Abstract
Prion diseases are closely associated with the conversion of the cellular prion protein (PrPC) to an abnormal conformer (PrPSc) [Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383]. Monoclonal antibodies that bind epitopes comprising residues 96–104 and 133–158 of PrPC potently inhibit this process, presumably by preventing heterodimeric association of PrPC and PrPSc, and suggest that these regions of PrPC may be critical components of the PrPC–PrPSc replicative interface. We reasoned that transplanting PrP sequence corresponding to these regions into a suitable carrier molecule, such as an antibody, could impart specific recognition of disease-associated forms of PrP. To test this hypothesis, polypeptides containing PrP sequence between residues 89–112 or 136–158 were used to replace the extended heavy chain complementarity-determining region 3 of an IgG antibody specific for the envelope glycoprotein of HIV-1. Herein the resulting engineered PrP-IgGs are shown to bind specifically to infective fractions of PrP in mouse, human, and hamster prion-infected tissues, but not to PrPC, other cellular components, or the HIV-1 envelope. PrPSc reactivity was abolished when the sequence of the PrP 89–112 and 136–158 grafts was mutated, scrambled, or N-terminally truncated. Our findings suggest that residues within the 89–112 and 136–158 segments of PrPC are key components of one face of the PrPC–PrPSc complex. PrPSc-specific antibodies produced by the approach described may find widespread application in the study of prion biology and replication and in the detection of infectious prions in human and animal materials.
Transmissible spongiform encephalopathies, including Creutzfeldt–Jakob disease (CJD) in humans and bovine spongiform encephalopathy (BSE) and scrapie in animals, are a family of neurodegenerative diseases caused by prions (1). The emergence in Europe of a new variant form of CJD (vCJD) is closely associated with the ingestion of BSE prion-tainted meat and has elevated concern over the threat that prions pose to the safety of food and blood products (2, 3). Although the number of vCJD cases is currently relatively small, the absence of a sensitive diagnostic test for prion infection has prevented an accurate assessment of how many of the millions of individuals likely exposed to BSE prions are currently incubating disease (4).
PrPSc, an abnormal conformer of the ubiquitous cellular prion protein (PrPC), is the major constituent of purified infectious prion preparations. During prion propagation, the formation of nascent prion infectivity is thought to proceed by means of a template-dependent process in which PrPSc self-replicates by driving the conformational rearrangement of PrPC. Exactly how the distinct PrPC and PrPSc conformers interact with one another, and possibly other auxiliary molecules (5, 6), in the prion replicative complex is unknown. However, the observation that different prion strains retain their characteristic properties over multiple passages indicates that prion propagation is a highfidelity process and suggests that molecular interactions between PrPC and PrPSc are extremely specific (1, 7).
High-affinity antibodies distinguishing between PrPC and PrPSc can be of value in studying the specific machinery of prion replication and in the diagnosis of prion infection. Monoclonal antibodies of the IgM class, recovered by immunizing Prnp0/0 mice with recombinant PrP preparations, have been reported (8, 9). However, the utility of these antibodies is likely to be limited by their relatively low affinity for PrP antigen (10) and reliance on the solvent exposure of hydrophobic epitope motifs that may be present in misfolded molecules other than PrP (9).
Recently, we reported that monoclonal antibody Fab fragments reacting with different epitopes of PrPC efficiently inhibit prion propagation in a scrapie prion-infected neuroblastoma cell line (11). The inhibitory effect we observed is most readily explained by Fab binding to cell-surface PrPC and thereby hindering the docking of PrPSc template or a cofactor critical for conversion of PrPC to PrPSc. Two of the antibody fragments used in these experiments, Fabs D18 and D13, possessed a particularly potent inhibitory effect, indicating that their PrPC epitopes, thought to span residues 133–157 and 96–104 (12), respectively, may play an important role in binding directly to PrPSc. Inhibition of PrPSc formation by an antibody recognizing the 143–151 segment of PrP (10, 13) and by synthetic PrP peptides (14, 15), further implicates the central region of PrP in formation of the PrPC–PrPSc interface. To further investigate this possibility, PrP sequence motifs corresponding to the epitopes of the inhibitory D18 and D13 Fabs were grafted into a recipient antibody scaffold. Here we report that the resulting motif-grafted antibodies bind specifically and with high affinity to disease-associated conformations of PrP.
