Abstract
We present an optimized system for rapid generation of Localization and Affinity Purification (LAP)-tagged mammalian stable cell lines that facilitates complex purification and interacting protein identification. The improved components of this method, including the flexibility of inducible expression, circumvent issues associated with toxicity, clonal selection, sample yields and time to data acquisition. We have applied this method to the study of cell cycle regulators and novel microtubule-associated proteins.
Keywords: High throughput, LAP tag, Mammalian cells, Proteomics, Systems biology
Mapping of protein interaction networks in mammalian cells has been hampered by the lack of a general high throughput tagging system that is compatible with current advances in cloning and cell line technologies. Historically, epitope tags have been added to the termini of proteins to allow their purification with anti-epitope antibodies or affinity resins. Tandem affinity purification (TAP) technology, a two-step affinity purification procedure initially used in yeast, utilizes a more stringent purification protocol that reduces the rate of false positive interactions and has been successfully adapted for use in other organisms [1]. The recent application of Localization and Affinity Purification (LAP) technology to metazoans has not only enabled researchers to tandem affinity purify proteins and identify interacting partners, but also to study the sub-cellular localization of proteins in live cells [2]. These methodologies have greatly advanced our understanding of the function of individual proteins and the networks in which they function. However, a number of issues have hindered current high throughput approaches to protein complex identification in mammalian cells: the cumbersome molecular biology needed to generate vectors applicable for LAP/TAP tagging; the length of time to generate and select clones of a stable cell line with the tagged protein of interest; the toxicity associated with constitutively overexpressing proteins, even at low expression levels; the production of sufficient material to identify interactors via mass spectrometry; and summing these together, the overall length of time to data acquisition. To address these issues, we developed a series of mammalian LAP/TAP-tag vectors that incorporate current advances in cloning (Gateway, Invitrogen) and mammalian cell line technologies (Flp-In, Invitrogen) to create a methodology that expedites protein tagging and identification of protein interactors, which we call gLAP-Flp.
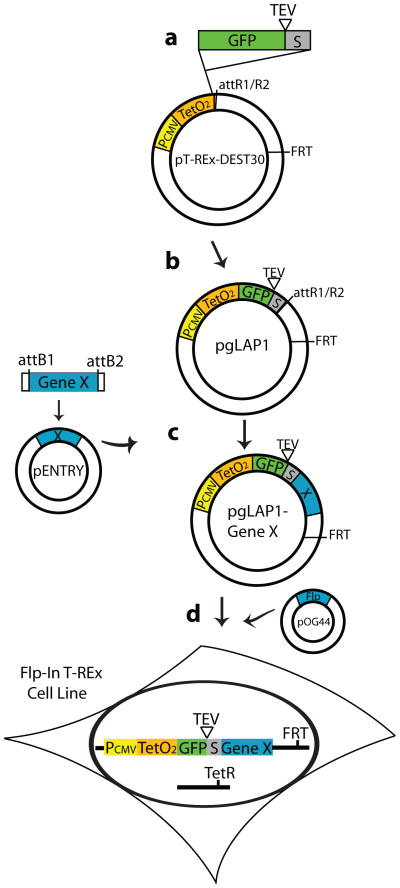
Gateway technology facilitates high throughput cloning of target sequences into various expression vectors for use in in vitro and in vivo systems [3]. The ORFeome community has to date cloned thousands of mammalian full-length open reading frames into Gateway-compatible vectors that are now available to researchers ([4–6], www.invitrogen.com, www.openbiosystems.com). With this resource in mind, we synthesized a LAP tag (EGFP-TEV-S-peptide) module for N-terminal protein tagging and inserted this module into pT-Rex-DEST30 (Invitrogen), upstream of the attR1/R2 Gateway cloning sites to create pgLAP1 (Fig. 1a). pgLAP1 contains a pCMV promoter (regulated by two downstream Tet operator sites) driving the expression of EGFP-TEV-S-peptide-tagged gene of interest (Fig. 1b), as well as an FRT site (see below). Gateway clones (in pENTRY vectors), or amplified PCR products that have been integrated into a DONR vector, can be swiftly inserted in-frame into pgLAP1 via the LR clonase reaction (Fig. 1c). For details on this and other Gateway conservative recombination reactions, see [3, 7]. Use of pgLAP1 bypasses two major time-consuming processes: first, the need to shuffle a gene of interest into two separate vectors, which was necessary for adapting the original LAP technology to mammalian systems; and second, cumbersome cloning using PCR or restriction enzymes. Gateway-based cloning facilitates the movement of large sets of clones into pgLAP1 simultaneously, making systems wide proteomics network studies feasible.
