Abstract
Translocation of viral double-stranded DNA (dsDNA) into the icosahedral prohead shell is catalyzed by TerL, a motor protein that has ATPase, endonuclease, and translocase activities. TerL, following endonucleolytic cleavage of immature viral DNA concatemer recognized by TerS, assembles into a pentameric ring motor on the prohead’s portal vertex and uses ATP hydrolysis energy for DNA translocation. TerL’s N-terminal ATPase is connected by a hinge to the C-terminal endonuclease. Inchworm models propose that modest domain motions accompanying ATP hydrolysis are amplified, through changes in electrostatic interactions, into larger movements of the C-terminal domain bound to DNA. In phage φ29, four of the five TerL subunits sequentially hydrolyze ATP, each powering translocation of 2.5 bp. After one viral genome is encapsidated, the internal pressure signals termination of packaging and ejection of the motor. Current focus is on the structures of packaging complexes and the dynamics of TerL during DNA packaging, endonuclease regulation, and motor mechanics.
Keywords: bacteriophage, virus assembly, DNA packaging, terminase, ATPase, molecular motor
INTRODUCTION
Bacteriophages probably form the largest biomass on Earth (1). Predominant among these are the tailed phages with an icosahedral capsid (head) in which the viral genome (chromosome) is packaged. How these viruses fill the head with DNA to near-crystalline density and release it into a new host cell at near-perfect efficiency has fascinated researchers for decades. Not surprisingly, more recent animal virus descendants such as the herpesviruses retain the basic aspects of the packaging mechanism (2). Phage DNA packaging has also been a good model to understand fundamental biological mechanisms, such as transduction of ATP energy into mechanical work, condensation and decondensation of DNA, and movement of protein and DNA templates against each other.
Progress in understanding phage DNA packaging mechanisms, especially the structures of motor components and the nature of the motor’s power stroke, has spawned numerous reviews, including general reviews by the present authors (3, 4) and by Casjens (5). The use of single-molecule techniques to study DNA packaging was reviewed by Chemla & Smith (6). There are recent focused reviews on bacteriophage T4 (7, 8), φ29 (9), SPP1 (10), and giant viruses (11). Here we emphasize progress since the last round of general reviews. Due to space limits, we refer to our previous reviews for earlier specific references (3, 4).
VIRUS ASSEMBLY: AN OVERVIEW
During infection of tailed bacteriophages and herpesviruses, an icosahedral prohead or procapsid shell of precise dimensions is assembled (Figure 1). A dodecameric portal protein initiates head assembly (Figure 1a), nucleating the coassembly of the major capsid protein and scaffold proteins to form the prohead. The 12-fold symmetric portal protein is located at a unique 5-fold vertex of the shell, creating a 12:5 symmetry mismatch that is conserved in tailed phages and herpesviruses (Figure 1b). The scaffold is removed, in some phages by a protease that is also part of the scaffold, followed by diffusion of cleaved peptides out of the immature prohead, yielding the mature prohead, generally an empty, rounded, thick-walled structure.
Figure 1.
Tailed bacteriophage assembly. A schematic illustration of the pathway of bacteriophage assembly, which includes (a) initiation of head assembly by the portal, (b) assembly of major and minor capsid proteins and scaffold proteins into an immature prohead, (c) maturation and expansion of the prohead, (d) packaging of the DNA, (e) assembly of decoration and/or neck (connector) proteins, and, finally, (f) attachment of the tail.
In parallel with prohead assembly, DNA replication and recombination create concatemeric progeny viral DNA, i.e., covalent end-to-end polymers of virus chromosomes. The terminase complex, consisting of large (TerL) and small (TerS) subunits, cuts the concatemers and generates the termini of the packaged genomes (the terms terminase and Ter are used interchangeably). Terminase recognizes a packaging signal on the concatemer and makes an endonucleolytic cut, generating a free end to which terminase remains bound. The DNA-bound terminase docks on the portal and assembles an oligomeric ring motor. Terminase then initiates DNA translocation, utilizing energy from ATP hydrolysis. After about 10–25% of the viral genome is packaged, the capsid protein undergoes a major conformational change resulting in expansion of the shell (12) (Figure 1c). The wall of the shell becomes thinner and the head more angular as the outer capsid dimensions increase by ~15% and the inner capsid volume by ~50%. The expanded prohead’s volume matches the size of the viral genome. In some phages, so-called decoration proteins bind to the surface of the expanded capsid, reinforcing the capsid structure. After encapsidating the viral genome, terminase makes another cut, terminating packaging and dissociating from the head while remaining bound to the newly generated concatemer end (Figure 1d). The motor-DNA complex then docks on another prohead and sponsors encapsidation of the next genome. In this way, the concatemer is processed, generating a series of DNA-filled heads.
The packing density of the encapsidated DNA approaches that of a crystal (500–550 mg/mL), creating an internal pressure of ~20 pN, sufficient to eject much of the DNA (13–17). DNA loss from the pressurized capsid is prevented by the portal, and by assembly of neck proteins on the portal that seal off the capsid (Figure 1e). Addition of a tail and tail fibers generates an infectious virion (Figure 1f). At the start of a new infection, the DNA is ejected into the new host cell through the channel created by the portal, neck, and tail.
Viral DNA packaging is a complex and dynamic process and must be precisely orchestrated such that the encapsidated genome can be efficiently delivered into a new host cell. For simplicity, we divide packaging into three blocks—initiation, translocation, and termination—and discuss what we have learned so far and the mechanisms that we still don’t know.
PACKAGING INITIATION
Viral DNA Recognition and Processing
Many viruses, including the cohesive end–containing cos phages (e.g., λ, HK97, P2), the T7/T3-like phages, and the herpesviruses, produce specific ends through endonuclease cutting (5). In cos phage λ, complementary 12-nt-long single-stranded cohesive ends are produced when terminase introduces nicks, staggered by 12 bp at cosN, the nicking site. The pac phages, such as P22 and SPP1, use a headful strategy (3, 5). That is, initiation cleavage occurs at or near a terminase recognition site called pac, followed by unidirectional DNA translocation into the head shell. When the shell is filled with slightly more than 100% of the viral DNA sequence (a headful), a nonspecific, double-strand DNA cleavage occurs to terminate packaging. As a result, the first cut is near a pac sequence, and all the subsequent cuts in the processive series will be sequence nonspecific. The T4-like phages use a variation of the pac strategy. Here, even the first cut does not occur at a strictly specific sequence. The resulting packaged DNA has about 3–10% terminal redundancy, depending on the phage. The redundant ends allow concatemer generation by recombination and replication. Differences in genome recognition notwithstanding, all these viruses employ a common mechanism: (a) terminase cutting at a sequence, often nonspecific ( pac phages), that is different from, but nearby, a recognition sequence; (b) translocation of one (cos phages) or slightly more than one ( pac phages) viral genome, a length that approximates the inner volume of the shell (headful); and (c) a second, packaging-dependent cut to terminate translocation (3).
Major exceptions to the above strategy are viruses with terminal proteins, such as φ29, adenoviruses, and their relatives. These viruses use a protein-primed DNA replication strategy, and hence concatemers are not produced. Instead, unit-length genomes with the terminal protein attached to the DNA 5′ end are the replication product as well as the packaging substrate. The phage φ29 packaging ATPase, TerL, not surprisingly lacks the endonuclease. Another unusual aspect of φ29 is the lack of a TerS and the presence of a 174-nt pRNA that, along with TerL, assembles as a packaging motor.
The Small Terminase Subunit, TerS
Viral genome recognition and end generation require collaboration by both terminase subunits. TerS and TerL form a hetero-oligomeric complex, the holoterminase. We have little information on the stoichiometry and structure of the complex. TerS selects the viral DNA concatemer from a pool that often includes host DNA. TerL makes the cut and docks one of the newly created ends at the special portal vertex of the head to initiate DNA packaging.
TerS proteins generally are small, ranging from about 140 to 180 amino acids, and consist of three domains (4): an N-terminal DNA-binding domain, a central oligomerization domain, and a C-terminal TerL-binding domain (Figure 2). TerS has two main functions: viral DNA recognition and TerL regulation. Atomic structures of TerS have been reported for cos phage λ (Escherichia coli); pac phages P22 (Salmonella spp.), Sf6 (Shigella spp.), and SPP1-like phage SF6 (Bacillus subtilis); and the T4 relative 44RR (Aeromonas salmonicida) (18–22).
Figure 2.
Structure of the small terminase, TerS. X-ray structures of the TerS oligomers from various phages are shown in rainbow colors, ranging from blue at the N terminus to red at the C terminus. Shown in the center are the top views (magenta). The subdomains are labeled in the Sf6 TerS. DNA modeled in the TerS of the SPP1-like phage SF6 and in that of phage Sf6 is shown (21, 25). In the Sf6 TerS-DNA model, the DNA clashed with the β-strands of the C-terminal β-barrel domain inside the channel.
TerS molecules assemble into gear-shaped oligomers of radially arrayed monomers. The number of subunits per oligomer varies from phage to phage over a range of 8 to 12. The N terminus is a small, globular, DNA-binding domain (DBD). Thus far there is genetic evidence only in cos phages λ and 21 that the DBD, a winged helix-turn-helix motif, specifically binds DNA (18). DBDs are tethered to the central cone-shaped oligomerization domain, which is made of two long α-helices forming an antiparallel coiled coil. A TerL specificity domain (the neck), which includes a parallel β-barrel motif, extends from the core and interacts with TerL’s N-terminal ATPase domain. Swapping of TerS neck domains—for example, between λ and 21, Sf6 and P22, or T4 and RB49 (18, 23, 24)—swaps TerL specificity.
There is controversy about whether the TerS-DNA complex involves wrapping (21, 25) or threading through the central channel (19) (Figure 2). The tethered DBDs extend out from the core. This arrangement may provide positional plasticity for the DNA to wrap around in a nucleosome-like complex. DNA footprinting (λ, SPP1) and cyclization (SPP1) studies of TerS-DNA complexes are consistent with the wrapping model (26, 27). With respect to the alternative model, threading, in Sf6 TerS the core channel at the narrowest point is only 17 Å wide, too narrow to accommodate the 23-Å-wide dsDNA (Figure 2) (20). However, the stoichiometry and architecture of TerS oligomers assembled in the presence of TerL and other viral components might be different, and the channel might be wider. A TerS-DNA cocrystal structure is needed to resolve the structure of the viral genome recognition complex.
The Headful Nuclease, TerL
TerLs are large 49–81-kDa packaging motor proteins with two globular domains joined by a hinge (28). The N-terminal domain contains the ATPase center that powers DNA translocation, which is discussed in detail below.