Methods
Preparation of Motif-Grafted Antibodies. Mouse PrP sequences corresponding to amino acid residues 89–112, 136–158, and 141–158 were independently grafted to replace heavy chain complementarity-determining region 3 (HCDR3) of antibody b12 (16), by using a two-step overlap extension PCR (17). Oligonucleotide primers were subjected to 2-fold polyacrylamide gel electrophoresis purification (Operon Technologies, Alameda, CA) and contained the following sequences: PelSeq (5′-ACCTATTGCCTACGGCAGCCG-3′), CG1d (5′-GCATGTACTAGTTTTGTCACAAGATTTGG-3′), MoPrP89–112 5′ (5′-CATAATCAGTGGAACAAGCCCAGCAAACCAAAAACCAACCTCAAGCATGTGGGCGGTTATATGGACGTCTGGGGCAAAGG-3′), MoPrP89–112 3′ (5′-GGGCTTGTTCCACTGATTATGGGTACCCCCTCCTTGGCCCCATCCACCCACTCTCGCACAATAATAAACAGC-3′), MoPrP136 –158 5′ (5′-GTTTATTATTGTGCGAGAGTGGGCGGGAGGCCCATGATCCATTTTGGCAACGAC-3′), MoPrP136–158 3′ (5′-GCGGTACATGTTTTCACGGTAGTAGCGGTCCTCCCAGTCGT TGCCA A A ATGGATCATGGGCCTG-3′), and MoPrP141–158 5′ (5′-GTTTATTATTGTGCGAGAGTGGGCGGGT T TGGCA ACGACTGGGAGGACCGCTAC-3′). A scrambled MoPrP136–158 graft was introduced into the b12 antibody by using the primers MoPrP 136–158 RAN 5′ (5′-ATCTACCATATGTTTAACGGCGAAAACCGTGACTACTGGTACGAGCGCGACGGCGGTTATATGGACGTCTGGGGC-3′) and MoPrP136–158 RAN 3′ (5′-TTCGCCGTTAAACATATGGTAGATGCGCATGTAGGGAGGCCTCCCGCCCACTCTCGCACA ATA ATA A ACAGT-3′). All PCRs were performed with Pfu DNA Polymerase (Stratagene) under the following conditions. Step 1: 94°C, 30 sec; 52°C, 1 min; 72°C, 1 min 30 sec; 35 cycles plus a 10-min incubation at 72°C. Step 2: 94°C, 30 sec; 50°C, 1 min; 72°C, 2 min; 10 cycles in the absence of flanking primers PelSeq and CG1d followed by 30 further cycles after addition of flanking primers, plus a 10-min incubation at 72°C. The resulting b12 PrP heavy chain fragments were inserted between the XhoI and SpeI sites of pComb3H, then subcloned into the pDR12 vector containing the parental b12 light chain gene for expression as human IgG1 in Chinese hamster ovary cells (18).
Immunoprecipitation. Whole brains from normal or RML or 79A scrapie prion-infected mice (killed 130–150 days after intracerebral inoculation) were homogenized at 10% (wt/vol) in Tris-buffered saline (TBS; 0.05 M Tris/0.2 M NaCl, pH 7.4) containing 1% Nonidet P-40 and 1% sodium deoxycholate (DOC), diluted in an equal volume of TBS, then rehomogenized and sonicated. Homogenates of normal or prion-infected brain were clarified at 500 g for 15 min at 4°C. A proportion of clarified prion-infected homogenate was digested with proteinase K (50 μg/ml) for 1 h at 37°C. PMSF was added to all samples to a final concentration of 2 mM. For each immunoprecipitation, antibody at a final concentration of 0.3–10 μg/ml was incubated for 2 h at room temperature with an aliquot of brain homogenate containing ≈1 mg of total protein in a reaction mixture adjusted to a final volume of 500 μl with assay buffer (TBS containing 3% Nonidet P-40 and 3% Tween 20). Tosyl-activated paramagnetic beads (Dynal) coupled either to polyclonal goat anti-human IgG F(ab′)2 (for detection of human PrP-grafted antibodies) or to polyclonal goat anti-mouse IgG F(ab′)2 (for detection of Fab D13 and IgG 6H4) were added to the antibody–homogenate mixture and incubated overnight at 4°C. Beads were then washed four times in washing buffer (TBS containing 2% Nonidet P-40 and 2% Tween 20) and once with TBS before separation by magnet. Pelleted beads were resuspended in 20 μl of loading buffer (150 mM Tris·HCl, pH 6.8/6% SDS/0.3% bromophenol blue/30% glycerol) and heated to 100°C for 5 min. Samples were then run on 12% SDS/PAGE gels and transferred onto nitrocellulose membranes. Membranes were blocked with 5% (wt/vol) nonfat dry milk in TBS containing 0.1% Tween 20 (TBST) for 1 h at room temperature, and blotted PrP was detected with Fab D13 or IgG 6H4 antibodies at 1 μg/ml. After five washes in TBST, blotted PrP protein was detected by incubation for 30 min at room temperature with a horseradish peroxidase-conjugated goat anti-mouse IgG (Pierce) diluted 1:10,000 in blocking buffer. Membranes were then washed five times in TBST and developed with enhanced chemiluminescence reagent (Amersham Pharmacia) onto film. For plasminogen (19) binding studies, 100 μg/ml biotinylated human plasminogen (Enzyme Research Laboratories, South Bend, IN) was incubated with 1 mg of brain homogenate, then captured onto streptavidin-coated agarose beads. The beads were spun briefly, washed, resuspended in loading buffer, heated, and repelleted; the bead eluate was examined for the presence of PrP by Western blotting as described above.