Figure 1.
Workflow for generation of LAP-tagged proteins and mammalian stable cell lines. (a) Synthesis of EGFP-TEV-S peptide module and cloning into pT-REx-DEST30. (b) Generation of pgLAP1 containing Gateway-compatible GFP-TEV-S-Peptide driven by a Tet-regulated pCMV promoter (contains two Tet operator sites). (c) Gateway in-frame cloning of gene of interest (X) into pgLAP1, to create Localization and Affinity Purification (LAP: GFP-TEV-S-Peptide) tagged gene of interest. (d) Generation of LAP-tagged stable cell lines by co-transfection of pgLAP1-Gene X with Flp recombinase expression vector (pOG44) into Flp-In T-REx (Tetracycline-Repressible Expression) FRT (Flp Recombination Target) containing cell lines.
Previous systems used to generate stable LAP-tagged genes of interest in mammalian cell lines were hampered by the need to use FACS (Fluorescence Activated Cell Sorting) or cloning rings to select individual clones, followed by a lengthy expansion of cells to generate sufficient material for proteomic studies. To simplify these steps, we used Flp-In and Flp-In T-REx cell lines available from Invitrogen to create clonal cell lines. Flp-In cell lines have an FRT (Flp Recombination Target) site integrated into a single genomic locus (Fig. 1d). pgLAP1 contains an FRT site and co-transfection with a vector expressing the Flp recombinase (pOG44, Invitrogen) enables pgLAP1 to recombine into a single site within the genome (Fig. 1d). Thus, the Flp-In method does not require researchers to work with viruses to obtain single integrants. After selecting integrants with hygromycin, the entire polyclonal population is continually grown until harvesting for protein purification. This method bypasses the need for FACS sorting and clonal selection, thereby dramatically reducing the time needed to generate a stable cell line and acquire sufficient quantities of tagged material.
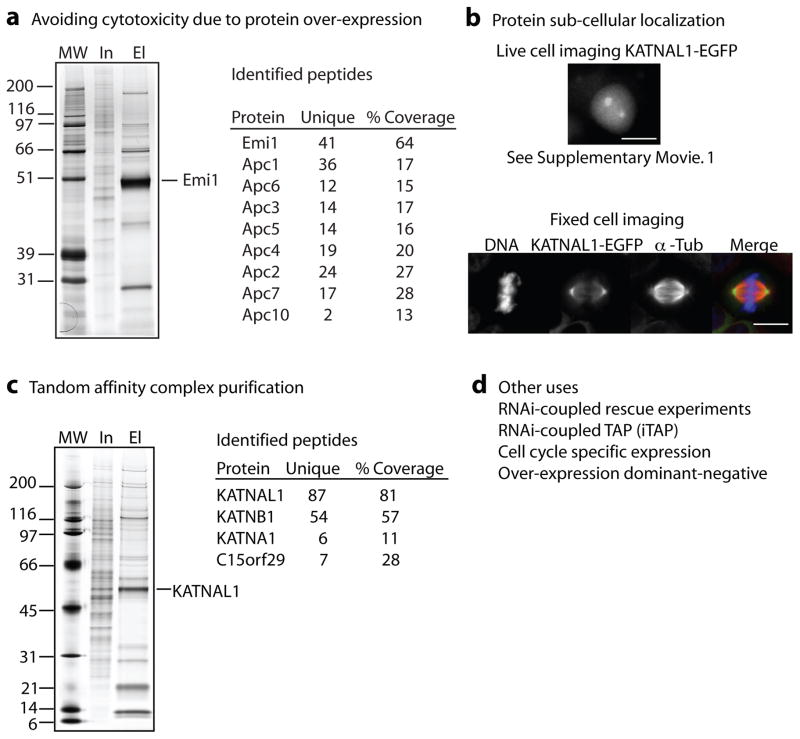
Protein over-expression can lead to protein mislocalization and non-native protein interactions resulting in identification of false interactions. In addition, protein over-expression, even slight, can often lead to toxicity, especially in the case of proteins essential for cell cycle progression. As an example we considered Emi1 (Early Mitotic Inhibitor 1). Generation of a stable epitope-tagged Emi1 cell line had been difficult due to its toxicity when over-expressed [8]. To address this issue, we employed our gLAP-Flp strategy to create a LAP-Emi1 293 Flp-In T-REx stable cell line. T-REx cell lines stably express the Tet repressor that binds to the Tet operator binding sites downstream of the pCMV promoter to repress protein expression. Upon tetracycline/doxycycline addition, protein expression ensues. Using this system, we transiently induced LAP-Emi1 expression and successfully purified it and many components of the Anaphase Promoting Complex (APC) (Fig. 2a, Supplementary Table. 1), an interaction we have previously described [9].