The C-terminal domain is the endonuclease that processes concatemeric DNA into genome-sized DNA molecules. The atomic structures of this domain from a number of viruses (29–35) show that it has the RNase H fold (36) that is also present in resolvases and integrases (Figure 3). The C-terminal domain includes a DNA-binding groove lined with basic amino acid residues that presumably interact with the DNA backbone phosphates. The position of the endonuclease-bound DNA can be modeled based on the known DNA-RNA hybrid–RNase H cocrystal structure (36). There is a deep valley at one end of this groove into which three acidic residues are arranged in a catalytic triad. These and a fourth conserved residue (Asp) coordinate two Mg2+ (or Mn2+) ions, forming the nuclease catalytic center (Figure 4). As in RNase H, one of the Mg2+ ions probably stabilizes the transition state and the other Mg2+ ion generates the OH− nucleophile for phosphodiester bond cleavage (29, 35, 37). Mutations at any of the catalytic residues of phage T4 TerL results in loss of DNA cleavage activity, but the packaging activity remains intact. Consequently, the mutant proteins cannot package circular DNA but can package the DNA if it is linearized (29). In SPP1, however, changing some of these residues leads to loss of both nuclease and DNA packaging activities (38).
Figure 3.
Structure of the headful packaging nuclease. X-ray structures of the TerL C domains from various viruses are shown (32–35, 38). A structural model of the T4 C domain–DNA complex is shown in surface view (top row, second from left). The side chains of conserved catalytic residues, including the catalytic triad acidic residues, are shown as red sticks and are enclosed in dashed ovals.
Figure 4.
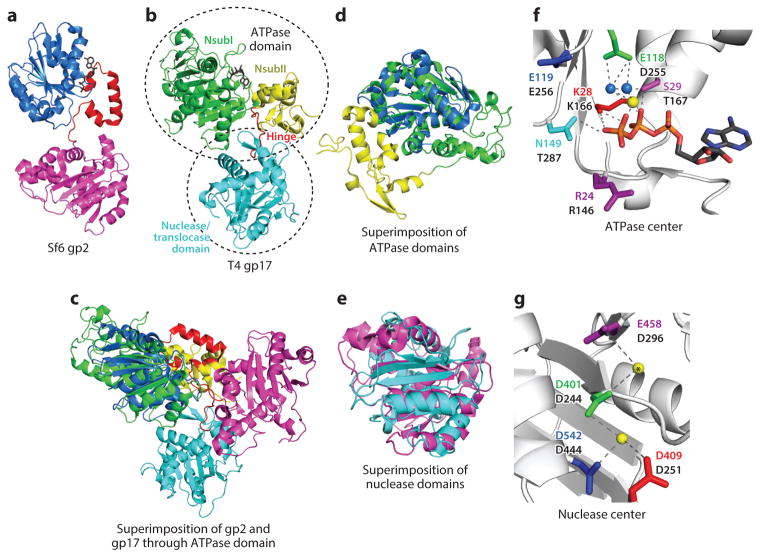
Structure of the large terminase. (a,b) X-ray structures of TerL proteins from phages Sf6 (gp2) (panel a) and T4 (gp17) (panel b) (31, 35). The gp2 subdomains are colored as follows: ATPase (blue; amino acids 1–172), hinge (red; amino acids 173–210), nuclease (magenta; amino acids 211–470). The gp17 subdomains are colored as follows: ATPase NsubI ( green; amino acids 62–313), ATPase NsubII ( yellow; amino acids 10–61 and 314–355), hinge (red; amino acids 356–364), nuclease (cyan; amino acids 365–567). ATP molecules are shown as gray sticks. (c–e) Superimposition of gp2 and gp17 full-length structures (panel c), ATPase domains (panel d), and nuclease domains (panel e). (f) The gp2 ATPase center is shown with ATP, Mg2+ ion ( yellow sphere), and two water molecules (blue spheres). Catalytic residues are shown as sticks and are numbered according to the gp2 sequence at the top, with equivalent gp17 residues labeled at the bottom. ( g) The gp17 nuclease center. Mg2+ ions are shown as yellow spheres. The position of a second Mg ion (asterisk) is modeled based on superimposition with SPP1 nuclease in which two Mn2+ ions are present in the crystal structure. Catalytic triad residues, shown as sticks, and a fourth conserved aspartic residue are numbered according to the gp17 sequence at the top, with equivalent gp2 residues labeled at the bottom.
TerL’s endonuclease activity must be tightly controlled such that the concatemer is cut only when packaging is initiated and terminated. It must remain inactive or inaccessible during translocation. Otherwise, partially packaged particles that are noninfectious will be produced. Several factors, such as TerS, the nucleotide status of the ATPase domain, and mobile elements in the linker, regulate the endonuclease (30, 32, 39, 40). However, more detailed structural and biochemical data are needed to understand how the switch from nuclease to translocation, and vice versa, operates.
Recently, a significant departure from the two-subunit terminase paradigm was found in phage HK97 and related phages. A third component, an HNH nuclease, is also required. The HK97 holoterminase alone cleaves cos DNA very poorly but cuts efficiently in the presence of the HNH nuclease (41). Similarly, the helper phage–dependent Staphylococcus pathogenicity island SaPIbov5 requires HNH-encoding helper phages for cos-dependent packaging (42). Mutants in the HNH’s putative nuclease catalytic site are unable to sponsor cos cleavage. Sequence analyses further suggest that many cos-cutting terminases encode HNH proteins. Herpes simplex virus (HSV) and human cytomegalovirus (HCMV) produce TerLs that form heterotrimers with two other proteins (43, 44). These viruses thus may have also evolved a tripartite terminase system.
The Holoterminase
Cleavage must occur on both strands of the DNA molecule, requiring a 2-fold rotationally symmetric TerL dimer. In addition, for cos phages, the 12-bp cohesive ends must be separated to generate free ends. It is widely accepted that a TerS-TerL holoterminase complex coordinates this whole process. In fact, in the absence of TerS or its recognition site, TerL loses specificity and behaves as a nonspecific endonuclease, even in the case of phage λ, in which its TerL otherwise makes precise cuts at the cosN sequence (43). The stoichiometry and structure of the terminase complex is unknown. The holoterminase might be an unstable and dynamic complex, as was thought in the case of phage T4, in which TerS (gp16) regulates TerL (gp17) activities but a TerS:TerL complex could not be isolated. However, TerS:TerL complexes could be purified from phages λ and P22 (19, 45). The λ complex is a tetramer of TerS2:TerL1 heterotrimers: [TerS2:TerL1]4. Tetramers are highly active for cos cleavage as well as for DNA packaging and do not require the E. coli integration host factor (IHF). IHF’s assistance otherwise is needed, both in vivo and in vitro, to generate maximal terminase activity (45). These observations suggest that [TerS2:TerL1]4 might be the functional holoterminase complex. Structural information on a holoterminase complex, preferably bound to DNA, is needed to further understand the end-generation mechanism.
DNA End Insertion
After cutting, the terminase complex must dock one of the newly generated ends to the prohead portal in order to initiate DNA translocation. Considerable changes in the holoterminase likely occur during this transition. An alternative view, that the initiation complex might sponsor DNA translocation (46), may not be plausible. For cutting, the TerL molecules must be in a 2-fold rotationally symmetric orientation in order to make contact with the same chemical elements of the antiparallel DNA strands. In contrast, for translocation, a parallel arrangement of TerL monomers in an oligomeric ring configuration appears to be essential. Indeed, the φ29 packaging motor tracks along—i.e., makes periodic contact with—one of the strands, the 5′→3′ strand (in the direction of packaging), during translocation (47, 48). Thus, the holoterminase complex used for cutting must be remodeled for DNA translocation (and again for packaging termination; see below).
Although the details are not known, TerL must remain bound to the end and assemble on the portal before the end slips out of the complex. Indeed, genetic studies with pac phage P1 and cos phage λ show that one of the ends, the pac-containing (P1) or cosB-containing (λ) end, is protected and used for initiating the processive packaging series, whereas the other end is unprotected and susceptible to RecBC exonuclease cleavage (49–51). It seems likely that additional free TerL molecules assemble on the DNA-holoterminase-portal complex to assemble an oligomeric packaging motor. However, much remains to be understood about this dynamic transition from cutting to packaging.
In vitro studies show that end insertion is an efficient, but promiscuous, process. It can occur by multiple pathways. For instance, in vitro DNA packaging does not require a concatemer substrate or DNA cutting. Packaging can occur from a free end of a short oligonucleotide and in some cases (e.g., T4) without even requiring TerS. Packaging can also occur by first assembling the TerL subunits on the portal and then capturing the end, or alternatively, terminase can first assemble on the DNA end and then dock on the portal. Surprisingly, the T4 prohead portal can capture a DNA end even in the absence of TerL; and when TerL is provided, the motor assembles around the bound DNA and, after ATP is added, translocates the DNA (52). These observations suggest that the structure of the phage packaging machine may have evolved to efficiently perform this seemingly difficult task of inserting the 23-Å-diameter DNA end into the ~30–35-Å-diameter portal channel. In vivo, however, where the relative concentrations of the packaging components are low and competing interactions are numerous, a dynamically active holoterminase complex is essential to seamlessly perform this transition.
Triggering Translocation
Although the end of a DNA molecule is inserted into the motor channel, it may still be in a dynamic state of dissociation and reassociation unless stabilized by translocating several base pairs of DNA into the capsid. In T4, lowering the motor’s rate of ATP hydrolysis either by lowering the ATP concentration or by mutating the ATP-binding site severely impaired packaging initiation (52). Thus, a rapid succession of ATPase firing, analogous to the cranking of an automobile engine, might be essential at the start of DNA packaging to overcome the dissociation of DNA from the motor. This would push the DNA past the ~100-Å-long (~30-bp) translocation channel into the capsid, making release no longer a significant barrier for translocation. The ATPase-stimulatory property of TerS might be essential at this step, considering that TerS is probably still associated with the TerL-DNA complex (holoterminase) during this transition.
DNA TRANSLOCATION
Anatomy of the Packaging Machine
The packaging machine consists of two essential components, the motor and the portal. As mentioned above, TerL assembles on the portal as an oligomeric motor, generates energy, and uses this energy to translocate DNA. The portal assists the motor and provides a channel through which DNA is transported into the capsid. Although the motor-portal complex functions in association with the capsid, the capsid as such is unlikely to be directly involved in the translocation mechanism. The TerS subunit is also required—probably to regulate motor function, but not in the mechanics of translocation because DNA can be efficiently packaged in vitro in the absence of TerS (53).
The motor
It is now clear that TerL is the motor protein that hydrolyzes ATP and translocates DNA into the capsid. As mentioned above, TerL consists of an N-terminal ATPase/motor domain and a C-terminal nuclease/translocase domain, as well as a hinge connecting the two domains (28) (Figure 4a,b). The T4 ATPase domain has two subdomains, a larger catalytic subdomain (NsubI) and a smaller regulatory subdomain (NsubII). Extensive mutagenesis and biochemical studies combined with bioinformatics analyses, in particular of phage T4 TerL (gp17), elucidated the functional motifs present in the TerL domains (3, 4). Atomic structures of T4 TerL provided structural context for the catalytic residues identified in these motifs (35). Subsequent atomic structures of the C domain from phages SPP1 and P22, HCMV, and HSV1 and the full-length structure of phage Sf6 TerL established that the fold and the overall structure of the ATPase and nuclease domains are well conserved in different TerL proteins despite having very low sequence similarity (30–34).