Immunoprecipitation in the presence of Triton X-100 was performed exactly as described above, except that the brain homogenization and reaction buffers contained 1% Triton X-100 rather than Nonidet P-40/DOC detergents. Samples of cerebral cortex from three cases of vCJD and sCJD types I and II and from two neurological control cases from patients with Alzheimer's disease and Lewy body dementia were selected from the National Creutzfeldt–Jakob Disease Surveillance Unit brain bank for this study. All tissues were obtained at autopsy with permission for retention and research. Diagnosis was confirmed by neuropathological examination of the brain and Western blot analysis of PrPSc. Samples were homogenized in buffers containing Nonidet P-40/DOC and Triton X-100 and used in immunoprecipitation assays as described above for mouse scrapie brain. Homogenates of hamster brains were prepared from healthy animals and from animals with clinical signs of scrapie after intracerebral inoculation with 263K prions.
Prion Infectivity Bioassay. Groups of eight CD-1 mice were inoculated intracerebrally with a 30-μl volume of PBS containing the equivalent of 1% of the total number of anti-human IgG-coupled paramagnetic beads used to perform a single immunoprecipitation experiment. Incubation times were recorded as the period taken to develop terminal scrapie sickness.
Nomenclature. Numbering of residues corresponds to Syrian hamster PrP throughout.
Results
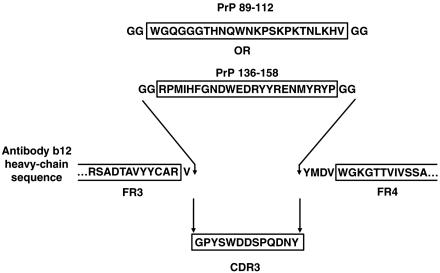
Motif-Grafted Antibodies Specifically Recognize Disease-Associated PrP Conformers. Relatively long recognition sequences have been grafted previously into the HCDR3 of the antibody molecule to generate desired binding properties (17). Therefore, we decided to graft mouse PrP sequences corresponding to amino acids 89–112 and 136–158 into the HCDR3 of IgG1 b12 (16), a human recombinant antibody specific for HIV-1 gp120, by use of overlap PCR. Antibody b12 was chosen as the recipient molecule for transplanted PrP sequence because the parental antibody possesses a relatively long HCDR3 (18 aa) that projects vertically from the surface of the antigen binding site (20). To maximally distance PrP sequence from the antibody surface, each graft was placed between the first N-terminal residue and four C-terminal residues of the parental HCDR3 (Fig. 1). In addition, two glycine residues were incorporated at each flank of the PrP sequence. The resulting PrP-IgGs (89–112 and 136–158) were expressed in Chinese hamster ovary cells and purified to homogeneity (18).
Fig. 1.
Schematic illustration of mouse PrP 89–112 or 136–158 sequence replacing IgG b12 HCDR3 sequence to yield PrP-IgGs 89–112 and 136–158. The N-terminal Val residue and four C-terminal residues (Tyr-Met-Asp-Val) of the original b12 HCDR3 are retained; two Gly residues are added to each flank of the grafted PrP sequence.
