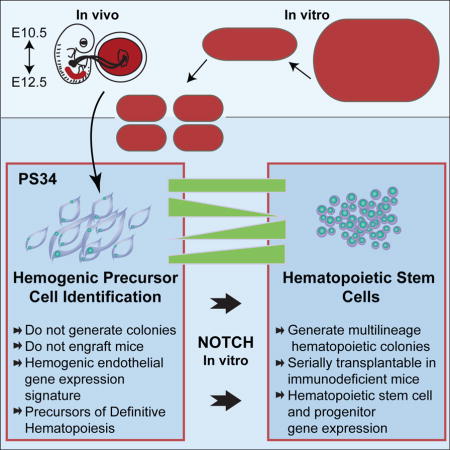
Summary
Definitive hematopoiesis emerges via an endothelial-to-hematopoietic transition in the embryo and placenta; however, the precursor cells to hemogenic endothelium are not defined phenotypically. We previously demonstrated that the induction of hematopoietic progenitors from fibroblasts progresses through hemogenic precursors that are Prom1+Sca1+CD34+CD45− (PS34CD45−). Guided by these studies we analyzed mouse placentas and identified a population with this phenotype. These cells express endothelial markers, are heterogeneous for early hematopoietic markers, and localize to the vascular labyrinth. Remarkably, global gene expression profiles of PS34CD45− cells correlate with reprogrammed precursors and establish a hemogenic precursor cell molecular signature. PS34CD45− cells are also present in intraembryonic hemogenic sites. After stromal co-culture, PS34CD45− give rise to all blood lineages and engraft primary and secondary immunodeficient mice. In summary, we show that reprogramming reveals a phenotype for in vivo precursors to hemogenic endothelium, establishing that direct in vitro conversion informs developmental processes in vivo.
Graphical abstract
Introduction
The seminal studies by Yamanaka and colleagues demonstrating that four pluripotency transcription factors (TFs) can change a somatic cell into an induced pluripotent stem cell (iPSC) caused a sea change in the stem cell field. They also motivated others to investigate whether combinatorial TF-mediated reprogramming strategies could change cell fate without traversing through pluripotency. Done successfully, this would provide alternatives for producing cell-types of interest for regenerative medicine (Xu et al., 2015). The development of iPSCs and directly reprogrammed cells caused a paradigm shift in the way we think about cell fate stability in multipotent and unipotent somatic cells. This led to the identification of minimal TF networks that kick-start lineage reprogramming (Pereira et al., 2012), and provided mechanistic insights into TF modes of action (Soufi et al., 2012; Wapinski et al., 2013). However, it is not known if direct lineage reprogramming recapitulates developmental lineage specification (Pereira et al., 2013), or if it leads to aberrant cell types without a clear in vivo equivalent (Doulatov and Daley, 2013).
We recently demonstrated direct reprogramming of mouse fibroblasts into hematopoietic stem progenitor cells (HSPC) with four TFs: Gata2, Gfi1b, cFos and Etv6 (Pereira et al., 2013). These TFs induce a dynamic process that progresses through hemogenic precursors (HPs). HP cells express Prominin1, Sca1, CD34, are CD45 negative and have a global transcriptional profile highly enriched in vascular and endothelial genes. The hematopoietic cells that emerge afterwards have a gene expression program highly similar to bona fide HSCs from aorta-gonad-mesonephros (AGM), placenta and early fetal liver. Transfer of the TFs to inducible lentiviral vectors and aggregation culture demonstrated that the programmed population contained multi-lineage clonogenic progenitors.
The major sites of definitive hematopoiesis in mid-gestation are the AGM and placenta with subsequent migration to the fetal liver and bone marrow where HSCs expand and mature, respectively (Dzierzak and Speck, 2008; Medvinsky et al., 2011; Mikkola and Orkin, 2006). In the AGM they are thought to “bud” directly from a small population of hemogenic endothelial (HE) cells (Bertrand et al., 2010a; Boisset et al., 2010; Zovein et al., 2008). An endothelial-to-hematopoietic transition (EHT) was suggested based on imaging experiments (Bertrand et al., 2010a; Boisset et al., 2010; Eilken et al., 2009; Kissa and Herbomel, 2010; Lancrin et al., 2009). The EHT remains poorly understood due to the lack of specific HP markers for prospective isolation (Medvinsky et al., 2011). Furthermore it is becoming increasingly apparent that this is not a single-step process (Boisset et al., 2014; Kieusseian et al., 2012; Rybtsov et al., 2014; Rybtsov et al., 2011; Taoudi et al., 2008). Emerging evidence suggests that hematopoietic lineage divergence from the embryonic endothelium may occur prior to E10.5 and before extensive formation of intra-aortic clusters (Rybtsov et al., 2011; Swiers et al., 2013). There is additional evidence to support this obtained during the in vitro differentiation of PSCs into hematopoietic cells (Ditadi et al., 2015).
Our direct reprogramming process appears to recapitulate developmental hematopoiesis and traverses through a HP cell with a specific phenotype (Pereira et al., 2013). Therefore, we asked if this information could provide insights into HSC ontogeny in vivo. We analyzed mouse placentas and embryos for the presence of cells with a PS34CD45− surface phenotype. We show that PS34CD45− cells can be isolated from mouse placentas and detected in the AGM. We have characterized the transcriptional profile of these cells by mRNAseq and generated a HP molecular signature. PS34CD45− cells acquire the capacity to generate multilineage colonies and to be serially transplanted after a short in vitro maturation step that involves activation of the Notch pathway.
In summary, we have isolated an early HP with a defined phenotype that can be matured in vitro into transplantable HSPCs. We therefore provide evidence that both the induction of hemogenic cells in vitro and the isolation of PS34CD45− cells can be used as a powerful platform to study HSC ontogeny.