Significant differences in the full-length structures of T4 and Sf6 TerL proteins are of note (Figure 4c). The 465 amino acid Sf6 TerL is smaller than the 610 amino acid T4 TerL (31, 35). This difference is due in part to the lack of a separate subdomain II (NsubII) in Sf6 TerL. However, Sf6 TerL has a larger hinge, which might carry out the regulatory function of T4 TerL’s NsubII, as follows: ATP binds in the crevice formed by the ATPase domain and the hinge in Sf6 TerL, whereas in T4 TerL, the crevice is between NsubI and NsubII (Figure 4a,b). In Sf6 TerL, the arginine finger that triggers ATP hydrolysis alternates between coordinating the β and γ phosphates of ATP and forming charge-charge interaction with a Glu residue in the hinge, whereas in T4 TerL, the interaction is with an equivalent residue in the NsubII domain. Thus, the differences in the TerL structures might reflect adaptations by different phages while preserving the basic functional and mechanistic features.
The power generator
The N-terminal domains of TerLs contain a conserved ATPase catalytic center (54), which provides the energy for DNA packaging (Figure 4d,e). The N-terminal domain of T4 and Sf6 TerLs has a RecA-type ATP-binding pocket, a six-stranded parallel β-sheet with interspersed α-helices (known as the Rossmann fold) that orient the residues of the catalytic motifs—Walker A, Walker B, catalytic carboxylate, coupling motif (C motif), and Q motif—into the pocket (3, 4) (Figure 4e). The TerL Walker A [G/A/S-XXXGK(T/S)] is one residue shorter than the canonical Walker A [GXXXXGK(T/S)]. The Walker A forms a phosphate-binding loop in which the Lys ε-amino group coordinates the β and γ phosphates of ATP and the Thr/Ser hydroxyl with the Mg-ATP complex. The Walker B motif is a β-strand of four hydrophobic amino acids terminated by an Asp residue that coordinates the Mg2+ of the Mg-ATP complex. The Asp-Mg2+ positions the β and γ phosphates of ATP for nucleophilic attack by the OH− nucleophile generated through splitting of a bound water molecule by the catalytic carboxylate, a Glu residue present next to the Walker B Asp. In addition, a hydrophobic or nonpolar amino acid next to Glu might also be essential (55). In phage λ gpA, mutations in or near the Walker B and the C motif reduce packaging velocity (56). These residues may be part of a velocity controller that controls the rate of ATP hydrolysis and, in turn, motor velocity. The C motif (aka motif III in helicases), a conserved amino acid triplet with Thr or Ser often present at the third position, couples ATP hydrolysis to DNA movement. TerL also contains an adenine-binding motif 10–25 residues upstream of the Walker A, later named the Q motif because Gln is a key residue (57, 58). These catalytic signatures form a network of interactions with ATP and fire ATP hydrolysis upon receiving appropriate cues from the packaging machine. Perturbations, even a loss of a single hydrogen bond or a change in the length of a side chain by a single carbon, could lead to lethality. For instance, Asp255Glu (Walker B) or Thr287Ala (C motif) in T4 gp17 results in near-complete loss of ATP hydrolysis and DNA packaging (59). The λ gpA mutant Tyr46Phe fails to package the full-length genome because of increased DNA slipping and lowered force generation (60). Finally, ATP hydrolysis is triggered by an arginine finger that fires ATP hydrolysis. The arginine finger of phage terminases is located as part of the Walker A motif and fires the ATPase of the same motor subunit (cis), but not that of the adjacent subunit (trans) as was found in ring-type helicases.
The DNA translocator
As described above, the structures of the TerL C-terminal domain contain a conserved nuclease catalytic center with a clearly delineated DNA-binding groove (Figures 2 and 4f,g). Mutational and structural evidence suggests that this domain is also responsible for DNA translocation (28, 31, 35). However, the DNA groove that does the translocation has not been established. In phage SPP1, some of the mutations in the nuclease center that failed to cut DNA also failed to package DNA, implying that the nuclease groove may also be involved in DNA translocation (38). The structural organization of Sf6 TerL is consistent with this hypothesis (31). If so, the nuclease activity must be turned off during translocation, and the groove function must switch from DNA cutting to DNA bind-release (see below), which probably requires significant reorganization of the groove. A second shallow DNA groove was predicted on the opposite side of the T4 C domain based on the structural data (35). The cryoelectron microscopy (cryo-EM) structure of the T4 prohead-motor complex places this groove in the DNA translocation channel in line with the portal channel, leading to the proposal that an alternative groove might be dedicated for DNA translocation. Mutations at one of the proposed residues of this groove (Arg406) resulted in loss of DNA translocation (35). However, this second groove has not been found in other TerL C domain structures. Further genetic, biochemical, and structural studies of TerL-DNA complexes are needed to resolve this question.
The portal
Although there is no significant amino acid sequence similarity, the overall shape, structure, and stoichiometry of portals are well conserved in phages and herpesviruses (4). Atomic structures of the dodecameric portal assembly have been determined from three different phage families; Podoviridae (φ29, P22), Siphoviridae (SPP1), and Myoviridae (T4) (Figure 5) (61–65). These show a common core structure. The cone-shaped structure consists of “crown” and “wing” domains at the wide end, positioned in the prohead interior, and a central “stem,” also called the core, with 24 long α-helices (2 per subunit) that bundle into a palisade and are slanted at a 30–40° angle relative to the central axis. A “stalk” or “clip” domain, which includes an αβ structure and the associated loops that protrude outside the prohead, forms the terminase docking site. Phage P22’s portal has a fifth domain attached to the crown (64), an ~120 amino acid long glutamine-rich α-helical barrel (66). Portal domains are connected with linkers or hinges at the most constricted regions of the portal channel. The length of the channel is 75–110 Å and the radius is 35–65 Å, sufficiently wide to accommodate the dsDNA.
Figure 5.
Structure of the portal assembly. X-ray structures of the portal assemblies from phages P22, SPP1, and φ29 and a cryoelectron microscopy (cryo-EM) structure of phage T4 portal assembly are shown in rainbow colors, ranging from blue at the N terminus to red at the C terminus (61, 62, 66). A cross section of the SPP1 portal (magenta) is shown to identify the subdomains; clip or stalk, stem, tunnel loop, wing, and crown.
Genetic and biochemical studies suggest that the portal, in addition to providing a channel for DNA transport, is actively engaged in the packaging process (63, 67–69). However, a widely considered model that involves rotation of the portal to translocate DNA has been ruled out (70, 71). Certain mutations in channel helices or cross-linking of the helices results in loss of DNA packaging activity (72). The 15–20 amino acid long tunnel loops, which connect the stem and wing domains, protrude into the channel and likely interact with the DNA. These loops are disordered and are not seen in the electron density of two of the three X-ray structures, and the density is weak in the cryo-EM structure of the T4 portal, suggesting conformational flexibility. Deletion of the loop does not affect DNA packaging per se, but the motor cannot complete the packaging of the viral genome. The last packaged DNA leaks out in these mutants, and the motor tries repeatedly to repackage the leaked DNA (63, 73). The phage particles produced have shorter genomes and are noninfectious. These observations suggest that the tunnel loops might restrain the packaged genome. In addition, these loops might also restrain the packaged DNA during translocation, which could explain the unidirectional translocation observed when the portal was employed as a passive nanopore (74). Finally, certain portal mutations cause underpackaging or overpackaging of DNA, or affect the efficiency of termination, implying that the portal senses the internal pressure and signals the motor to terminate packaging when the head is full (68, 69, 75, 76). The portal thus seems to act as a communicator and regulator of the packaging motor.
Motor stoichiometry
TerL assembles on the portal into an oligomeric motor. Cryo-EM structures of prohead-motor complexes from phages φ29 and T4 show a ring of extra density attached to the protruding clip domain of the dodecameric portal (Figure 6a,b, panel i) (35, 48, 77). This density corresponds to a pentamer of gp17 in T4 (Figure 6a, panels ii and iii) and a pentamer of the pRNA-gp16 complex in φ29 (Figure 6b, panels ii and iii) (77). In T4, the X-ray structures of the individual gp17 domains could be fitted into the density, with the N-terminal ATPase domain facing the portal and the C-terminal nuclease/translocase domain forming the bottom portion of the ring. Negative-stain electron microscopy structures of the phage T7 TerL and portal-TerL complex also showed a pentameric TerL ring stacked under a dodecameric portal. Fitting of TerL structural models generated using the T4 atomic structures gave the same domain orientation relative to the portal as the T4 motor (78).
Figure 6.

Structure of the DNA packaging machine. (a) (i) Cryoelectron microscopy (cryo-EM) structure of the phage T4 prohead-gp17 complex (35). gp17 assembles as a pentamer (boxed) at the special portal vertex. (ii) Enlarged view of the pentameric motor. (iii) The X-ray structures of the N-terminal ATPase domain ( green and yellow) and the C-terminal nuclease domain (cyan) are fitted into the cryo-EM density, and DNA is inserted in the center of the channel. The phage SPP1 portal is fitted into the cryo-EM density of the portal. (b) (i) Cryo-EM structure of the φ29 prohead-pRNA-gp16 complex (9). (ii) Enlarged and (iii) cross-section views of the portal (cyan), pRNA (magenta), and gp16 TerL ATPase ( purple) complex assembled at the special vertex.
Alternative stoichiometries have been reported for phage λ TerL (gpA) and φ29 pRNA, as follows (45, 79). Negative-stain electron microscopy and analytical ultracentrifugation studies of an active λ holoterminase complex showed a tetramer of protomers: [TerS2:TerL1]4. Probability analysis of wild-type gpA and a packaging-deficient gpA mutant assembling into active or inactive complexes suggested a motor stoichiometry of four or five TerL subunits (46). In the case of φ29, a hexameric stoichiometry was deduced by counting the number of steps needed to completely photobleach fluorescently labeled pRNA bound to proheads. On the other hand, the recent sub-nanometer asymmetric cryo-EM reconstruction of the φ29 prohead-pRNA complex performed without imposing any symmetry has verified the pentamer stoichiometry of the φ29 packaging motor (48).
From mechanistic considerations, the stoichiometry of the motor might have to be the same in all phages, given that the dodecameric portal stoichiometry and 10.5-bp helical pitch of DNA are fixed. Although there is consensus for a pentameric packaging motor, more high-resolution cryo-EM structures and single-molecule fluorescence experiments that allow direct counting of motor subunits in the actively packaging machines are needed to definitively establish motor stoichiometry.