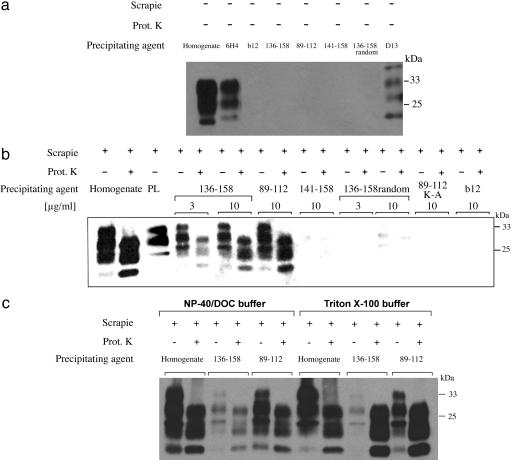
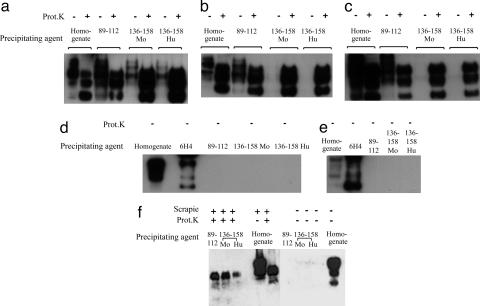
To study the reactivity of the PrP-IgG molecules against PrPC, PrPSc, and PrP 27–30, we first performed immunoprecipitation experiments using brain homogenates prepared from normal mice and from mice infected with the RML or 79A strains of scrapie prions. Precipitated PrP was detected by Western blot (Fig. 2). As positive controls, we used Fab D13 (12) and IgG 6H4 (8) to precipitate PrPC from normal mouse brain homogenates (Fig. 2a) and plasminogen (19) to precipitate PrPSc from prion-infected brain samples (Fig. 2b). Reaction of PrP-IgG 89–112 or 136–158 with PrPC in normal mouse brain was not detected when the antibodies were used at a final concentration of 10 μg/ml (Fig. 2a). However, at the same or lower concentrations, each of these IgGs immunoprecipitated three PrP bands from undigested and proteinase K-digested prion-infected brain homogenates (Fig. 2b). These bands correspond in size to the di-, mono-, and unglycosylated forms of PrPSc and PrP 27–30, the proteinase-resistant core of PrPSc in which the N-terminal portion of the protein between residues 23 and 90 has been enzymatically degraded. The data do not directly preclude the possibility that a fraction of PrP immunoprecipitated from undigested scrapie prion-infected brain homogenate included PrPC that is bound to PrPSc. However, in experiments using prion-infected tissues, the quantity of immunoprecipitated PrP appeared to be independent of proteinase K digestion, suggesting that the majority of the PrP species precipitated by the antibody in the absence of proteinase K treatment was PrPSc rather than PrPC. Under identical experimental conditions, the parental b12 IgG did not react with PrPC, PrPSc, or PrP 27–30 (Fig. 2 a and b). Moreover, IgGs containing PrP sequence no longer recognized gp120 (the target antigen of the parental b12 antibody), did not bind to any other protein when used to probe Western blots of mouse brain homogenate, and were completely unreactive with PrPSc after its denaturation to a PrPC-like conformation by heating in the presence of SDS (data not shown). Therefore, we conclude that grafted PrP sequence composed of residues 89–112 or 136–158 endows specific antibody recognition of PrPSc and that these disease-associated epitopes are retained in PrP 27–30.
Fig. 2.