Results
Sca1, Prom1 and CD34 Mark Hemogenic Precursor Cells in Midgestation Mouse Placentas
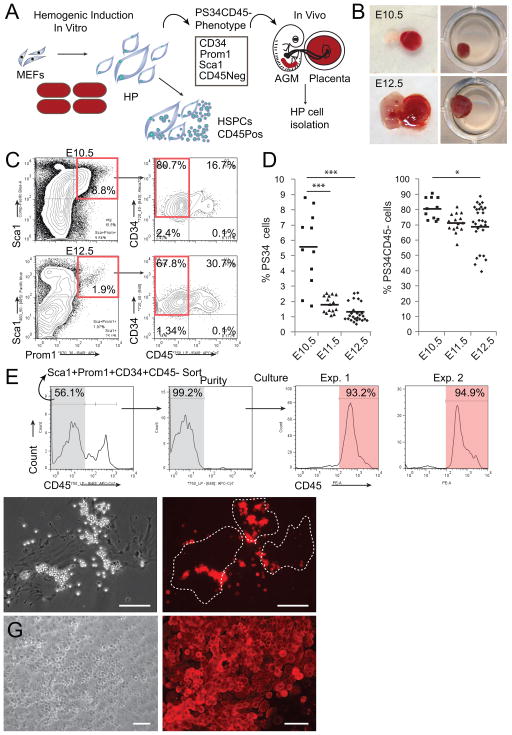
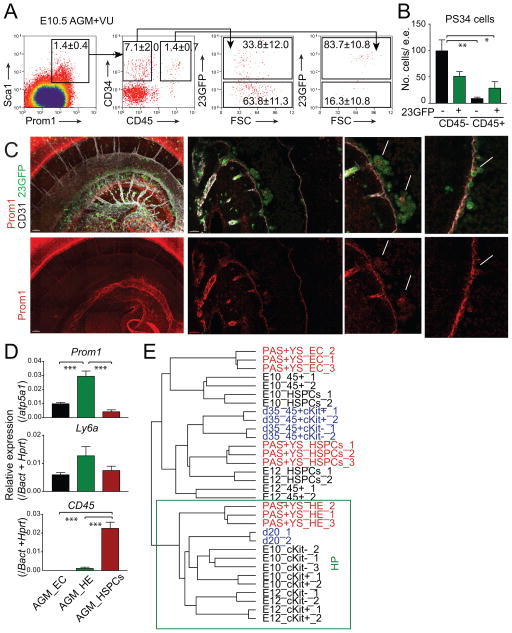
We asked if cells with the PS34 hemogenic phenotype identified during in vitro reprogramming were present in mouse placentas at E10.5, E11.5 and E12.5, times before and at the peak of HSC activity (Gekas et al., 2005; Ottersbach and Dzierzak, 2005) (Figure 1A). Placentas were isolated, freed of umbilical cord and maternal decidua, dissociated to single cells and analyzed by flow cytometry (Figure 1B and 1C). We identified a large population of Sca1+ cells in placenta while Prom1 was restricted to a smaller population (0.6–8.8%). Prom1+Sca1+ cells co-express CD34 and the majority do not express the pan-hematopoietic marker CD45 (Figure 1C). This represents the same cell surface phenotype previously identified in vitro by introduction of transcription factors (TFs) into fibroblasts (Pereira et al., 2013). The PS34 population is more abundant at E10.5 and declines at E11.5 and E12.5 (5.5±2.4%, 1.8±0.5% and 1.3±0.6%, respectively, Figure 1D). The percentage of CD45− PS34 cells decreases with gestation time (80.6±7.0%, 71.1±6.2% and 69.7±14.2%, respectively; Figure 1D), consistent with the developmental progression of hemogenic cells. We next sorted PS34CD45− cells and cultured them on gelatin-coated dishes. Interestingly, after 7 days we observed the emergence of CD45+ cells (Figure 1E and 1F), in association with large adherent flat cells (Figure 1F). Sorted PS34CD45+ cells did not survive in these culture conditions (not shown). When plated on AFT024 HSC-supportive stroma (Moore et al., 1997) PS34CD45− cells generate large hematopoietic colonies (not shown) and CD45+ cobblestone-like colonies after 4 weeks (Figure 1G). These results highlight the utility of TF-mediated programming to provide insights into developmental processes and suggest that the PS34CD45− phenotype may be used to isolate HPs in vivo.
Figure 1. Prominin1, Sca1 and CD34 Mark HP Cells in Midgestation Mouse Placentas.
(A) Strategy to isolate HP cells in vivo using information generated from in vitro hemogenic induction.
(B) E10.5 and E12.5 placentas were isolated and dissociated to a single cell suspension and (C) analyzed for expression of Prom1, Sca1, CD34 and CD45, red boxes highlight populations of interest.
(D) Percentage of Prom1+Sca1+CD34+ cells (PS34, left panel) as well as the percentage of the subpopulation of PS34CD45− cells (right panel) from litters at E10.5, E11.5 and E12.5 (each mark represents a single placenta, n=11–28). *p<0.05; ***p<0.001.
(E) PS34CD45− cells were sorted and plated on gelatin-coated dishes in Myelocult with SCF, IL-3 and Flt3l for 7 days, stained for CD45 and analyzed by flow cytometry or (F) immunofluorescence. Dashed lines highlight large adherent cells associated with round non-adherent CD45+ cells.
(G) PS34CD45− cells were cultured on AFT024 stroma for 4 weeks; cobblestone areas are stained for CD45. Scale bar = 100 μm.