Figure 2.
Multiple uses of LAP-tagged stable cell lines. (a) Avoidance of toxic effects associated with over-expression of cell cycle regulatory proteins, such as Emi1, by tetracycline-regulated transient protein expression. Tandem affinity purification of LAP-Emi1, identifies its interaction with the Anaphase Promoting Complex/Cyclosome (APC/C). For a and c, MW= molecular weight, In= input, El= eluate, for peptide information see Supplementary Table. 1. (b) Protein sub-cellular localization of KATNAL1, a novel microtubule-associated protein, via in vivo time-lapse fluorescence microscopy and post fixation immunofluorescence. Bar = 10 μm. (c) Purification of LAP-KATNAL1 and identification of interacting proteins by mass spectrometry. (d) Other uses: RNAi-coupled rescue experiments and TAP (iTAP), cell cycle specific expression, and over-expression.
In addition, we have successfully used this system to determine the localization and interacting proteins of novel mitotic microtubule-associated proteins. Recently, we identified a hypothetical katanin p60-like protein KATNAL1 (NP_115492.1) as a protein binding to microtubules in mitosis (JZT, unpublished results). Katanin is a microtubule-severing enzyme, consisting of a catalytic p60 subunit and a regulatory p80 subunit, that controls mitotic and meiotic spindle length, cilia biogenesis, and axonal growth [10–12]. We generated a LAP-KATNAL1 293 Flp-In T-REx stable cell line and analyzed its sub-cellular localization by live-cell and fixed-cell fluorescence microscopy. KATNAL1 localized to the mitotic organizing center (MTOC) in interphase and to mitotic spindle poles throughout mitosis (Fig. 2b, Supplementary Movie. 1, and Supplementary Fig. 1). This localization is in agreement with the localization of the canonical p60 and p80 subunits in human cells [13]. Additionally, we were able to detect KATNAL1 localization to the central spindle in late anaphase (Supplementary Fig. 1). Purification of LAP-KATNAL1 uncovered an interaction with KATNB1, the p80 subunit of katanin, an uncharacterized p80-like protein (c15orf29, NP_078989.1) and KATNA1 (Fig. 2c, Supplementary Table. 1). Thus, it is likely that multiple p60/p80 heterodimers exist that regulate microtubule-severing, not just the previously studied canonical katanin complex. To our knowledge, this is the first characterization of KATNAL1.