Specific immunoprecipitation of PrPSc and PrP 27–30 from prion-infected mouse brain. (a) Normal brain. IgG b12 (lane 3) and PrP-IgGs 136–158 (lane 4), 89–112 (lane 5), 141–158 (lane 6), and 136–158 random (lane 7) were incubated with supernatant from a centrifuged homogenate prepared from whole brains of normal mice. Antibodies were precipitated with polyclonal goat anti-IgG F(ab′)2 linked to paramagnetic beads. Precipitates were analyzed on Western blot for the presence of PrP. PrPC was detected by directly blotting 5 μl of a 5% clarified normal brain homogenate (lane 1) and was specifically precipitated by the control anti-PrPc antibodies 6H4 and D13 (lanes 2 and 8). No PrPC was detected after immunoprecipitation with IgG b12 or any of the PrP-IgGs. All antibodies were used at a final concentration of 10 μg/ml. (b) Prion-infected brain. PrPSc and PrP 27–30 specifically immunoprecipitated from a centrifuged homogenate of undigested and proteinase K-digested RML prion-infected mouse brain. PrPSc and PrP 27–30 present after blotting of 5 μl of 5% clarified prion-infected brain homogenate are shown in the first and second lanes, respectively. PrPSc precipitated by plasminogen (100 μg/ml) is also shown (lane 3). Equivalent PrP bands are present after immunoprecipitation with PrP-IgGs 136–158 (lanes 4–7) and 89–112 (lanes 8 and 9). No or trace amounts of PrP were precipitated by PrP-IgGs 141–158, 136–158 random, 89–112 K-A, or the parental b12 IgG molecule, indicating that specificity for PrPSc and PrP 27–30 critically depends on the grafted PrP 89–112 and 136–158 sequence motifs. (c) Immunoprecipitation of PrPSc and PrP 27–30 in the presence of Nonidet P-40/DOC and Triton X-100. A prion-infected mouse brain was bisected laterally; one hemisphere was homogenized in buffer containing 1% Nonidet P-40 and 1% DOC, and the other hemisphere was homogenized in buffer containing 1% Triton X-100. Directly blotting 5 μl of 5% homogenates of each of these brain samples indicated that roughly equal amounts of PrPSc were present in each of these preparations (lanes 1 and 2 and lanes 7 and 8). IgGs 89–112 and 136–158 (10 μg/ml) precipitated very similar amounts of PrPSc from both brain homogenates (lanes 3 and 5 and lanes 9 and 11), but significantly greater amounts of PrP 27–30 were immunoprecipitated from Triton X-100 homogenate (lanes 10 and 12) than from homogenate prepared by using 1% Nonidet P-40 and 1% DOC (lanes 4 and 6). Comparative densitometric analysis of the PrP bands we observed indicated that the motif-grafted PrP 89–112 and 136–158 antibodies precipitated not more than 25–30% of the input PrPSc or PrP 27–30 from brain homogenate prepared by using either Nonidet P-40/DOC or Triton X-100 buffer.
To further demonstrate that the PrP grafts imparted specificity for disease-associated PrP conformations, molecules were constructed in which (i) the amino acids comprising the 136–158 graft were scrambled and (ii) the four lysine residues contained within the 89–112 graft were mutated to alanine residues. The resulting antibodies, termed PrP 136–158 random and 89–112 K-A, showed no or trace reactivity with PrPSc and PrP 27–30 when used in an immunoprecipitation assay at a final concentration of 10 μg/ml and no reactivity when used at a concentration of 3 μg/ml (Fig. 2b). Interestingly, specificity for PrPSc and PrP 27–30 was also lost when the PrP 136–158 graft was N-terminally truncated to residues 141–158 (Fig. 2b). This finding suggests that PrP sequence between residues 136 and 140 (inclusive) may be of critical importance in PrPC–PrPSc interactions. In support of this conclusion, a single Syrian hamsterspecific substitution at position 138 of mouse PrP has previously been shown to significantly inhibit production of proteinase K-resistant PrP (21). Furthermore, a natural dimorphism at the equivalent position of goat PrP is linked with increased resistance of the host to infection with both sheep and bovine prions (22).
Specific interaction between plasminogen and PrPSc depends on the presence of detergent that disrupts membrane rafts (23). To determine whether the binding interactions between IgGs 89–112 and 136–158 and PrPSc and PrP 27–30 were affected by detergent conditions, parallel immunoprecipitation experiments were performed in which prion-infected mouse brain homogenate was prepared by using either Nonidet P-40 and DOC (reagents disrupting membrane rafts) or Triton X-100 (a detergent preserving raft architecture). The results indicate that reactivity of the PrP-grafted antibodies with PrPSc is unaffected by detergent conditions and that binding to PrP 27–30 is significantly enhanced in the presence of Triton X-100 (Fig. 2c). Under equivalent conditions, IgG b12 bound to neither PrPSc nor PrP 27–30 (data not shown). Similarly, IgGs 89–112 and 136–158 did not recognize PrPC in normal mouse brain extracted in the presence of Triton X-100 (data not shown).