Portal-motor interface
Portal structures established that the loops of the clip domain that protrude at the bottom of the portal assembly are positioned to interact with the packaging motor. Indeed, genetic and biochemical studies mapped the motor-binding site in the T4 and SPP1 portals to residues present in these loops (63, 80). Furthermore, a peptide corresponding to these residues of the T4 portal bound gp17 and inhibited DNA packaging. However, the region of TerL that interacts with the portal is controversial. Early genetic and biochemical studies showed that a portal-binding site is present in the last 15 amino acids of λ gpA and T3 gp19 (4). The putative binding motif maps to LYWEDD (amino acids 571–576) and LSGEDE (amino acids 636–641) in gpA and gp19, respectively. But in phage T4, the C-terminal 33 amino acids are not essential for DNA packaging. In addition, second-site suppressors, i.e., compensating mutations that alleviate the defects of the T4 portal (gp20), map to the central (Ser336Asn) and C-terminal (Ser583Asn) regions of gp17 (81). Both the N-terminal ATPase domain and the C-terminal nuclease domain inhibit in vitro DNA packaging, presumably by competing with the full-length gp17 for portal binding (28). In the cryo-EM structure of the T4 prohead-gp17 complex, the N-terminal ATPase domain structure fits into the density facing the portal, whereas the C-terminal nuclease/translocase domain structure fits into the density of the distal lobe (Figure 6a, panels ii and iii). Mutagenesis data showed that a helix-loop-helix peptide in the central region of gp17 contains the portal-binding site, and a peptide corresponding to this region inhibits DNA packaging (82). Several heat-sensitive mutations have been mapped to this region, and changing the loop residues of the helix-loop-helix leads to loss of DNA packaging (82). These studies are consistent with the presence of a portal-binding site in the central region of gp17. On the other hand, fluorescence resonance energy transfer studies suggested that a fluorescent probe attached to the C terminus of gp17 is closer in distance to the N terminus of the portal than is another fluorescent probe attached to the N terminus of gp17, which led to the prediction of an opposite orientation (83). In phage SPP1, the C-terminal domain but not the N-terminal domain of TerL (gp2) bound to the prohead portal (38). However, the latter observation would not rule out the possibility that the binding site might be in the central region of TerL. A high-resolution cryo-EM structure of the prohead-motor complex is necessary to establish the orientation of motor domains as well as to define the residues that interact with the portal.
Packaging Dynamics
Optical tweezers technology analyzes the dynamic aspects of DNA translocation at the single-molecule level in real time (6). Prohead-motor complexes are tethered to a microsphere (2 μm in diameter) with anticapsid antibodies. DNA molecules, biotinylated at one end, are tethered to a second, streptavidin-coated microsphere (Figure 7a). The packaging machine, suspended between the two beads, is assembled either by directly tethering an ATPγS-stalled packaging complex or by bringing the prohead-motor bead and the DNA bead close to each other. In the latter case, the motor captures the free end of a DNA molecule on the DNA bead and begins packaging when ATP is provided. The beads are then pulled apart by applying a small amount of force (5 pN) to fully extend the DNA. The force generated by the motor can then be measured at piconewton resolution, and the motor velocity, defined as the reduction of DNA tether length with time, can be measured at a resolution of less than 1 nm in 0.1 s. In addition, slips and pauses can be detected and quantified.
Figure 7.
Analysis of packaging dynamics at the single-molecule level in real time. (a) (i) The dual optical tweezers setup to analyze the dynamics of DNA packaging using single packaging motors. (ii) Reduction of tether length when the motor packages DNA. (b) The dwell-burst translocation cycle of the phage φ29 DNA packaging motor.
Force, rate, and processivity
Studies with the phage φ29, T4, and λ packaging motors have revealed that the packaging motor is exceedingly powerful, generating forces of ~60 pN, 20–25 times the force generated by the myosin motor. Such high forces are probably essential to overcome the strong repulsive forces in the condensed DNA as well as the forces that resist DNA bending and the entropic penalty to confine the highly negatively charged polymer in a nanoscale compartment.
The velocity of the packaging motor varies among the different phages. The φ29 motor packages the 19.3-kb viral genome at 100–150 bp/s, the phage λ motor packages the 48.5-kb genome at ~600 bp/s, and the phage T4 motor packages the 170-kb genome at ~900 bp/s (13, 84, 85). These are the initial rates when the capsid is nearly empty and there is negligible pressure buildup inside the capsid. Rates as high as 2,000 bp/s have been recorded for the T4 motor, making it the fastest known packaging motor. Velocity differences might allow phages to package differently sized genomes in the same amount of time, ~3–5 min. Rapid DNA packaging within this time window might be an important aspect of phage evolution, to maximize the number of completed phages in the ~25–30-min infection cycle. As an example of the consequence of slow assembly, the Thr194Met mutation in λ’s TerL, which slows the translocation rate about 8-fold, is lethal (56, 86).
Motor velocity decreases with applied external force (6). Remarkably, the T4 motor can translocate at a speed of ~380 bp/s even when a large amount of force, 40 pN, is applied, giving the motor a power density of ~5,000 kW/m3. If the nanoscale motor were scaled up to a macromotor, the T4 packaging motor would be approximately twice as powerful as that of a typical automobile engine.
A fit motor must be highly processive and must not dissociate from the packaging complex until the entire viral genome is encapsidated. Otherwise, noninfectious, partially packaged virus particles will be produced. Indeed, phage packaging motors exhibit high processivity. One way to assess processivity is by measuring the frequency of pauses, when the motor makes random stops for a fraction of a second but remains bound to the capsid and then resumes packaging. The pause frequency of the φ29 motor in the initial stages of packaging is approximately once per 12 kb, but it rises sharply during the late stages of head filling (see below). The T4 motor, on the other hand, exhibits a higher pausing frequency, approximately once per 3 kb. This might be a feature that allows the T4 motor to overcome the numerous DNA roadblocks, such as nicks, branches, and bound proteins, that stem from T4’s high level of recombination.
The free energy of ATP hydrolysis, when expressed in units of force and displacement, is equivalent to ~120 pN/nm. Because all three phage packaging motors studied generate ~60 pN of force, the motor would translocate 2 nm, or approximately 6 bp, if 100% of the free energy were to be converted into DNA translocation. An ~2-bp step size (87, 88) therefore gives an estimated ~30% efficiency for the conversion of ATP free energy into mechanical motion. Of the remaining 70%, a fraction likely supports conformational transitions and the rest is probably released as heat.
Dwell and burst
High-resolution measurements of φ29 packaging showed that each translocation cycle is separated into two distinct phases, dwell and burst (89) (Figure 7b). In the dwell phase, the motor pauses and loads the subunits with ATP, and in the burst phase, it translocates DNA in 10-bp increments. ATPs bind to the motor during the dwell phase, one ATP to each subunit, with no apparent cooperativity. The dwell phase is thus very sensitive to ATP concentration, and the time the motor spends in this phase sharply increases at low ATP concentration. In contrast, the burst phase is not affected by ATP concentration, as it is initiated only after all the motor subunits are fully loaded with ATP. Firing in rapid succession by the motor subunits completes the burst phase, and the motor enters another dwell phase to reload ATP. Consequently, the motor velocity greatly decreases with decreasing ATP concentration even though the actual translocation rate during the burst remains unchanged.
Step size
The oligomeric ring structure of the packaging motor implies that each ATPase takes turns hydrolyzing ATP and translocating a certain length of DNA into the phage head. The step size is equal to the number of DNA base pairs translocated per ATP hydrolyzed. Initial calculations from ensemble (bulk) assays gave a step size of ~2 bp/ATP for the phage φ29 and T3 motors (87, 88). This value is probably an underestimate, because in bulk assays, some of the ATPases in the reaction mixture “idle,” hydrolyzing ATP but not packaging DNA.
By slowing down φ29 DNA packaging to <10 bp/s by using low ATP concentrations and applying high external force, it was possible to resolve each burst into sub-bursts (47). Unexpectedly, there are only four sub-bursts instead of five, each corresponding to translocation of 2.5-bp, a nonintegral number (47). Although ATP hydrolysis was not measured, these data compel one to conclude that each sub-burst is powered by firing of one ATPase subunit. This means that, in the φ29 pentameric motor, the step size is 2.5 bp and only four of the five ATPase subunits translocate DNA, whereas the fifth ATPase subunit may have a special regulatory role (see below).
DNA grips
Gripping of DNA is essential for translocation and retention of DNA inside the capsid. In the dwell-burst mechanism, the motor takes a significant pause after each burst to load ATPs. During this time, the DNA must be tightly bound to prevent its release from the capsid. TerL and the portal loops play significant roles in retention, based on mutant behavior. In addition, the motor must grip the DNA in order to push it into the capsid, and also probably during transfer from one subunit to the next. Otherwise, the 5–8 pN of force applied in single-particle experiments could pull some of the packaged DNA out of the capsid. In the infected cell, the internal capsid pressure accumulated due to DNA packaging would do the same. Thus, regardless of the mechanism, there might be at least two DNA coordinated grip-release events in each translocation cycle (Figure 8) that occur for different reasons and probably on different timescales and potentially involve different components of the packaging machine.
Figure 8.

Schematic of the inchworm DNA translocation mechanism. The hands represent the motor parts that grip and release the DNA. One hand grips (panel i) and pushes DNA into the capsid (panel ii) and then releases. The second hand grips the DNA (panel iii) to prevent the release of translocated DNA while it is handed over to the first hand to repeat the process (panel i′).
Packaging experiments in φ29 using substrates with short stretches (~10–20 bp) of modified DNA are instructive (90). The modifications include neutral DNA containing methylphosphonates instead of phosphates, abasic DNA lacking purine/pyrimidine bases, single-stranded DNA, or an unstructured non-DNA polymer. Packaging was more sensitive to insertions in the 5′→3′ strand than in the 3′→5′ strand (relative to the direction of packaging), indicating that the motor might grip (track) the 5′→3′ strand. Surprisingly, however, the motor can package through these aberrant regions as long as the insert length is ≤10 bp. Packaging was greatly reduced, or failed, if the aberrant insert was 11 bp or longer. This means that the motor must engage with the canonical dsDNA once every 10 bp, which aligns with the dwell phase after each 10-bp burst. Thus, the motor might come into phase with a periodic element of DNA, such as backbone phosphates, while being reloaded with ATP. On the other hand, the burst phase does not appear to require specific interactions with DNA, because the motor can traverse up to 10 bp of unnatural DNA or even a non-DNA polymer without stalling. Whether these results apply to other phage motors remains to be seen. The φ29 TerL C domain is smaller and lacks the nuclease center. It is not known whether φ29 TerL has a DNA groove similar to the one found in other TerL C domains (Figure 3). Hence, there might be differences in the DNA gripping features of different phage motors. Furthermore, the DNA binding affinity is modulated by the nucleotide state of the ATPase. For instance, the phage λ TerL Tyr46Phe mutant with an altered Q motif frequently loses grip on the DNA, resulting in frequent slips and loss of processivity (60).