Flp-In LAP-tagged stable cell lines have multiple uses in addition to avoiding toxic over-expression effects, identifying protein sub-cellular localization, and tandem affinity purification (Fig. 2d). It is also possible to couple RNAi treatment with an RNAi-resistant LAP-tagged gene to test protein function or to enhance inclusion of the LAP-tagged protein into endogenous complexes for purification [14] (Fig. 2d). Additionally, the method can be used to test dominant-negative phenotypes caused by over-expression or to express a protein of interest in a cell cycle-specific manner with synchronized cells.
In addition to the pgLAP1 vector previously described, we have created a series of additional vectors to accommodate a number of specific tagging and expression needs. For instance, the EGFP moiety is ~26 kDa and its large size may render some LAP-tagged proteins non-functional. To create a smaller epitope tag, we engineered a vector with a small TAP tag (pgLAP2, Flag-TEV-S peptide) that may circumvent problems encountered with a larger tag (Supplementary Methods). Additionally the strongly expressing CMV promoter may not be compatible with constitutive expression lines or proteins with dominant or potentially toxic activities. With this in mind, we have engineered LAP (pgLAP3, EGFP-TEV-S peptide) and TAP (pgLAP4, Flag-TEV-S peptide) vectors which utilize the constitutive, low expressing EF1α promoter (Supplementary Methods). We have also created vectors with a C-terminal LAP tag (pgLAP5, S peptide – PreScission Protease-EGFP) (Supplementary Methods). We have made these vectors available to the research community (www.addgene.org/Peter_Jackson) and hope that this system will expedite the identification of protein complexes and interaction networks.
Supplementary Material
Acknowledgments
We thank A. Eldridge for critical reading of the manuscript and A. Desai, I. Cheeseman, F. McNally, and members of the Jackson lab for helpful discussions. This work was supported by Genentech Inc, National Institute of General Medical Sciences grants RO1 GM60439 and RO1 GM54811 (P.K.J), NIH Medical Scientist Training Program (J.J.M), Stanford Cancer Biology Postdoctoral Fellowship (J.Z.T) and The Leukemia and Lymphoma Society Postdoctoral Fellowship (J.Z.T).
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Puig O, Caspary F, Rigaut G, Rutz B, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 2.Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE. 2005;2005:pl1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- 3.Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site- specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brasch MA, Hartley JL, Vidal M. ORFeome cloning and systems biology: standardized mass production of the parts from the parts-list. Genome Res. 2004;14:2001–2009. doi: 10.1101/gr.2769804. [DOI] [PubMed] [Google Scholar]
- 5.Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamesch P, Li N, Milstein S, Fan C, et al. hORFeome v3. 1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–315. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- 8.Margottin-Goguet F, Hsu JY, Loktev A, Hsieh HM, et al. Prophase destruction of Emi1 by the SCF(betaTrCP/Slimb) ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev Cell. 2003;4:813–826. doi: 10.1016/s1534-5807(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 9.Miller JJ, Summers MK, Hansen DV, Nachury MV, et al. Emi1 stably binds and inhibits the anaphase-promoting complex/cyclosome as a pseudosubstrate inhibitor. Genes Dev. 2006;20:2410–2420. doi: 10.1101/gad.1454006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baas PW, Karabay A, Qiang L. Microtubules cut and run. Trends Cell Biol. 2005;15:518–524. doi: 10.1016/j.tcb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 11.McNally K, Audhya A, Oegema K, McNally FJ. 2006 Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma N, Bryant J, Wloga D, Donaldson R, et al. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J Cell Biol. 2007;178:1065–1079. doi: 10.1083/jcb.200704021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNally FJ, Okawa K, Iwamatsu A, Vale RD. Katanin, the microtubule- severing ATPase, is concentrated at centrosomes. J Cell Sci. 1996;109(Pt 3):561–567. doi: 10.1242/jcs.109.3.561. [DOI] [PubMed] [Google Scholar]
- 14.Forler D, Kocher T, Rode M, Gentzel M, et al. An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat Biotechnol. 2003;21:89–92. doi: 10.1038/nbt773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.