PS34 Cells Express Endothelial and Hematopoietic Markers
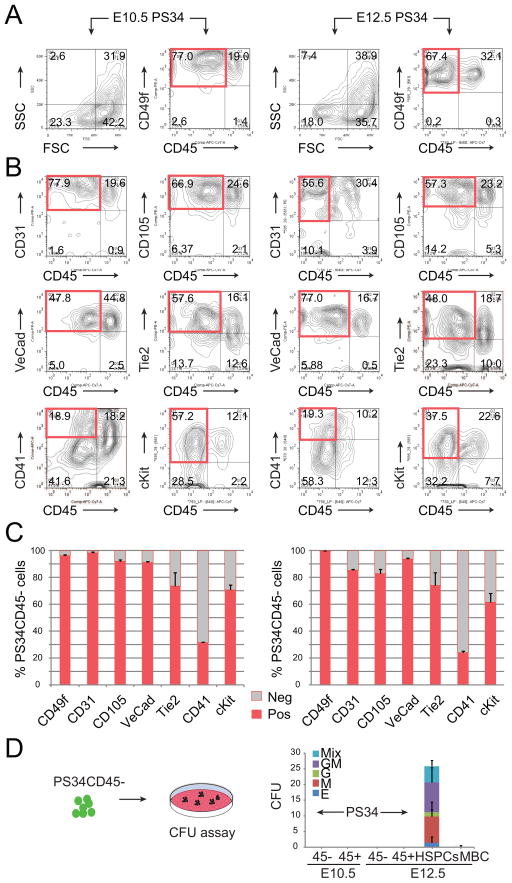
We next addressed whether PS34 cells express endothelial and hematopoietic markers associated with HE cells. We analyzed the expression of CD49f (Integrin-α6), CD31 (Pecam1), CD105 (Endoglin), VE-Cadherin, Tie2, CD41 (Integrin-α2b) and cKit within the PS34 population from E10.5 and E12.5 placentas (Figure 2 and S1). Both at E10.5 and E12.5 most of the PS34CD45− cells express the endothelial markers VE-Cadherin, CD31, CD105 and Tie2 (Figure 2B and 2C). PS34CD45− cells also express CD49f (Figure 2A and 2C), a marker previously identified on in vitro reprogrammed hemogenic cells (Pereira et al., 2013). PS34CD45+ cells also express VE-Cadherin, CD31, CD105, Tie2 and CD49f consistent with the retention of endothelial markers by emergent HSPCs (Taoudi et al., 2008). Interestingly, PS34CD45− cells are heterogeneous for the early hematopoietic markers CD41 (31.5±0.3% at E10.5 and 24.1±1.1% at E12.5) and cKit (70.9±3.6% at E10.5 and 61.4±6.9% at E12.5) showing that the PS34 marker combination captures a heterogeneous population of cells undergoing EHT (Figure 2B and 2C). This is consistent with the reported placental expression of CD41 (Rhodes et al., 2008) and the requirement for cKit signaling during HE cell specification (Marcelo et al., 2013). We next addressed whether PS34 cells have hematopoietic progenitor activity in vitro by performing colony-forming unit (CFU) assays in methylcellulose. Both PS34CD45− and PS34CD45+ at E10.5 and E12.5 cells do not generate CFU in contrast to the E12.5 cKit+CD34+CD45+ HSPC population (Figure 2D). Similarly, early AGM HE does not contain CFU (Boisset et al., 2014; Swiers et al., 2013). The lack of CFU in our precursor populations was confirmed by plating >7,000 placental PS34CD45− cells (Table S1). These data confirm that the PS34CD45− phenotype marks HPs and excludes all downstream committed hematopoietic progenitors and HSCs. This contrasts with other endothelial markers (for example VE-cadherin, CD31 and Tie-2) whose expression is retained by HSPCs. These data also highlight the regulation of CD41 in the placenta, which, in contrast to the AGM region, does not mark HSPCs (Robin et al., 2011).
Figure 2. PS34CD45− Cells Express Endothelial and Hematopoietic Markers.
E10.5 (left panels) and E12.5 (right panels) placentas were dissociated to a single cell suspension and analyzed. Prom1+Sca1+CD34+ cells are displayed for (A) FCS/SSC, CD49f and (B) CD31, CD105, VE-Cadherin, Tie2, CD41 and cKit expression. Red boxes highlight the phenotype Prom1+Sca1+CD45− cells.
(C) Quantification of the percentage of PS34CD45− cells positive for CD49f, CD31, CD105, VE-Cadherin, Tie2, CD41 and cKit (mean ± SD, n=2). See also Figure S1.
(D) PS34CD45− and PS34CD45+ cells were isolated from placentas at E10.5 and E12.5 and assayed for hematopoietic colony formation. HSPCs (CD45+ckit+CD34+) and mature blood cells (CD45+cKit−CD34−) were included as controls. Color code represents the type of hematopoietic colony (mean ± SD, n=2). See also Table S1.
PS34 Cells Originate from Embryonic Tissue and Localize to the Vascular Labyrinth
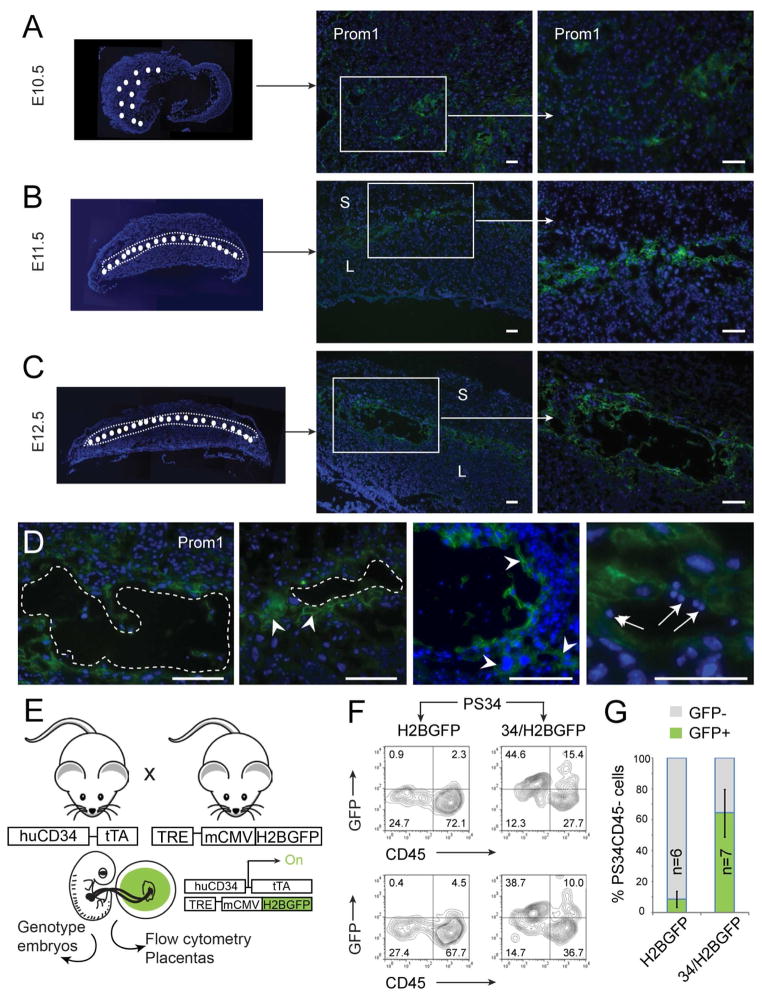
Placental HSCs emerge in association with the placental vascular labyrinth and originate from fetal tissue (Gekas et al., 2005; Ottersbach and Dzierzak, 2005; Rhodes et al., 2008). The expression of CD34 is restricted to the vascular labyrinth (Ottersbach and Dzierzak, 2005). Placental Prom1+Sca-1+ cells express CD34 (Figure 1C) suggesting that these precursor cells would also localize to the vascular labyrinth. To define the localization of Prom1+ cells in developing mouse placentas we analyzed transversal sections at E10.5, E11.5 and E12.5. Prom1+ cells are detected in clusters in