DNA rotation
DNA is likely held as a rigid rod in the ~100-Å-long motor channel and moved linearly into the prohead by translational motion, although some eccentricity is expected due to its symmetry mismatch with the motor and the portal. However, rotation of DNA is essential for two reasons. First, as the packaging motor pumps DNA into the capsid, it needs to be bent (wound) in order to conform to the dimensions of the capsid. This would introduce torsion, which must be released by rotation of DNA outside the capsid. Such release could probably occur spontaneously during translocation, for example during the burst phase in φ29, when the DNA is not held tightly by the motor. Second, the helical pitch of the B-form DNA is 10.4–10.5 bp, not exactly 10 bp, and it varies with the DNA sequence. The 5-fold symmetry of the packaging motor and the 10-bp burst of each translocation cycle mean that the DNA returns to the same motor subunit exactly every 10 bp, not every 10.5 bp. This means that either the motor or the DNA must rotate by 14–18° after (or during) each translocation cycle in order to bring the motor into phase with the DNA. By attaching a third bead to a nicked strand in the middle of the packaging DNA substrate and measuring its angle relative to the DNA axis, it was found that the bead rotates in the left-handed direction by ~14° per burst cycle or 1.4° per base pair of DNA packaged (15) (Figure 7b). Further, the amount of rotation increased to ~48° per cycle at the last stage of DNA packaging, when the capsid is nearly 100% full. At this point, the burst decreased to 9 bp from 10 bp, requiring 48° of rotation. These observations fit with the 2.5-bp step size and pentamer motor stoichiometry but do not account for the additional rotation that might be needed to relieve the torsion introduced due to winding of the DNA inside the capsid. It is also unclear why the DNA rotates in one direction only.
Motor coordination
The dwell-burst mechanism of the φ29 motor requires strict coordination between the motor subunits. All the motor subunits must be loaded with ATP before firing (burst) can begin. It also imposes division of labor among the motor subunits. Because only four of the five subunits fire, the DNA always returns to the same fifth subunit after translocating one helical turn. The motor then enters the dwell phase, reloading the ATP fuel while the fifth subunit holds onto the DNA. This means that some feature of the motor must make the fifth subunit a special subunit. This functional asymmetry might be due to as-yet-unknown asymmetry of the motor ring structure, or it may simply be that this subunit happens to be at the right place at the right time after each dwell phase to interact with the DNA.
A model for motor coordination and a special role for the fifth subunit stems from the pausing behavior of the motor in the presence of nonhydrolyzable ATP analogs (91) (Figure 7b). The model predicts that the motor subunits during DNA translocation exist in a nucleotide-bound state, either with ADP or ATP, but never in an empty (apo) state. At the start of a new translocation cycle, all five subunits of the motor will be in the ADP state as a result of ATP hydrolyses in the previous burst cycle, in which ADP remains bound to the motor while Pi is released. ATP binds to the special subunit, causing a conformational change that leads to tight interactions with the backbone phosphates of the DNA. This might also be the point at which the DNA rotates by 14–18° to come into phase with the special subunit. This conformational transition is then communicated to the next subunit, which then binds ATP, releasing its ADP. However, no cooperativity—i.e., change in ATP binding affinity of the next subunit—is involved. This interlaced ATP-ADP exchange is repeated in sequence until all the motor subunits are loaded with ATP (Figure 7b). The special subunit then hydrolyzes ATP, reverting to the ADP conformation, in which the motor has the least affinity to DNA, and thus breaking contacts with the DNA. The burst phase is then triggered, with the four remaining subunits hydrolyzing ATP in rapid succession. Release of Pi molecules is coupled to translocation of 4 × 2.5 bp of DNA. Another dwell phase starts as the special subunit captures ATP and makes tight contacts with the DNA.
The above dwell-burst cycle repeats in a strictly coordinated fashion, with four subunits translocating DNA and the fifth subunit regulating the precise timing of the start of the burst cycle. This division of labor among the motor subunits, including the regulatory role for one of the subunits, repeats during genome packaging unless there is a pause or slip during the burst phase or a nonhydrolyzable ATP analog is bound to one of the motor subunits. Here, the special role might switch to another subunit in order to recover from the pause and restart another series of dwell-burst translocation cycles. In total, ~2,000 such cycles would lead to the encapsidation of the 19.3-kb φ29 genome.
Alternative models
Whether a strictly coordinated dwell-burst mechanism is common to other phage or viral packaging motors is not known. In the case of the phage λ motor, ensemble assays showed that a single ATPase mutant subunit can poison the packaging motor (46). Probability calculations are consistent with strict coordination of the λ packaging motor, but this does not necessarily mean that the dwell-burst mechanism is operational.
Data from the phage T4 packaging motor argue against a strict dwell-burst mechanism. T4’s packaging rate does not decrease sharply with decreasing ATP concentration, as observed in φ29 (92). Instead, the motor randomly pauses and the frequency of pausing increases with decreasing ATP concentration. The paused motor unpackages, slowly releasing the packaged DNA, a phenomenon thus far observed only with the T4 motor. Consequently, at an ATP concentration of 25 μM, no net packaging occurs even though the motor is still packaging at a rate of ~300 bp/s because frequent pausing and unpackaging release the packaged DNA.
Although the dwell-burst mechanism cannot be excluded, this behavior is inconsistent with such a mechanism. In φ29, the dwells (pauses) are evenly spaced and the dwell time increases at low ATP concentration, but no random pauses are observed (47). During the dwell, the φ29 motor makes strong contacts with the DNA (through the special motor subunit), whereas the T4 motor makes weak contacts as it unpackages DNA when paused. Whereas the dwell duration is sensitive to ATP concentration in φ29, it is independent of ATP concentration in T4 (92). Thus, the T4 motor does not seem to be as strictly coordinated as the φ29 motor. It might belong to a different category of motors that may have sacrificed motor coordination for speed during evolution, for instance by giving up the dwell phase, in order to package ~8 times more DNA in the same period of time. This is consistent with the assignment, based on sequence analyses, that the φ29 packaging ATPase belongs to the HerA-FtsK superfamily of ATPases, a lineage that is distinct from the lineage of terminases from T4 and other large tailed bacteriophages (93). However, both these lineages belong to the broad ASCE (additional strand conserved E) division P-loop NTPases but probably evolved at different time points from an ancestral ASCE ATPase and then clustered into independent groups during the evolution of tailed bacteriophages.
Translocation Mechanism
Many models have been proposed to explain the mechanism by which the energy from ATP hydrolysis is used to drive DNA translocation into the capsid (reviewed in 3). However, it remains an open question. Recent atomic structures of the packaging proteins and analyses of single packaging machines combined with mutational studies have generated specific and testable hypotheses.
Inchworm
There is now a consensus that DNA translocation occurs by an inchworm-type mechanism (Figure 8). One part of the packaging machine (the motor) grips the DNA (panel i) and translates about 2 bp of DNA into the capsid (panel ii) and releases. Another part of the machine (motor or portal) then grips the translocated DNA (panel iii) while the motor returns to grip the DNA to repeat the process (panel i′).
Perhaps the most detailed model for this type of mechanism has been proposed for T4 based on structural and biochemical data (35, 94) (Figure 9). In this model, the T4 motor subunit (gp17) exists in two conformational states, “relaxed” (“extended”) and “tensed” (“compact”). In the extended state (cryo-EM structure), the N- and C-terminal domains of gp17 are separated by ~7 Å through a flexible hinge. Binding of ATP to the N-terminal ATPase subdomain I (NsubI) and of DNA to the C-terminal nuclease/translocase domain (Figure 9a) orients the arginine finger (Arg162) into the ATPase catalytic site, triggering ATP hydrolysis (Figure 9b). Electrostatic repulsion between the negatively charged products ADP (3–) and Pi (3–) drives them apart, causing the regulatory subdomain (NsubII) to rotate by 6° (X-ray structure) (Figure 9c). Five complementary charge pairs and hydrophobic surfaces between the N and C domains align and attract each other, creating a >2,000-Å interacting interface and causing the C-terminal domain–DNA complex to move ~7 Å closer to the N-terminal ATPase domain (X-ray structure). Consequently, ~2 bp of DNA is translocated into the capsid (Figure 9d). Product (ADP, Pi) release causes NsubII to rotate back to its original position, misaligning the charge pairs and returning the C domain to the extended state (Figure 9e). The DNA is now in register with the adjacent gp17 subunit, which, already loaded with ATP, takes its turn to bind to DNA and repeat the translocation cycle. In this way, fueled by ATP chemical energy, alternating conformational states of the motor generate electrostatic force that drives DNA motion.
Figure 9.
The inchworm-type translocation mechanism proposed for phage T4 DNA packaging. Shown is the sequence of steps (a–e) in a single power stroke that results in translocation of 2 bp of DNA. A schematic illustration of the gp17-DNA complex is shown in the center, with various parts of the motor labeled (94).
This model is supported by genetic and biochemical analyses, particularly the functional behaviors of mutants in the arginine finger, C motif, and hinge region (35, 95). Perhaps the best evidence came from the analysis of charged-pair residues that are part of the electrostatic force generator (94). Switching some of these residues to the opposite charge resulted in impaired force generation. When 50 pN of external force was applied, the velocity of the mutant motors dropped to near zero, whereas the wild-type motor still packaged at a rate of ~100 bp/min. Molecular dynamics simulations showed that these measurements correlated with the computed free energy differences between the extended and compact states of the motor (96).
The dwell-burst mechanism as well as the mechanochemical details of the φ29 packaging motor fit well with the inchworm-type DNA packaging mechanism (9). The details might vary, however. For instance, electrostatic interactions between a positively charged loop of the motor subunit and the DNA backbone, similar to those proposed for the phage P4 single-stranded RNA packaging motor (97), might work as a lever to push the DNA into the capsid.
An inchworm-type mechanism was also inferred from the atomic structures of the phage Sf6 TerL protein (gp2) bound to ATP, ATPγS, or ADP. Even though there are structural differences between T4 TerL and Sf6 TerL (Figure 4), the C-terminal domain of Sf6 TerL, like its counterpart from T4 TerL, is predicted to bind DNA. The C domain–DNA complex is proposed to move by ~7 Å (or 2 bp) when the ATP is hydrolyzed by the N-terminal ATPase domain. Like in T4, translocation is powered by the motor protein alternating between two conformational states, which involves changes in electrostatic interactions (31).
DNA crunching
The classic inchworm mechanism involves simple translational motion of DNA. Although subtle (and transient) changes in DNA structure might occur when the motor grips and releases DNA, or when the motor exerts high forces on the DNA in order to translocate it, this model does not require a major structural change in the B-form DNA. Alternative inchworm models have been proposed that invoke a major structural alteration, such as DNA compression during each translocation step (98). In this type of model, the DNA is packaged by the inchworm mechanism except that the ATP hydrolysis energy is converted into DNA torsional energy, which then drives DNA motion. This requires pushing of DNA by the motor while at the same time requiring the portal (or another part of the motor) to tightly grip the DNA to prevent its motion. Consequently, the force applied by the motor compresses the DNA in the motor channel (referred to as “crunching”), resulting in partial unwinding or conversion to an A form. The portal’s grip is then released while the motor’s grip is retained, resulting in relaxation of the DNA back to the B form and entry of ~2 bp into the capsid. A number of observations on the T4 motor are consistent with this model (83, 98, 99). For instance, intercalating compounds tightly bound to DNA are expelled during DNA translocation, which might be due to DNA crunching. However, alternative explanations are possible, and it is unclear what advantages crunching would provide over simple translational movement of DNA; in addition, as mentioned above, crunching would require more complicated coordination of the DNA grips.