PrP Fractions Recognized by Motif-Grafted Antibodies Contain Prion Infectivity. To ascertain whether the PrP fractions specifically recognized by IgGs 89–112 and 136–158 contained prion infectivity, aliquots of anti-human IgG-coupled paramagnetic beads used in the immunoprecipitation procedure were implanted intracerebrally into groups of eight CD-1 mice. All mice receiving beads incubated with prion-infected brain homogenates in the presence of either IgG 89–112 or 136–158 developed scrapie with mean incubation times of 180–190 days postinoculation (Table 1). In contrast, at 365 days postinoculation, prion disease had developed in only 1 of 16 mice receiving equivalent quantities of anti-human IgG-coupled beads incubated with prion-infected brain homogenates in the absence of IgGs 89–112 and 136–158 (Table 1). These data indicate that IgGs 89–112 and 136–158 specifically precipitate prion infectivity.
Table 1. IgGs 89-112 and 136-158 specifically precipitate prion infectivity.
| Precipitating agent | IgG 89-112 | IgG 136-158 | Anti-IgG-coupled paramagnetic beads alone | |||
|---|---|---|---|---|---|---|
| Proteinase K treatment | + | - | + | - | + | - |
| No. of mice developing scrapie | 8/8 | 8/8 | 8/8 | 8/8 | 1/8 | 0/8 |
| Mean incubation time to disease, days ± SEM | 181 ± 0.31 | 188 ± 0.73 | 188 ± 0.85 | 190 ± 0.85 | 221 | >365 |
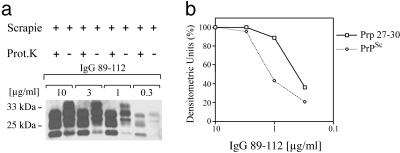
Binding Affinities for Disease-Associated PrP Isoforms Lie in the nM Range. Of the PrP-grafted antibody preparations we tested in this study, IgG 89–112 appeared to possess the greatest affinity for disease-associated PrP conformers. To estimate the affinity of this molecule for mouse PrPSc and PrP 27–30, a series of immunoprecipitation experiments were performed with decreasing concentrations of antibody. The relative amounts of PrP precipitated at each antibody concentration were visualized by immunoblot and quantitated by densitometric analysis (Fig. 3). Plotting densitometry values against antibody concentration yielded a titration curve from which antibody concentrations producing 50% maximum binding signals against PrPSc and PrP 27–30 could be determined and used to estimate binding constants for these antigens. The results indicate that IgG 89–112 possesses apparent affinities of ≈2 nM for mouse PrP 27–30 and 7 nM for mouse PrPSc. Similar analysis of the affinity of IgG 136–158 for mouse PrP 27–30 and PrPSc antigens indicated a binding constant in the 20–25 nM range (data not shown). Using the immunoprecipitation assay, we additionally determined that IgG 89–112 was able to precipitate detectable levels of PrP 27–30 from a 0.01% (wt/vol) homogenate of proteinase K-treated RML prion-infected brain, whereas PrPC immunoprecipitated by 6H4 could not be detected when normal mouse brain was diluted beyond a 1% (wt/vol) homogenate (data not shown).
Fig. 3.
Titration of IgG 89–112 reactivity with PrPSc and PrP 27–30. (a) PrPSc and PrP 27–30 immunoprecipitated by different concentrations (0.3–10 μg/ml) of IgG 89–112 from undigested and proteinase K-digested RML prion-infected mouse brain homogenates. (b) Densitometric measurement of PrPSc and PrP 27–30 bands identified in the immunoblot in a. Values are given as densitometric units, where 100% is equivalent to the intensity of the bands immunoprecipitated at an antibody concentration of 10 μg/ml.
Species Reactivity. Immunoprecipitation experiments were performed to determine whether IgGs 89–112 and 136–158 reacted with disease-associated PrP found in prion-infected human and hamster brains (Fig. 4). In this series of experiments, an antibody containing a graft of human PrP sequence equivalent to that of mouse PrP 136–158 was also included (IgG 136–158 Hu). IgG 89–112 recognized disease-associated PrP conformers from human brain homogenates infected with sporadic CJD (sCJD) (types I and II) and variant CJD (vCJD) prions (Fig. 4 a–c). IgGs 136–158 Mo and 136–158 Hu recognized PrP 27–30 from all of the human brain samples but precipitated PrPSc only from the sCJD type I tissue. Notably, the IgG 136–158 Hu reagent also bound well to mouse PrPSc and PrP 27–30 (data not shown). The pattern of reactivity we observe suggests that in some tissues a proteinase K labile molecule, possibly PrPC, may effectively compete with the relatively lower-affinity IgG 136–158 reagent for binding to PrPSc. Neither IgG 89–112 nor IgG 136–158 (Mo or Hu) recognized PrP in the brain tissues of donors diagnosed with Alzheimer's disease or Lewy body dementia (Fig. 4 d and e). Both the 89–112 and 136–158 antibodies reacted well with PrP 27–30 from proteinase K-treated scrapie prion-infected hamster brain homogenate but not with hamster PrPC (Fig. 4f).