E10.5 placentas (Figure 3A). At E11.5 and E12.5 Prom1+ cells are localized to the vascular labyrinth and interestingly, restricted to a layer at the maternal-fetal interface (Figure 3B and 3C). Prom1+ cells localize to the blood vessel lining or surrounding areas, consistent with the homogeneous expression of endothelial markers by PS34 cells (Figure 2). Higher magnification shows that Prom1+ cells are large in size and often found in association with clusters of small hematopoietic cells (Figure 3D). In addition, Prom1 was not detected in E13.5 fetal liver sections (not shown) as previously reported (Zhu et al., 2009), confirming the specificity of Prom1 to HPs but not HSCs.
Figure 3. PS34 Cells Originate from Fetal Tissue and Localize to the Vascular Labyrinth.
Placentas from (A) E10.5, (B) E11.5 and (C) E12.5 were stained with antibodies against CD133 (Prom1, green). The left panels show composite low magnification pictures of transversal sections of placentas. The region of the labyrinth that contains Prom1+ cells is highlighted (white circles).
(D) High magnification of vasculature showing representative Prom1+ cells (arrowheads) and associated blood cells (arrows). Blood vessels are highlighted with dotted lines. Scale bar = 100 μm. S, spongiotrophoblast layer; L, labyrinth.
(E) Strategy to confirm the fetal origin of PS34 cells. Mouse placentas and embryos were isolated from crosses of huCD34-rtTA with TRE-H2BGFP transgenic mice. Embryos and placentas were isolated at E12.5. Embryos were genotyped and double-transgenic and single transgenic placentas were selected for analysis.
(F, G) Flow cytometry analysis of PS34 cells derived from double transgenic (34/H2BGFP) or single transgenic (H2BGFP) placentas. Two examples and the quantification of GFP+PS34CD45− cells in 6–7 independent placentas are shown (mean ± SD).
In order to rigorously prove the embryonic origin of PS34 cells we crossed heterozygous huCD34tTA and TetO-H2BGFP mice (Qiu et al., 2014). At E12.5 both embryos and placentas were isolated. The embryos were genotyped and single TetO-H2BGFP (controls) and double transgenic (huCD34tTA x TetO-H2BGFP) were selected for analysis (Figure 3E). Only embryonic tissue can have both transgenes, therefore, the expression of GFP is indicative of fetal origin and activity of the huCD34 reporter. Placentas were dissociated, stained for Prom1, Sca1, CD34 and CD45 and analyzed for the expression of GFP (Figure 3F). PS34CD45− cells express GFP (64.5±15.3%) demonstrating their fetal origin (Figure 3F and 3G). Together, these data show that PS34 cells originate from fetal tissue, localize at the site of HSC emergence in the placenta and display a phenotype consistent with their hemogenic function.
Transcriptional Profiling of Placental Hemogenic Cells Demonstrates a Developmental Progression
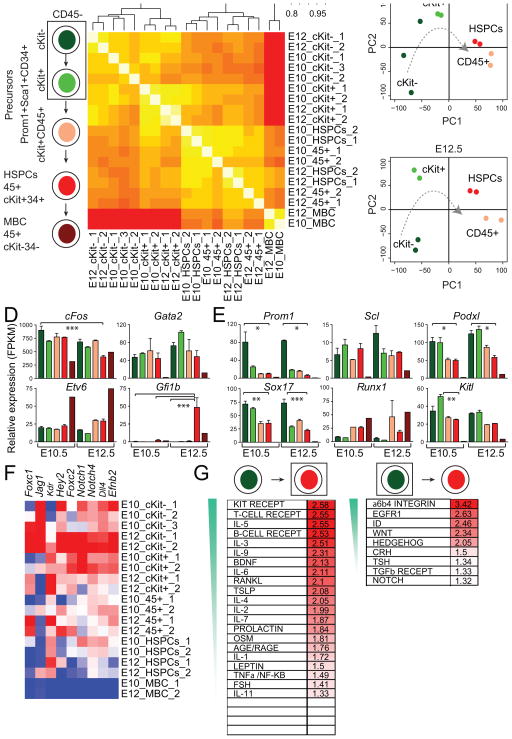
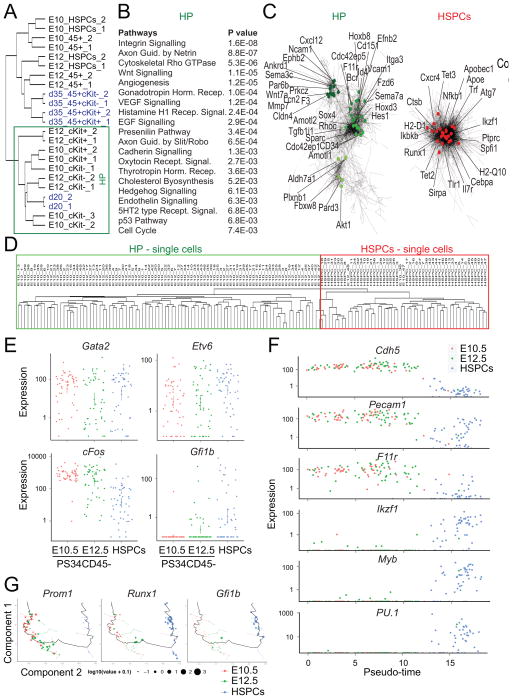
To better define the precursor and emergent hematopoietic cells in the placenta, we performed mRNA sequencing (mRNAseq) on PS34 populations at E10.5 and E12.5. Two or three biological replicates were sorted from PS34 subsets including the cKit−CD45−, cKit+CD45− and cKit+CD45+ populations (Figure 4A and S2A). We separated the PS34CD4− cells by cKit expression because of the heterogeneous expression of cKit by HPs (Figure 2B and 2C) and the requirement for cKit signaling during HE cell specification (Marcelo et al., 2013; Ruiz-Herguido et al., 2012). Placental HSPCs (CD45+cKit+CD34+ cells) and mature blood cells (CD45+cKit−CD34−) were profiled as controls. Spearman rank correlation of the dataset shows that replicas correlate with each other and, as expected, the most distant cell type was the mature blood cell population (Figure 4B). CD45− (cKit− and cKit+) and CD45+ cells (PS34 CD45+ and