Another more explicit model for DNA structural changes as drivers of translocation has been proposed. In this model, the DNA alternates between two conformational states, the low-energy B form and the high-energy A form (100). Dehydration converts B-form DNA (10.4 bp/turn and 3.4 Å/bp) into A-form DNA (11 bp/turn and 2.6 Å/bp). Consequently, the length of the DNA is reduced by 23%, or 2.5 bp/turn, equivalent to the step size of the φ29 motor. In each translocation cycle, the upper, capsid-proximal region of the motor grips the DNA and initiates dehydration of one turn of DNA, utilizing the energy from ATP hydrolysis. The length of the DNA will be reduced by ~2.5 bp, which means that 2.5 bp more DNA will enter the motor channel. Then a lower, capsid-distal component of the motor grips the DNA, initiating rehydration of DNA and converting the A form back into B form. The DNA now elongates, translocating ~2.5 bp DNA into the capsid. Four such steps, equivalent to one burst in φ29, would translocate 10 bp, or approximately one helical turn, of DNA into the capsid. In this “scrunchworm” model, the motor does not directly do the pushing of the DNA. Instead, it provides energy for the interconversion of two energetically different DNA conformers and coordinates the grip-release states with the dehydration-hydration cycles. However, there is no structural evidence to suggest that the motor channel can be occluded from the highly aqueous physiological environment in order for the dehydration to occur, which in the T4 motor must happen on millisecond timescales.
PACKAGING TERMINATION
A recent revelation from single-molecule studies is that the mechanism of DNA packaging includes active monitoring of capsid filling and signaling of the motor to regulate function (14, 15). This feedback system is indeed critical to ensure that each head receives the full complement of the viral genome, to time the termination of packaging and ejection of the motor, and to stabilize the packaged DNA.
One of the signaling mechanisms involves recognition of cues from the genome sequence for phages such as λ. Phage λ makes sequence-specific termination cuts at cos, and cutting is tightly linked to head filling, in addition to sequence specificity. If an ectopic cos is placed in the middle of the λ chromosome, the motor passes over cos without cutting and the cut is made only at a cos present between 78% and 105% of the wild-type genome length (headful packaging) (101). To terminate packaging, the cosQ and cosN subsites are required (102). The cosQ packaging signal is located ~17 bp upstream of cosN. The cosQ site enables recognition of cosN. If cosQ is deleted, translocation continues past the downstream cosN, resulting in a full head with a protruding DNA that blocks neck and tail addition, a lethal event (103). Examination of the bypassed cosN of a null cosQ mutant shows that the top strand of cosN—i.e., the 5′→3′ strand in the direction of packaging—has been properly nicked, but not the bottom cosN strand (104). This result indicates that cosQ is required to present a TerL that is properly oriented to nick the bottom strand of cosN. It is proposed that cosQ reconfigures a motor subunit, or recruits a TerL from solution. Thus, as at initiation, there is reason to think that major conformational changes of terminase occur during the termination process.
Another signaling mechanism is the internal pressure of the packaged DNA (14). At 70% capsid filling, the internal pressure increases by only about 1 pN (in φ29), too small a force to affect motor velocity. But the motor velocity is reduced to 50%. If the motor was stalled for a few seconds by adding ATPγS and then was restarted with ATP, the motor velocity would be the same as its initial velocity (zero filling). This means that the motor function is affected not by the load on the motor but by allosteric regulation of the packaged DNA. The packaged DNA relaxes to a minimum-energy state, a slow process in the confined quarters of the capsid. This process is apparently communicated to the motor to slow down motor velocity and to give time for the DNA to relax to an energy minimum.
In the last stage of filling, the internal pressure raises sharply to 20–23 pN, applying a considerable load on the motor (14, 15). The ~23-pN internal pressure is 3–5-fold lower than the originally predicted value (13) that did not take into consideration the allosteric regulation. Consequently, at the late stage, both allosteric regulation and the 20–23-pN additional load cause severe changes in the stepping behavior of the motor. The packaging velocity is greatly reduced, and the motor makes frequent pauses and slips. At nearly 100% filling, the dwell phase of φ29 increases by 6-fold, burst duration by 7.5-fold, and pause frequency and duration by 5-fold and 30-fold, respectively. Ultimately, a point is reached at which there would be no net packaging, which likely signals ejection of the motor from the packaging complex.
The portal may function as a signal transducer between the packaged DNA and the motor. Certain portal mutants of phage P22 overpackage DNA (late termination) (68, 76), whereas other portal mutants of SPP1 terminate prematurely (76). Portal mutations in λ affect the efficiency of cosQ-dependent cleavage (69, 75). In phage P22, a DNA strand is wrapped around the portal assembly (105), and differences were observed in the portal structures of the prohead and the finished phage. Mutations in the tunnel loops of the φ29 and T4 portals lead to either release of the packaged DNA or underpackaging of genomic DNA. These observations suggest that portal interactions with the DNA through the crown and wing domains, with the capsid through the wing domain, and with the motor through the clip domain transduce signals to regulate motor function. This might be particularly critical at the late stage of packaging. At 100% filling, a portal conformational change may trigger the events of termination: (a) motor reorganization, (b) termination DNA cleavage, (c) motor dissociation, and (d) motor binding to the newly created concatemer end. There is currently little evidence as to the temporal order of these events. Top-strand nicking of cosN by a λ cosQ mutant indicates that motor dissociation from the portal is not obligatory for that endonucleolytic action (104). Alternatively, the portal change may eject the motor from the filled head while the tunnel loops restrain the DNA from leaking out. Freed from the head, terminase reorganizes into a cutting enzyme and makes a double-strand cut to terminate packaging. Neck proteins assemble on the portal and seal off the packaged head (106–110). These events ensure transfer of a full genome complement into the head and then into a new host cell. The terminase that is still bound to the newly generated DNA end assembles on a new prohead and sponsors packaging of the next chromosome along the concatemer.
CONCLUSIONS AND FUTURE DIRECTIONS
Recent major progress in understanding viral DNA packaging stems from the application of powerful biophysical, structural, and microscopic techniques that permit researchers to probe DNA translocation at the atomic level and from combining these results with the traditionally strong genetic and biochemical approaches. X-ray crystallography and high-resolution cryo-EM have generated the structures of packaging motor components, TerS, TerL, the portal, and the prohead-motor complex from several phages. Such structures have provided a good understanding of these components and a general understanding of their interactions. Though we understand the ATPase center well, much remains to be learned about the mechanics of the motor during the act of moving DNA. Single-molecule studies using optical tweezers methodologies generated quantitative information about the kinetics and energetics of translocation, and some details about the power stroke. Future progress requires understanding the dynamic changes that occur during the dramatic transitions that accompany the initiation, translocation, and termination stages of the packaging process. Single-molecule fluorescence studies and structural analyses of packaging complexes will likely play important roles in elucidating these mechanisms.
Acknowledgments
The authors thank many colleagues for assistance with the preparation of figures: Victor Padilla-Sanchez and Zhang Liang (The Catholic University of America); Bonnie Draper (St. Andrews University); Leonor Oliveira and Paulo Tavares (Unité de Virologie Moléculaire et Structurale, CNRS); Fred Antson (University of York); and Paul Jardine (University of Minnesota). The authors are grateful to Shelley Grimes and Paul Jardine (University of Minnesota); Marc Morais (University of Texas Medical Branch); Leonor Oliveira and Paulo Tavares (Unité de Virologie Moléculaire et Structurale, CNRS); Michael Rossmann (Purdue University); and Carlos Catalano, David Oritz, and Jay Yang (University of Colorado) for critically reading the manuscript and providing thoughtful suggestions. The research in the authors’ laboratories has been funded by the National Science Foundation (V.B.R.: MCB-0923873, MCB-1158495) and the National Institutes of Health (V.B.R.: AI081726; M.F.: GM51611 and GM88186). Special thanks to our present and former lab members for their contributions, and our apologies to colleagues for not being able to cite their work due to space limitations.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Venigalla B. Rao, Email: rao@cua.edu.
Michael Feiss, Email: michael-feiss@uiowa.edu.