Fig. 4.
IgGs 89–112, 136–158 Mo, and 136–158 Hu specifically recognize disease-associated PrP conformers from prion-infected human and hamster brain homogenates. (a–c) PrPSc and PrP 27–30 immunoprecipitated from centrifuged homogenates of undigested and proteinase K-digested sCJD (types I and II) and vCJD brains. (d and e) No PrP was immunoprecipitated from equivalently prepared homogenates of brains of patients with Lewy body dementia and Alzheimer's disease. (f) IgGs 89–112, 136–158 Mo, and 136–158 Hu immunoprecipitate PrP 27–30 from proteinase K-treated prion-infected hamster brain homogenate but not PrPC from normal hamster brain. The PrP-grafted antibodies precipitated not more than 30% of the input PrPSc or PrP 27–30.
Discussion
Although a considerable body of evidence indicates that direct interaction between PrPC–PrPSc is a critical step in the formation of nascent prion infectivity, the specific nature of this bimolecular complex remains unknown. By grafting segments of PrP sequence recognized by potently inhibitory Fab fragments into a recipient IgG protein scaffold, we have identified two independent constrained PrP peptide motifs that possess remarkably high intrinsic specificity and affinity for epitopes found exclusively on PrPSc and PrP 27–30. The hybrid PrP–antibody reagents we created bind disease-associated PrP isoforms produced by different prion strains and in different species. Moreover, antibody binding to these misassembled PrP isoforms is robust, largely independent of detergent conditions, and demonstrably tied to prion infectivity. Based on the findings of this study, we suggest that the 89–112 and 136–158 regions of PrP are critical components of the PrPC–PrPSc interface. Further evaluation of the relative importance of individual PrPC residues to the PrPC–PrPSc interaction will require the production of additional antibodies containing truncated and mutated PrP sequence. Furthermore, in situ randomization of antibody-grafted PrP sequences, followed by selection against infectious prion particles, should evolve molecules possessing ultra-high affinity for PrPSc that will be invaluable in the drive toward effective prion diagnostics. Data forthcoming from these studies can be used experimentally to directly determine, through the use of novel PrP transgenes, how the kinetic properties of PrPC–PrPSc interactions modulate prion pathogenesis in vivo. Finally, screening for small molecules competing with IgGs 89–112 and 136–158 for binding to PrPSc may yield candidate drugs capable of potently inhibiting prion replication.
Acknowledgments
This work is dedicated to the memory of Valeriano Moroncini, M.D. We thank Chris Birkett, Estelle Leclerc, and Erica Ollman Saphire for advice and Surachai Supattapone (Dartmouth Medical School, Hanover, NH) for hamster brain samples. This work was supported by a grant from the National Institutes of Health (to R.A.W.) and a Biology of the Transmissible Spongiform Encephalopathies award from the Biotechnology and Biological Sciences Research Council (to J.P.B.).
Abbreviations: DOC, sodium deoxycholate; CJD, Creutzfeldt–Jakob disease; HCDR3, heavy chain complementarity-determining region 3.
