A, Ikeda H, Motoyama K, Higashi T, Hirayama F, Uekama K. Inhibitory effect of siRNA complexes with polyamidoamine dendrimer/alpha-cyclodextrin conjugate (generation 3, G3) on endogenous gene expression. Eur J Pharm Sci. 2011 Oct 9;44(3):375–384. doi: 10.1016/j.ejps.2011.08.019. [DOI] [PubMed] [Google Scholar]
- [51].Cun D, Foged C, Yang M, Frokjaer S, Nielsen HM. Preparation and characterization of poly(DL-lactide-co-glycolide) nanoparticles for siRNA delivery. Int J Pharm. 2010 May 5;390(1):70–75. doi: 10.1016/j.ijpharm.2009.10.023. [DOI] [PubMed] [Google Scholar]
- [52].Naito M, Ishii T, Matsumoto A, Miyata K, Miyahara Y, Kataoka K. A phenylboronatefunctionalized polyion complex micelle for ATP-triggered release of siRNA. Angew Chem Int Ed Engl. 2012 Oct 22;51(43):10751–10755. doi: 10.1002/anie.201203360. [DOI] [PubMed] [Google Scholar]
- [53].Hu Y, Atukorale PU, Lu JJ, Moon JJ, Um SH, Cho EC, Wang Y, Chen J, Irvine DJ. Cytosolic delivery mediated via electrostatic surface binding of protein, virus, or siRNA cargos to pH-responsive core-shell gel particles. Biomacromolecules. 2009 Apr 13;10(4):756–765. doi: 10.1021/bm801199z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Convertine AJ, Diab C, Prieve M, Paschal A, Hoffman AS, Johnson PH, Stayton PS. pH-responsive polymeric micelle carriers for siRNA drugs. Biomacromolecules. 2010 Nov 8;11(11):2904–2911. doi: 10.1021/bm100652w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Prencipe G, Tabakman SM, Welsher K, Liu Z, Goodwin AP, Zhang L, Henry J, Dai HJ. PEG Branched Polymer for Functionalization of Nanomaterials with Ultralong Blood Circulation. Journal of the American Chemical Society. 2009 Apr 8;131(13):4783–4787. doi: 10.1021/ja809086q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Gref R, Domb A, Quellec P, Blunk T, Muller RH, Verbavatz JM, Langer R. The controlled intravenous delivery of drugs using PEG-coated sterically stabilized nanospheres. Advanced Drug Delivery Reviews. 2012 Dec;64:316–326. doi: 10.1016/0169-409X(95)00026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Trewyn BG, Slowing II, Giri S, Chen HT, Lin VSY. Synthesis and functionalization of a mesoporous silica nanoparticle based on the sol-gel process and applications in controlled release. Accounts of Chemical Research. 2007 Sep;40(9):846–853. doi: 10.1021/ar600032u. [DOI] [PubMed] [Google Scholar]
- [58].Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, Soh J, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Date H, Lam WL, Minna JD, Gazdar AF. PIK3CA mutations and copy number gains in human lung cancers. Cancer Research. 2008 Sep 1;68(17):6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen YY, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang LL, Soda M, Kikuchi A, Igarashi T, Nakagawara A, Hayashi Y, Mano H, Ogawa S. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008 Oct 16;455(7215):971–U956. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- [60].Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983 Dec 16;65(1-2):55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- [61].Borenfreund E, Puerner JA. Toxicity determined in vitro by morphological alterations and neutral red absorption. Toxicol Lett. 1985 Feb-Mar;24(2-3):119–124. doi: 10.1016/0378-4274(85)90046-3. [DOI] [PubMed] [Google Scholar]
- [62].Riddell RJ, Clothier RH, Balls M. An evaluation of three in vitro cytotoxicity assays. Food Chem Toxicol. 1986 Jun-Jul;24(6-7):469–471. doi: 10.1016/0278-6915(86)90095-5. [DOI] [PubMed] [Google Scholar]
- [63].Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- [64].Parodi A, Haddix SG, Taghipour N, Scaria S, Taraballi F, Cevenini A, Yazdi IK, Corbo C, Palomba R, Khaled SZ, Martinez JO, Brown BS, Isenhart L, Tasciotti E. Bromelain Surface Modification Increases the Diffusion of Silica Nanopartides in the Tumor Extracellular Matrix. Acs Nano. 2014 Oct;8(10):9874–9883. doi: 10.1021/nn502807n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Martinez JO, Boada C, Yazdi IK, Evangelopoulos M, Brown BS, Liu XW, Ferrari M, Tasciotti E. Short and Long Term, In Vitro and In Vivo Correlations of Cellular and Tissue Responses to Mesoporous Silicon Nanovectors. Small. 2013 May 27;9(9-10):1722–1733. doi: 10.1002/smll.201201939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Minardi S, Pandolfi L, Taraballi F, De Rosa E, Yazdi IK, Liu XW, Ferrari M, Tasciotti E. PLGA-Mesoporous Silicon Microspheres for the in Vivo Controlled Temporospatial Delivery of Proteins. Acs Applied Materials & Interfaces. 2015 Aug 5;7(30):16364–16373. doi: 10.1021/acsami.5b03464. [DOI] [PubMed] [Google Scholar]