HSPCs) generate two very distinct clusters. PS34CD45− cells cluster by phenotype (cKit− and cKit+ samples) and not by gestation time suggesting that the PS34 phenotype captures HPs regardless of gestation time. The sample order in the clusters from cKit− to cKit+ to CD45+ suggests the developmental progression captured within the PS34 compartment. Principal component analysis (PCA) placed PS34CD45+ and HSPCs very distant from CD45− HPs consistent with the striking phenotypic changes during EHT and the acquisition of CD45 expression. PS34 cKit− and PS34 cKit+ are closely related but clearly distinguished by principal component 2 (Figure 4C).
Figure 4. Gene Expression Analyses Capture a Developmental Program.
(A) Hemogenic PS34 cells from E10.5 and E12.5 placentas were fractioned into CD45−cKit−, CD45−cKit+ and CD45+cKit+ populations. HSPCs and mature blood cells (MBC) were also sorted for comparison.
(B) Global gene-expression levels were profiled by mRNAseq (biological replicates: 1, 2 and 3). Spearman rank correlation heatmap and hierarchical clustering dendrogram of the expression profiles is displayed.
(C) PCA shows the relative distances between samples and a hypothetical temporal trajectory at E10.5 (upper panel) and E12.5 (lower panel).
(D) Expression levels of programming factors and (E) EHT-associated genes are shown as FPKM mean values ± SD. Bar colors coordinate with isolated populations in (A). *p<0.05; **p<0.01, ***p<0.001.
(F) Heatmap showing expression of arterial markers. Red designates increased expression and blue decreased expression relative to the mean.
(G) GSEA for PS34CD45−cKit− cells (right panel) and HSPCs (left panel) of NetPath-annotated signaling pathways. Pathways are ordered according to the normalized enrichment score (NES) and only enriched pathways are shown (FDR < 0.3, NES > 1.3). See also Figure S2.
Alignment of reads at individual gene loci and quantification by fragments per kilobase of exon per million fragments mapped (FPKM) values (Figure 4) and heat maps (Figure S2B) confirm the expression of endothelial and hematopoietic markers by PS34 cells. We next addressed the pattern of TF expression during EHT. Gata2 and cFos used for the conversion of fibroblasts into hemogenic cells (Pereira et al., 2013) were highly expressed in placental PS34 cells (Figure 4D). Etv6 was expressed but also seemed to be expressed in mature blood cells while Gfi1b is only expressed in HSPCs consistent with the role of Gfi1b at the later stages of EHT in repressing endothelial genes (Lancrin et al., 2012). Transcriptional regulators associated with HSCs are expressed at different stages during EHT (Figure S2C): some of the TFs are expressed early (e.g. cFos, HoxB4, Erg, Lmo2, Id1 and Msi2) and decrease upon maturation. Other TFs such as Gata2, Tal1, Sox4 are continuously expressed from HP cells to HSPCs; and other TFs are only expressed at later stages (e.g. Gfi1b, Myb, Cebpα, Pu.1, Nfe2 and Ikzf1). The expression of Sox17 was recently identified in HE cells from both the mouse and human (Clarke et al., 2013; Nakajima-Takagi et al., 2013; Nobuhisa et al., 2014). Remarkably Sox17 is expressed at the highest levels in PS34CD45− cKit− cells with a pattern reminiscent of Prom1 expression (Figure 4E). However Sox17 continues to be expressed at lower levels in HSPCs while Prom1 expression decreases more markedly suggesting that this gene product more specifically labels HPs and possibly, excludes downstream HSPCs. As expected from their roles in the specification of definitive hematopoiesis, Scl/Tal1 and Runx1 are also expressed in PS34 cell populations (Figure 4E). The expression of other reported markers of hemogenic cells such as Podxl (Hara et al., 1999) and Kitl (Pereira et al., 2013) is also enriched in PS34CD45− hemogenic cells (Figure 4E).