References
- 1.Prusiner, S. B. (1998) Proc. Natl. Acad. Sci. USA 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruce, M. E., Will, R. G., Ironside, J. W., McConnell, I., Drummond, D., Suttie, A., McCardle, L., Chree, A., Hope, J., Birkett, C., et al. (1997) Nature 389, 498–501. [DOI] [PubMed] [Google Scholar]
- 3.Hill, A. F., Desbruslais, M., Joiner, S., Sidle, K. C., Gowland, I., Collinge, J., Doey, L. J. & Lantos, P. (1997) Nature 389, 448–450, 526. [DOI] [PubMed] [Google Scholar]
- 4.Aguzzi, A. (2001) Nat. Med. 7, 289–290. [DOI] [PubMed] [Google Scholar]
- 5.Kaneko, K., Zulianello, L., Scott, M., Cooper, C. M., Wallace, A. C., James, T. L., Cohen, F. E. & Prusiner, S. B. (1997) Proc. Natl. Acad. Sci. USA 94, 10069–10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zulianello, L., Kaneko, K., Scott, M., Erpel, S., Han, D., Cohen, F. E. & Prusiner, S. B. (2000) J. Virol. 74, 4351–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caughey, B. (2001) Trends Biochem. Sci. 26, 235–242. [DOI] [PubMed] [Google Scholar]
- 8.Korth, C., Stierli, B., Streit, P., Moser, M., Schaller, O., Fischer, R., Schulz-Schaeffer, W., Kretzschmar, H., Raeber, A., Braun, U., et al. (1997) Nature 390, 74–77. [DOI] [PubMed] [Google Scholar]
- 9.Paramithiotis, E., Pinard, M., Lawton, T., LaBoissiere, S., Leathers, V. L., Zou, W. Q., Estey, L. A., Lamontagne, J., Lehto, M. T., Kondejewski, L. H., et al. (2003) Nat. Med. 9, 893–899. [DOI] [PubMed] [Google Scholar]
- 10.Heppner, F. L., Musahl, C., Arrighi, I., Klein, M. A., Rulicke, T., Oesch, B., Zinkernagel, R. M., Kalinke, U. & Aguzzi, A. (2001) Science 294, 178–182. [DOI] [PubMed] [Google Scholar]
- 11.Peretz, D., Williamson, R. A., Kaneko, K., Vergara, J., Leclerc, E., Schmitt-Ulms, G., Mehlhorn, I. R., Legname, G., Wormald, M. R., Rudd, P. M., et al. (2001) Nature 412, 739–743. [DOI] [PubMed] [Google Scholar]
- 12.Williamson, R. A., Peretz, D., Pinilla, C., Ball, H., Bastidas, R. B., Rozenshteyn, R., Houghten, R. A., Prusiner, S. B. & Burton, D. R. (1998) J. Virol. 72, 9413–9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enari, M., Flechsig, E. & Weissmann, C. (2001) Proc. Natl. Acad. Sci. USA 98, 9295–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chabry, J., Caughey, B. & Chesebro, B. (1998) J. Biol. Chem. 273, 13203–13207. [DOI] [PubMed] [Google Scholar]
- 15.Chabry, J., Priola, S. A., Wehrly, K., Nishio, J., Hope, J. & Chesebro, B. (1999) J. Virol. 73, 6245–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burton, D. R., Pyati, J., Koduri, R., Sharp, S. J., Thornton, G. B., Parren, P. W., Sawyer, L. S., Hendry, R. M., Dunlop, N., Nara, P. L., et al. (1994) Science 266, 1024–1027. [DOI] [PubMed] [Google Scholar]
- 17.McLane, K. E., Burton, D. R. & Ghazal, P. (1995) Proc. Natl. Acad. Sci. USA 92, 5214–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maruyama, T., Rodriguez, L. L., Jahrling, P. B., Sanchez, A., Khan, A. S., Nichol, S. T., Peters, C. J., Parren, P. W. & Burton, D. R. (1999) J. Virol. 73, 6024–6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer, M. B., Roeckl, C., Parizek, P., Schwarz, H. P. & Aguzzi, A. (2000) Nature 408, 479–483. [DOI] [PubMed] [Google Scholar]
- 20.Saphire, E. O., Parren, P. W., Pantophlet, R., Zwick, M. B., Morris, G. M., Rudd, P. M., Dwek, R. A., Stanfield, R. L., Burton, D. R. & Wilson, I. A. (2001) Science 293, 1155–1159. [DOI] [PubMed] [Google Scholar]
- 21.Priola, S. A. & Chesebro, B. (1995) J. Virol. 69, 7754–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldmann, W., Martin, T., Foster, J., Hughes, S., Smith, G., Hughes, K., Dawson, M. & Hunter, N. (1996) J. Gen. Virol. 77, 2885–2891. [DOI] [PubMed] [Google Scholar]
- 23.Shaked, Y., Engelstein, R. & Gabizon, R. (2002) J. Neurochem. 82, 1–5. [DOI] [PubMed] [Google Scholar]