LITERATURE CITED
- 1.Hendrix RW, Smith MC, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. PNAS. 1999;96:2192–97. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rixon FJ, Schmidt MF. Structural similarities in DNA packaging and delivery apparatuses in herpesviruses and dsDNA bacteriophages. Curr Opin Virol. 2014;5:105–10. doi: 10.1016/j.coviro.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu Rev Genet. 2008;42:647–81. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 4.Feiss M, Rao VB. The bacteriophage DNA packaging machine. Adv Exp Med Biol. 2012;726:489–509. doi: 10.1007/978-1-4614-0980-9_22. [DOI] [PubMed] [Google Scholar]
- 5.Casjens SR. The DNA-packaging nanomotor of tailed bacteriophages. Nat Rev Microbiol. 2011;9:647–57. doi: 10.1038/nrmicro2632. [DOI] [PubMed] [Google Scholar]
- 6.Chemla YR, Smith DE. Single-molecule studies of viral DNA packaging. Adv Exp Med Biol. 2012;726:549–84. doi: 10.1007/978-1-4614-0980-9_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black LW, Rao VB. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv Virus Res. 2012;82:119–53. doi: 10.1016/B978-0-12-394621-8.00018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Black LW. Old, new, and widely true: the bacteriophage T4 DNA packaging mechanism. Virology. 2015;479–80:650–56. doi: 10.1016/j.virol.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morais MC. The dsDNA packaging motor in bacteriophage φ29. Adv Exp Med Biol. 2012;726:511–47. doi: 10.1007/978-1-4614-0980-9_23. [DOI] [PubMed] [Google Scholar]
- 10.Oliveira L, Tavares P, Alonso JC. Headful DNA packaging: bacteriophage SPP1 as a model system. Virus Res. 2013;173:247–59. doi: 10.1016/j.virusres.2013.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Chelikani V, Ranjan T, Kondabagil K. Revisiting the genome packaging in viruses with lessons from the “giants”. Virology. 2014;466–67:15–26. doi: 10.1016/j.virol.2014.06.022. [DOI] [PubMed] [Google Scholar]
- 12.Hendrix RW, Johnson JE. Bacteriophage HK97 capsid assembly and maturation. Adv Exp Med Biol. 2012;726:351–63. doi: 10.1007/978-1-4614-0980-9_15. [DOI] [PubMed] [Google Scholar]
- 13.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight φ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–52. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 14.Berndsen ZT, Keller N, Smith DE. Continuous allosteric regulation of a viral packaging motor by a sensor that detects the density and conformation of packaged DNA. Biophys J. 2015;108:315–24. doi: 10.1016/j.bpj.2014.11.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu S, Chistol G, Hetherington CL, Tafoya S, Aathavan K, et al. A viral packaging motor varies its DNA rotation and step size to preserve subunit coordination as the capsid fills. Cell. 2014;157:702–13. doi: 10.1016/j.cell.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sao-Jose C, de Frutos M, Raspaud E, Santos MA, Tavares P. Pressure built by DNA packing inside virions: enough to drive DNA ejection in vitro, largely insufficient for delivery into the bacterial cytoplasm. J Mol Biol. 2007;374:346–55. doi: 10.1016/j.jmb.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 17.Evilevitch A, Fang LT, Yoffe AM, Castelnovo M, Rau DC, et al. Effects of salt concentrations and bending energy on the extent of ejection of phage genomes. Biophys J. 2008;94:1110–20. doi: 10.1529/biophysj.107.115345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Beer T, Fang J, Ortega M, Yang Q, Maes L, et al. Insights into specific DNA recognition during the assembly of a viral genome packaging machine. Mol Cell. 2002;9:981–91. doi: 10.1016/s1097-2765(02)00537-3. [DOI] [PubMed] [Google Scholar]
- 19.Roy A, Bhardwaj A, Datta P, Lander GC, Cingolani G. Small terminase couples viral DNA binding to genome-packaging ATPase activity. Structure. 2012;20:1403–13. doi: 10.1016/j.str.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H, Finch CJ, Sequeira RD, Johnson BA, Johnson JE, et al. Crystal structure of the DNA-recognition component of the bacterial virus Sf6 genome-packaging machine. PNAS. 2010;107:1971–76. doi: 10.1073/pnas.0908569107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buttner CR, Chechik M, Ortiz-Lombardia M, Smits C, Ebong IO, et al. Structural basis for DNA recognition and loading into a viral packaging motor. PNAS. 2012;109:811–16. doi: 10.1073/pnas.1110270109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun S, Gao S, Kondabagil K, Xiang Y, Rossmann MG, Rao VB. Structure and function of the small terminase component of the DNA packaging machine in T4-like bacteriophages. PNAS. 2012;109:817–22. doi: 10.1073/pnas.1110224109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bhardwaj A, Casjens SR, Cingolani G. Exploring the atomic structure and conformational flexibility of a 320 Å long engineered viral fiber using X-ray crystallography. Acta Crystallogr D. 2014;70:342–53. doi: 10.1107/S1399004713027685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao S, Rao VB. Specificity of interactions among the DNA-packaging machine components of T4-related bacteriophages. J Biol Chem. 2011;286:3944–56. doi: 10.1074/jbc.M110.196907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao H, Kamau YN, Christensen TE, Tang L. Structural and functional studies of the phage Sf6 terminase small subunit reveal a DNA-spooling device facilitated by structural plasticity. J Mol Biol. 2012;423:413–26. doi: 10.1016/j.jmb.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chai S, Lurz R, Alonso JC. The small subunit of the terminase enzyme of Bacillus subtilis bacteriophage SPP1 forms a specialized nucleoprotein complex with the packaging initiation region. J Mol Biol. 1995;252:386–98. doi: 10.1006/jmbi.1995.0505. [DOI] [PubMed] [Google Scholar]
- 27.Higgins RR, Becker A. Interaction of terminase, the DNA packaging enzyme of phage λ, with its cos DNA substrate. J Mol Biol. 1995;252:31–46. doi: 10.1006/jmbi.1995.0473. [DOI] [PubMed] [Google Scholar]
- 28.Kanamaru S, Kondabagil K, Rossmann MG, Rao VB. The functional domains of bacteriophage T4 terminase. J Biol Chem. 2004;279:40795–801. doi: 10.1074/jbc.M403647200. [DOI] [PubMed] [Google Scholar]
- 29.Alam TI, Draper B, Kondabagil K, Rentas FJ, Ghosh-Kumar M, et al. The headful packaging nuclease of bacteriophage T4. Mol Microbiol. 2008;69:1180–90. doi: 10.1111/j.1365-2958.2008.06344.x. [DOI] [PubMed] [Google Scholar]
- 30.Smits C, Chechik M, Kovalevskiy OV, Shevtsov MB, Foster AW, et al. Structural basis for the nuclease activity of a bacteriophage large terminase. EMBO Rep. 2009;10:592–98. doi: 10.1038/embor.2009.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, Christensen TE, Kamau YN, Tang L. Structures of the phage Sf6 large terminase provide new insights into DNA translocation and cleavage. PNAS. 2013;110:8075–80. doi: 10.1073/pnas.1301133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roy A, Cingolani G. Structure of P22 headful packaging nuclease. J Biol Chem. 2012;287:28196–205. doi: 10.1074/jbc.M112.349894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nadal M, Mas PJ, Blanco AG, Arnan C, Sola M, et al. Structure and inhibition of herpesvirus DNA packaging terminase nuclease domain. PNAS. 2010;107:16078–83. doi: 10.1073/pnas.1007144107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selvarajan Sigamani S, Zhao H, Kamau YN, Baines JD, Tang L. The structure of the herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. J Virol. 2013;87:7140–48. doi: 10.1128/JVI.00311-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun S, Kondabagil K, Draper B, Alam TI, Bowman VD, et al. The structure of the phage T4 DNA packaging motor suggests a mechanism dependent on electrostatic forces. Cell. 2008;135:1251. doi: 10.1016/j.cell.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 36.Rychlik MP, Chon H, Cerritelli SM, Klimek P, Crouch RJ, Nowotny M. Crystal structures of RNase H2 in complex with nucleic acid reveal the mechanism of RNA-DNA junction recognition and cleavage. Mol Cell. 2010;40:658–70. doi: 10.1016/j.molcel.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 38.Cornilleau C, Atmane N, Jacquet E, Smits C, Alonso JC, et al. The nuclease domain of the SPP1 packaging motor coordinates DNA cleavage and encapsidation. Nucleic Acids Res. 2013;41:340–54. doi: 10.1093/nar/gks974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh-Kumar M, Alam TI, Draper B, Stack JD, Rao VB. Regulation by interdomain communication of a headful packaging nuclease from bacteriophage T4. Nucleic Acids Res. 2011;39:2742–55. doi: 10.1093/nar/gkq1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho AG, Gual A, Lurz R, Tavares P, Alonso JC. Bacillus subtilis bacteriophage SPP1 DNA packaging motor requires terminase and portal proteins. J Biol Chem. 2003;278:23251–59. doi: 10.1074/jbc.M301805200. [DOI] [PubMed] [Google Scholar]
- 41.Kala S, Cumby N, Sadowski PD, Hyder BZ, Kanelis V, et al. HNH proteins are a widespread component of phage DNA packaging machines. PNAS. 2014;111:6022–27. doi: 10.1073/pnas.1320952111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quiles-Puchalt N, Carpena N, Alonso JC, Novick RP, Marina A, Penades JR. Staphylococcal pathogenicity island DNA packaging system involving cos-site packaging and phage-encoded HNH endonucleases. PNAS. 2014;111:6016–21. doi: 10.1073/pnas.1320538111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heming JD, Huffman JB, Jones LM, Homa FL. Isolation and characterization of the herpes simplex virus 1 terminase complex. J Virol. 2014;88:225–36. doi: 10.1128/JVI.02632-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borst EM, Kleine-Albers J, Gabaev I, Babic M, Wagner K, et al. The human cytomegalovirus UL51 protein is essential for viral genome cleavage-packaging and interacts with the terminase subunits pUL56 and pUL89. J Virol. 2013;87:1720–32. doi: 10.1128/JVI.01955-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maluf N, Gaussier H, Bogner E, Feiss M, Catalano C. Assembly of bacteriophage λ terminase into a viral DNA maturation and packaging machine. Biochemistry. 2006;45:15259–68. doi: 10.1021/bi0615036. [DOI] [PubMed] [Google Scholar]
- 46.Andrews BT, Catalano CE. Strong subunit coordination drives a powerful viral DNA packaging motor. PNAS. 2013;110:5909–14. doi: 10.1073/pnas.1222820110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moffitt JR, Chemla YR, Aathavan K, Grimes S, Jardine PJ, et al. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–50. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao S, Saha M, Zhao W, Jardine PJ, Zhang W, et al. Insights into the structure and assembly of the bacteriophage 29 double-stranded DNA packaging motor. J Virol. 2014;88:3986–96. doi: 10.1128/JVI.03203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang H, Masters M. Bacteriophage P1 pac sites inserted into the chromosome greatly increase packaging and transduction of Escherichia coli genomic DNA. Virology. 2014;468–70:274–82. doi: 10.1016/j.virol.2014.07.029. [DOI] [PubMed] [Google Scholar]
- 50.Furth ME, Wickner SH. Lambda DNA replication. In: Hendrix RW, Roberts JW, Stahl FW, Weisberg RA, editors. Lambda II. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1983. pp. 145–73. [Google Scholar]
- 51.Sternberg N, Coulby J. Recognition and cleavage of the bacteriophage P1 packaging site ( pac). I Differential processing of the cleaved ends in vivo. J Mol Biol. 1987;194:453–68. doi: 10.1016/0022-2836(87)90674-7. [DOI] [PubMed] [Google Scholar]
- 52.Vafabakhsh R, Kondabagil K, Earnest T, Lee KS, Zhang Z, et al. Single-molecule packaging initiation in real time by a viral DNA packaging machine from bacteriophage T4. PNAS. 2014;111:15096–101. doi: 10.1073/pnas.1407235111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao VB, Black LW. Cloning, overexpression and purification of the terminase proteins gp16 and gp17 of bacteriophage T4; construction of a defined in vitro DNA packaging system using purified terminase proteins. J Mol Biol. 1988;200:475–85. doi: 10.1016/0022-2836(88)90537-2. [DOI] [PubMed] [Google Scholar]