HE cells activate an arterial identity program characteristic of sites such as the roof of the dorsal aorta in the embryo (Wilkinson et al., 2009). We asked whether the expression of the arterial genes Foxc1, Jag1, Kdr, Hey2, Foxc2, Notch1, Notch4, Dll4 and Efnb2 was activated in PS34 cells. The expression of this gene set was mostly enriched in PS34CD45− cells (Figure 4F). We used gene set enrichment analysis (GSEA) to compare the transition from PS34CD45−cKit− precursors to HSPCs with published gene sets (Figure 4G and S2D and Table S2). GSEA showed significant enrichment for Long Term (LT) and Short Term (ST) HSC gene sets in PS34CD45−cKit− cells (Figure S2D, 3 out 3 ST-HSC and 3 out of 3 LT-HSC gene sets; false discovery rate < 0.25). This result suggests that many HSC signature genes are already expressed in HPs. Other enriched and down-regulated gene sets included Hedgehog signaling, Tight Junction, Blood Clotting cascade, and endothelial mesenchymal transition (EMT) (EMT_DN). Conversely, in HSPCs, gene sets for hematopoietic progenitors and genes that are upregulated during EMT (EMT_UP) are enriched. We next determined whether immune signaling pathways were enriched in the PS34CD45−cKit− precursors and HSPCs. Curated immune signaling pathways (21 out of 32) were enriched in HSPCs including cKit, IL-3, and B and T cell receptors signaling, in agreement with hematopoietic specification (Figure 4G). In PS34CD45−cKit− precursors only 9 out of 32 immune pathways were enriched as expected in endothelial-like precursor cells. The enriched signaling pathways include α6β4 Integrin, Egfr1, Id, Hedgehog, Wnt and Notch, consistent with their roles in hemogenesis and the pathways previously identified in HP cells programmed in vitro (Pereira et al., 2013).
A Gene Expression Signature for Definitive Hemogenic Progenitor Cells
Due to the remarkable similarities between placental HP cells and in vitro programmed cells, we compared gene expression datasets to generate a robust signature for HPs. We reasoned that the overlap between programmed cells and placental-derived precursors may better illustrate the genes and pathways required for HP cell function (Figure 5). Interestingly, PS34CD45− cells, in particular PS34CD45−cKit− cells at E10.5 and E12.5 showed a robust clustering with programmed cells isolated at day 20 (Figure 5A). In contrast, programmed CD45+cKit+ and CD45+cKit− cells show similarities with HSPCs and PS34CD45+ cells from the placenta especially at E12.5 (Figure 5A). We used K-means clustering of the integrated gene list to extract clusters of genes representative of HPs and HSPCs. 10 clusters were identified using this analysis: 6 overrepresented in HPs and 4 enriched in HSPCs (Figure S3A). This analysis revealed the dynamic nature of hemogenic cells as they progress from cKit− to cKit+ (Figure S3A). We performed pathway analysis using the PANTHER classification system. Interestingly, HPs are enriched for integrin signaling, cytoskeletal regulation by Rho GTPases, Wnt signaling, angiogenesis, Vegf signaling, and others (Figure 5B). In contrast, HSPCs are enriched for pathways involved in immune cell function such as Inflammation, Toll receptor signaling, and B and T cell activation (Figure S3B). We also used co-expression clustering to confirm gene allocation to HE or HSPCs clusters (Figure 5C and S3C). Co-expression clustering generated two distinct gene networks, which overlapped with the HP and HSPCs clusters generated using K-means clustering (Figure S3C). As previously observed, the HP cluster was more heterogeneous than the HSPCs cluster (Figure 5C). Genes identified in the HP cluster include F11r, recently shown to be expressed in HSC precursors and required for HSC generation in zebrafish through Notch signaling (Kobayashi et al., 2014). Other identified genes include the polarity regulators Pard3, Par6b, Amotl1, Prkcz and Amotl2; Notch signaling mediator Hes1; semaphorins Sema3c and Sema7a and aldehyde dehydrogenase Aldh1a1. The complete list of genes and the allocation to the network sub-clusters is provided in Table S2.
Figure 5. Hemogenic Gene Expression Signature Defined by Integration of Placental Hemogenic Cells and Induced Hemogenic Cells.
(A) Hierarchical clustering showing the integration of gene-expression data from programmed cells [blue, data from (Pereira et al., 2013)] with placental HPs and HSPCs. HP cell clustering is highlighted.
(B) Pathway enrichment analysis was performed on genes enriched in the HP cluster. Enriched terms and correspondent P values are shown.
(C) Co-expression networks show the HP cell network (left panel) and HSPCs enriched network (right panel). Specific genes of the network are highlighted.
(D) Non-supervised hierarchical clustering showing the integration of genome-wide gene-expression data from placenta-derived single cells. Clustering between PS34CD45− at E10.5 and E12.5 (green) and HSPCs (red) are highlighted.
(E) Expression levels of programming factors in placental single cells. Each dot represent FPKM values for individual cells.
(F) Expression levels of endothelial and hematopoietic genes in placental single cells ordered with Monocle software (Pseudo-time). Each dot represents FPKM values for individual cells.
(G) Cell expression profiles (dots) in a two-dimensional independent component space. Lines connecting points represent edges of the minimum-spanning tree constructed by Monocle. Solid black line shows pseudo-time ordering. See also Figure S3.
Transcription regulator prediction using ENCODE Chip-sequencing data focused on genes activated in HPs (Figure S3D, left panel) showed highest enrichment for Gata2 targets (p value = 1.02E-07) and the components of the AP-1 TF complex cFos, cJun and JunD. This result suggests cooperation between Gata2 and AP-1 in the specification of HP identity. In contrast, in HSPCs a completely different set of transcriptional regulators was identified including Ikzf1 (p value = 1.41E-38), Pu.1 (p value = 3.35E-23) and Nfkb1 (p value = 2.023E-17) (Figure S3E, right panel). Analysis of genes up-regulated in the HSPC cluster using the Mouse Genome Informatic (MGI) mouse mutant phenotype database showed that genetic perturbations caused largely hematopoietic phenotypes (Figure S3F, right panel). In contrast, HP up-regulated genes impact blood vessel and embryo development (Figure S3F, left panel). We also performed comprehensive gene ontology (GO) analysis in HPs and HSPCs using DAVID clustering (Figure S3F, Table S2 and S3). The top molecular function, biological process and cellular component categories in HP were cytoskeletal regulation, adherens and tight junctions, cellular adhesion, embryonic morphogenesis, cell motion and angiogenesis and small GTPase regulator activity (Table S3). These data are consistent with the recent implication of cellular adhesion and migration during hemogenesis (Lie et al., 2014). GO categories in HPs and HSPCs are completely distinct. We provide the full lists of terms and genes included for both HP and HSPCs (Figure S3F and Table S2). To complete the molecular profiling we performed single cell mRNA-Seq analysis of the placental PS34CD45− population (at E10.5 and E12.5) and HSPCs (at E12.5). We have profiled 48 cells for each population, a total of 144 single cells. Using genome-wide unsupervised hierarchical clustering we show that PS34CD45− cells cluster together independently of their gestation time (E10.5 and E12.5) and HSPCs form a separate cluster (Figure 5D). Regarding programming factor expression we show that Gata2 and Etv6 are similarly expressed in the 3 populations profiled. However this analysis revealed that the expression of cFos decreases from E10.5 PS34CD45− to E12.5 PS34CD45− to HSPCs (Figure 5E). This result is much more evident at the single cell than population analysis (Figure 4D). We also observed that Gfi1b although not detected in the PS34CD45− populations could be detected in some PS34CD45− cells at E12.5 and further increased in the HSPC. These data supports the role of Gfi1b in the later stages of EHT. We then ordered those cells according to gene expression to reconstruct the trajectory of endothelial-to-hematopoietic transition (Figure 5F). Endothelial genes are downregulated (Cdh5, Pecam1, F11r) and hematopoietic genes upregulated (Ikzf1, Sfpi1, Myb) according to the pseudo-ordering of cells. When reconstructing the trajectory using principal component analysis and pseudo-ordering we can observe the overlap between Prom1 expression and genes expressed at later stages of EHT such as Runx1 and Gfi1b (Figure 5G). Using single cell data we confirmed the embryonic and endothelial arterial program of PS34 cells by addressing the frequency of cells expressing endothelial markers (CD31/Pecam1, CD34 – 98–100%), cytokeratins expressed by trophoblasts (Krt7, Krt19 and Krt17 – 0–15%), the trophoblast TF Hand1 (2–4%), the venous marker Nr2f1 (2%) and the arterial marker Efnb2 (81–96%) (Table S4). Together these analyses provide a molecular profile for the HP cell program and EHT.
A Cohort of AGM-Derived PS34 Cells Activate the +23GFP Runx1 Reporter
We next asked whether PS34 cells are present in intra-embryonic sites of HSC emergence. The AGM and the vitelline/umbilical arteries (VU) were isolated at E10.5, dissociated to single cells and analyzed (Figure 6A and 6B). Indeed, we identified a small population of Prom1+Sca1+ cells (1.4±0.4%) that contains CD45−CD34+ cells (7.1±2.0%, Figure 6A). We asked whether a cohort of intra-embryonic PS34 cells activates the Runx1 23GFP reporter that marks cells undergoing EHT (Swiers et al., 2013). While the PS34CD45+ population is mainly positive for 23GFP (83.7±10.8%), the PS34CD45− population is both 23GFP+ (33.8±12.0%) and 23GFP− (63.8±11.3%) (Figure 6A). This result suggests that the PS34 phenotype captures the early stages of EHT (PS34 23GFP− cells), prior to the activation of the 23GFP reporter. The total numbers of cells per embryo equivalent (e.e.) for both PS34CD45− 23GFP− and PS34CD45− 23GFP+ population are roughly 100 and 50 cells, respectively (Figure 6B). Whole mount imaging of embryos for the endothelial marker CD31, Prom1 and the 23GFP reporter showed the presence of CD31+Prom1+23GFP+ cells both in intra-aortic clusters and as part of the endothelial layer of the aorta (Figure 6C). To confirm that Prom1 and Ly6a (that encodes Sca-1) are expressed in embryo cells undergoing EHT, we isolated 23GFP+ HE and performed qRT-PCR using the Fluidigm Biomark system. Indeed, 23GFP+ HE expresses the highest levels of Prom1 and Ly6a and low levels of CD45 when compared to HSPCs and VE-cadherin+CD31+ endothelial cells (EC) (Figure 6D). We next asked if HE cells from the conceptus (Swiers et al., 2013) share the placental and programmed HP signature. We integrated genome-wide expression data from PS34 cells from the placenta, programmed cells as well as conceptus HE, EC and HSPCs. Remarkably 23GFP HE correlated with both placental and programmed HP (Figure 6E). These data confirm that the PS34 hemogenic phenotype can also be used to isolate HPs at intra-embryonic sites of definitive hematopoiesis. To analyze the contribution of Prom1+ cells to adult hematopoietic lineages we crossed homozygous Prom1-CreERT2 mice with R26StopYFP mice and activated Cre with a 4-hydroxytamoxifen injection at E10.5 to mark some of the Prom1-expressing cells during development (Figure S4A). If Prom1+ cells are precursors of HSCs, blood YFP+ cells will include multiple cell lineages in the adult. We analyzed the presence of YFP positive cells in the peripheral blood of the progeny at 2 and 15 months of age. We identified YFP-positive cells in peripheral blood only in 4-hydroxytamoxifen injected animals (Figure S4B). The lineage composition of the YFP+ cells confirmed that these cells include both myeloid (Gr1+Mac1+), lymphoid (CD19+ and CD4+/CD8+) and erythroid cells (Figure S4C and S4D). Bone marrow analyses of 15-month old mice confirmed the presence of granulocyte-macrophage progenitors (GMP), common myeloid progenitors (CMP) and megakaryocyte-erythroid progenitors (MEP) (Figure S4E). As an additional control we injected 4-hydroxytamoxifen in adult heterozygous mice (Prom1-CreERT2 x R26StopYFP) and could not detect YFP-positive cells in the peripheral blood after 2 months (not shown). These results confirm that Prom1 marks definitive hemogenic precursors during gestation that contribute to the adult myeloid and lymphoid lineages.
Figure 6. PS34 Cells Are Found in Intra-Embryonic Hemogenic Sites.
(A) E10.5 AGM+VU regions were isolated, dissociated to a single cell suspension and analyzed by flow cytometry. PS34CD45− and PS34CD45+ cells are gated and displayed for the expression of the 23GFP Runx1 reporter. Percentages are shown as mean ± SD, n=3.
(B) Number of PS34 cells per embryo equivalent (e.e.) (mean ± SD, n=3).