- 54.Mitchell MS, Matsuzaki S, Imai S, Rao VB. Sequence analysis of bacteriophage T4 DNA packaging/terminase genes 16 and 17 reveals a common ATPase center in the large subunit of viral terminases. Nucleic Acids Res. 2002;30:4009–21. doi: 10.1093/nar/gkf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kondabagil K, Dai L, Vafabakhsh R, Ha T, Draper B, Rao VB. Designing a nine cysteine-less DNA packaging motor from bacteriophage T4 reveals new insights into ATPase structure and function. Virology. 2014;468–70:660–68. doi: 10.1016/j.virol.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsay JM, Sippy J, DelToro D, Andrews BT, Draper B, et al. Mutations altering a structurally conserved loop-helix-loop region of a viral packaging motor change DNA translocation velocity and processivity. J Biol Chem. 2010;285:24282–89. doi: 10.1074/jbc.M110.129395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tanner NK, Cordin O, Banroques J, Doere M, Linder P. The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol Cell. 2003;11:127–38. doi: 10.1016/s1097-2765(03)00006-6. [DOI] [PubMed] [Google Scholar]
- 58.Kondabagil K, Draper B, Rao VB. Adenine recognition is a key checkpoint in the energy release mechanism of phage T4 DNA packaging motor. J Mol Biol. 2012;415:329–42. doi: 10.1016/j.jmb.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 59.Mitchell MS, Rao VB. Functional analysis of the bacteriophage T4 DNA-packaging ATPase motor. J Biol Chem. 2006;281:518–27. doi: 10.1074/jbc.M507719200. [DOI] [PubMed] [Google Scholar]
- 60.Tsay JM, Sippy J, Feiss M, Smith DE. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. PNAS. 2009;106:14355–60. doi: 10.1073/pnas.0904364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Simpson A, Tao Y, Leiman P, Badasso M, He Y, et al. Structure of the bacteriophage φ29 DNA packaging motor. Nature. 2000;408:745–50. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, et al. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–94. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Padilla-Sanchez V, Gao S, Kim HR, Kihara D, Sun L, et al. Structure-function analysis of the DNA translocating portal of the bacteriophage T4 packaging machine. J Mol Biol. 2014;426:1019–38. doi: 10.1016/j.jmb.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Olia AS, Prevelige PE, Jr, Johnson JE, Cingolani G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat Struct Mol Biol. 2011;18:597. doi: 10.1038/nsmb.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun L, Zhang X, Gao S, Rao PA, Padilla-Sanchez V, et al. Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution. Nat Commun. 2015;6:7548. doi: 10.1038/ncomms8548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang J, Lander GC, Olia A, Li R, Casjens S, et al. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in P22. Structure. 2011;19:496–502. doi: 10.1016/j.str.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Isidro A, Henriques AO, Tavares P. The portal protein plays essential roles at different steps of the SPP1 DNA packaging process. Virology. 2004;322:253–63. doi: 10.1016/j.virol.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 68.Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, et al. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol. 1992;224:1055–74. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- 69.Cue D, Feiss M. Genetic evidence that recognition of cosQ, the signal for termination of phage λ DNA packaging, depends on the extent of head filling. Genetics. 1997;147:7–17. doi: 10.1093/genetics/147.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hugel T, Michaelis J, Hetherington CL, Jardine PJ, Grimes S, et al. Experimental test of connector rotation during DNA packaging into bacteriophage φ29 capsids. PLOS Biol. 2007;5:e59. doi: 10.1371/journal.pbio.0050059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]
- 72.Cuervo A, Vaney MC, Antson AA, Tavares P, Oliveira L. Structural rearrangements between portal protein subunits are essential for viral DNA translocation. J Biol Chem. 2007;282:18907–13. doi: 10.1074/jbc.M701808200. [DOI] [PubMed] [Google Scholar]
- 73.Grimes S, Ma S, Gao J, Atz R, Jardine PJ. Role of φ29 connector channel loops in late-stage DNA packaging. J Mol Biol. 2011;410:50–59. doi: 10.1016/j.jmb.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jing P, Haque F, Shu D, Montemagno C, Guo P. One-way traffic of a viral motor channel for double-stranded DNA translocation. Nano Lett. 2010;10:3620–27. doi: 10.1021/nl101939e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wieczorek D, Didion L, Feiss M. Alterations of the portal protein of bacteriophage λ suppress mutations in cosQ, the site required for termination of DNA packaging. Genetics. 2002;161:21–31. doi: 10.1093/genetics/161.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tavares P, Santos MA, Lurz R, Morelli G, de Lencastre H, Trautner TA. Identification of a gene in Bacillus subtilis bacteriophage SPP1 determining the amount of packaged DNA. J Mol Biol. 1992;225:81–92. doi: 10.1016/0022-2836(92)91027-m. [DOI] [PubMed] [Google Scholar]
- 77.Morais MC, Koti JS, Bowman VD, Reyes-Aldrete E, Anderson DL, Rossmann MG. Defining molecular and domain boundaries in the bacteriophage φ29 DNA packaging motor. Structure. 2008;16:1267–74. doi: 10.1016/j.str.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dauden MI, Martin-Benito J, Sanchez-Ferrero JC, Pulido-Cid M, Valpuesta JM, Carrascosa JL. Large terminase conformational change induced by connector binding in bacteriophage T7. J Biol Chem. 2013;288:16998–7007. doi: 10.1074/jbc.M112.448951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schwartz C, De Donatis GM, Fang H, Guo P. The ATPase of the phi29 DNA packaging motor is a member of the hexameric AAA+ superfamily. Virology. 2013;443:20–27. doi: 10.1016/j.virol.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Oliveira L, Cuervo A, Tavares P. Direct interaction of the bacteriophage SPP1 packaging ATPase with the portal protein. J Biol Chem. 2010;285:7366–73. doi: 10.1074/jbc.M109.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin H, Rao VB, Black LW. Analysis of capsid portal protein and terminase functional domains: interaction sites required for DNA packaging in bacteriophage T4. J Mol Biol. 1999;289:249–60. doi: 10.1006/jmbi.1999.2781. [DOI] [PubMed] [Google Scholar]
- 82.Hegde S, Padilla-Sanchez V, Draper B, Rao VB. Portal-large terminase interactions of the bacteriophage T4 DNA packaging machine implicate a molecular lever mechanism for coupling ATPase to DNA translocation. J Virol. 2012;86:4046–57. doi: 10.1128/JVI.07197-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dixit AB, Ray K, Thomas JA, Black LW. The C-terminal domain of the bacteriophage T4 terminase docks on the prohead portal clip region during DNA packaging. Virology. 2013;446:293–302. doi: 10.1016/j.virol.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fuller DN, Raymer DM, Rickgauer JP, Robertson RM, Catalano CE, et al. Measurements of single DNA molecule packaging dynamics in bacteriophage λ reveal high forces, high motor processivity, and capsid transformations. J Mol Biol. 2007;373:1113–22. doi: 10.1016/j.jmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fuller DN, Raymer DM, Kottadiel VI, Rao VB, Smith DE. Single phage T4 DNA packaging motors exhibit large force generation, high velocity, and dynamic variability. PNAS. 2007;104:16868–73. doi: 10.1073/pnas.0704008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Duffy C, Feiss M. The large subunit of bacteriophage λ’s terminase plays a role in DNA translocation and packaging termination. J Mol Biol. 2002;316:547–61. doi: 10.1006/jmbi.2001.5368. [DOI] [PubMed] [Google Scholar]
- 87.Morita M, Tasaka M, Fujisawa H. DNA packaging ATPase of bacteriophage T3. Virology. 1993;193:748–52. doi: 10.1006/viro.1993.1183. [DOI] [PubMed] [Google Scholar]
- 88.Guo P, Peterson C, Anderson D. Prohead and DNA-gp3-dependent ATPase activity of the DNA packaging protein gp16 of bacteriophage φ29. J Mol Biol. 1987;197:229–36. doi: 10.1016/0022-2836(87)90121-5. [DOI] [PubMed] [Google Scholar]
- 89.Chemla YR, Aathavan K, Michaelis J, Grimes S, Jardine PJ, et al. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–92. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 90.Aathavan K, Politzer AT, Kaplan A, Moffitt JR, Chemla YR, et al. Substrate interactions and promiscuity in a viral DNA packaging motor. Nature. 2009;461:669–73. doi: 10.1038/nature08443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chistol G, Liu S, Hetherington CL, Moffitt JR, Grimes S, et al. High degree of coordination and division of labor among subunits in a homomeric ring ATPase. Cell. 2012;151:1017–28. doi: 10.1016/j.cell.2012.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kottadiel VI, Rao VB, Chemla YR. The dynamic pause-unpackaging state, an off-translocation recovery state of a DNA packaging motor from bacteriophage T4. PNAS. 2012;109:20000–5. doi: 10.1073/pnas.1209214109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burroughs AM, Iyer LM, Aravind L. Comparative genomics and evolutionary trajectories of viral ATP dependent DNA-packaging systems. Genome Dyn. 2007;3:48–65. doi: 10.1159/000107603. [DOI] [PubMed] [Google Scholar]
- 94.Migliori AD, Keller N, Alam TI, Mahalingam M, Rao VB, et al. Evidence for an electrostatic mechanism of force generation by the bacteriophage T4 DNA packaging motor. Nat Commun. 2014;5:4173. doi: 10.1038/ncomms5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Draper B, Rao VB. An ATP hydrolysis sensor in the DNA packaging motor from bacteriophage T4 suggests an inchworm-type translocation mechanism. J Mol Biol. 2007;369:79–94. doi: 10.1016/j.jmb.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 96.Migliori AD, Smith DE, Arya G. Molecular interactions and residues involved in force generation in the T4 viral DNA packaging motor. J Mol Biol. 2014;426:4002–17. doi: 10.1016/j.jmb.2014.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–55. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 98.Ray K, Sabanayagam CR, Lakowicz JR, Black LW. DNA crunching by a viral packaging motor: compression of a procapsid-portal stalled Y-DNA substrate. Virology. 2010;398:224–32. doi: 10.1016/j.virol.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage T4 terminase by DNA structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harvey SC. The scrunchworm hypothesis: Transitions between A-DNA and B-DNA provide the driving force for genome packaging in double-stranded DNA bacteriophages. J Struct Biol. 2015;189:1–8. doi: 10.1016/j.jsb.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feiss M, Siegele DA. Packaging of the bacteriophage lambda chromosome: dependence of cos cleavage on chromosome length. Virology. 1979;92:190–200. doi: 10.1016/0042-6822(79)90224-1. [DOI] [PubMed] [Google Scholar]
- 102.Cue D, Feiss M. Bacteriophage λ DNA packaging: DNA site requirements for termination and processivity. J Mol Biol. 2001;311:233–40. doi: 10.1006/jmbi.2001.4840. [DOI] [PubMed] [Google Scholar]
- 103.Cue D, Feiss M. A site required for termination of packaging of the phage λ chromosome. PNAS. 1993;90:9290–94. doi: 10.1073/pnas.90.20.9290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cue D, Feiss M. Termination of packaging of the bacteriophage λ chromosome: cosQ is required for nicking the bottom strand of cosN. J Mol Biol. 1998;280:11–29. doi: 10.1006/jmbi.1998.1841. [DOI] [PubMed] [Google Scholar]
- 105.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–95. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 106.Lhuillier S, Gallopin M, Gilquin B, Brasiles S, Lancelot N, et al. Structure of bacteriophage SPP1 head-to-tail connection reveals mechanism for viral DNA gating. PNAS. 2009;106:8507–12. doi: 10.1073/pnas.0812407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.White HE, Sherman MB, Brasiles S, Jacquet E, Seavers P, et al. Capsid structure and its stability at the late stages of bacteriophage SPP1 assembly. J Virol. 2012;86:6768–77. doi: 10.1128/JVI.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Maxwell KL, Yee AA, Booth V, Arrowsmith CH, Gold M, Davidson AR. The solution structure of bacteriophage λ protein W, a small morphogenetic protein possessing a novel fold. J Mol Biol. 2001;308:9–14. doi: 10.1006/jmbi.2001.4582. [DOI] [PubMed] [Google Scholar]
- 109.Maxwell KL, Davidson AR, Murialdo H, Gold M. Thermodynamic and functional characterization of protein W from bacteriophage λ: The three C-terminal residues are critical for activity. J Biol Chem. 2000;275:18879–86. doi: 10.1074/jbc.M001178200. [DOI] [PubMed] [Google Scholar]
- 110.Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J Mol Biol. 1984;172:523–43. doi: 10.1016/s0022-2836(84)80021-2. [DOI] [PubMed] [Google Scholar]