(C) Representative whole mount immunofluorescence of an E10.5 (37 somite pairs) 23GFP (green) embryo combined with antibodies against Prom1 (red) and CD31 (white). Left panels show a 3D reconstruction made with 75 confocal images (2.5 μm z-sections); middle and right panels show single-plane z-sections. Arrows highlight Prom1+CD31+23GFP+ cells localized to intra-aortic clusters. Ao, Aorta.
(D) Fluidigm qRT-PCR analysis of Prom1, Ly6a and CD45 in E10.5 AGM+VU endothelial (EC: CD144+(VE-Cadherin)Ter119−CD45−CD41−23GFP−), hemogenic endothelium (HE: CD144+Ter119−CD45−CD41−23GFP+) and HSPCs (CD144+Ter119−CD45−CD41+23GFP+) (mean ± SD, n=3). *p<0.05; **p<0.01, ***p<0.001.
(E) Hierarchical clustering showing the integration of gene-expression data from programmed cells [blue, (Pereira et al., 2013)], E8.5 conceptus (PAS+YS) cells [red, (Swiers et al., 2013)] and placental derived precursors. The box highlights HP cell clustering.
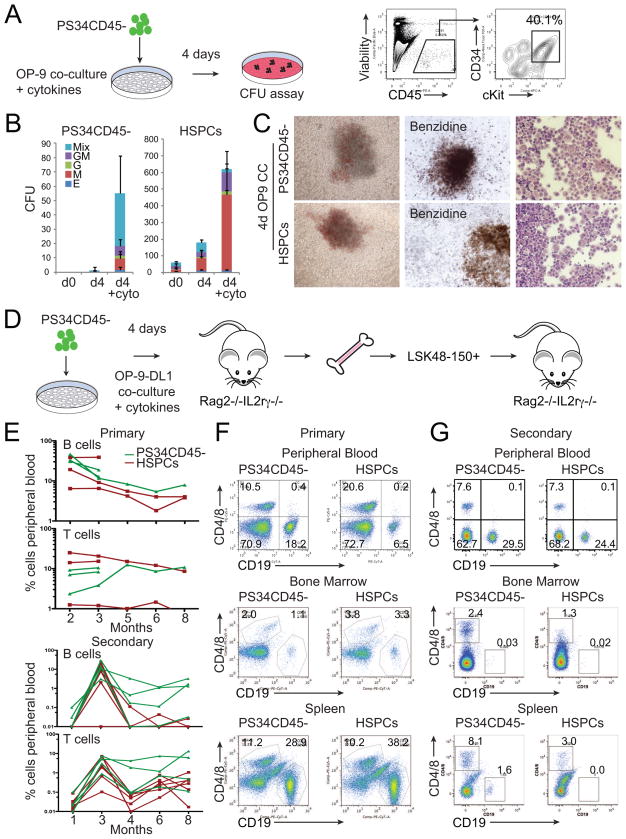
Upon Maturation in Co-Cultures PS34CD45− Generate Multi-lineage Hematopoietic Colonies
We hypothesized that placental PS34CD45− cells would require maturation in order to acquire clonogenic activity. Therefore, we performed co-cultures of PS34CD45− cells with OP-9 stromal cells and cytokines. After 4 days of co-culture, cells with an HSPC phenotype (CD45+CD34+cKit+) were generated and subsequently tested in CFU assays (Figure 7A). PS34CD45− derived cells generated large mixed colonies that were similar to those from HSPCs (Figure 7B and 7C). The presence of hematopoietic cytokines (SCF, IL-3, IL-6, FltL3 and TPO) was required to generate large numbers of hematopoietic colonies (>50 CFU per plate, Figure 7B). As controls, we plated freshly isolated PS34CD45−, which are unable to generate colonies and HSPCs, which do generate colonies (Figure 7B). Mixed colonies generated from both PS34CD45− and HSPCs were large, contained hemoglobinized erythroid cells and displayed mixed cellular morphologies (Figure 7C). Using flow cytometry and cellular morphology we confirmed that mixed colonies were composed of CD45+ cells that contain CD41+ megakaryocytes (Figure S5A and S5B), Gr1+ granulocytes and Mac1+ macrophages (Figure S5C and S5D), characteristic of definitive hematopoietic colonies. Functionally, both PS34CD45− and HSPC-derived macrophages were able to phagocytose FITC-coated beads (Figure S5E). Another hallmark of definitive hematopoiesis is the expression of adult β-globin by erythroid cells. We flow sorted TER119+ erythroid cells (Figure S5F) from mixed colonies and analyzed the transcriptome by mRNASeq. Similarly to HSPCs, PS34CD45− derived TER119+ cells express Hbb-b1 and Hbb-b2 adult genes but not Hbb-bh2, Hbb-bh2 and Hbb-γ embryonic and fetal genes; thereby, confirming the definitive nature of the erythroid cells (Figure S5G).
Figure 7. Placental Hemogenic Cells Engraft Immunodeficient Mice after OP9-DL1 Co-culture.
(A) E12.5 PS34CD45− cells were co-cultured on OP-9 in the presence of cytokines (SCF, Fltl3L, IL-3, IL-6 and TPO) for 4 days. FACS analysis shows the generation of CD45+CD34+cKit+ cells that were tested for clonogenic activity.
(B) Number and type of colonies generated at day 0, and after co-culture on OP9 of PS34CD45− or HSPCs (CD45+cKit+CD34+) with or without cytokines (mean ± SD, n=3).
(C) Pictures of representative mixed colonies (left), Benzidine hemoglobin staining (middle) and modified Giemsa staining (right).
(D) E12.5 placentas were dissociated and PS34CD45− cells isolated and cultured in gelatin-coated dishes or co-cultured with OP-9 or OP9-DL1 cells in the presence of cytokines for 4 days. Co-cultures were dissociated and transplanted intravenously into Rag2−/−/IL2Rγc−/− mice.
(E) Mice were bled from 2–8 (experiment 1) or 2–3 (experiment 2) months after transplantation (upper panel) with PS34CD45− co-cultured with OP9-DL1 (green lines). Additional mice were transplanted with freshly isolated HSPCs (red lines). After 14 weeks or 8 months of primary transplant, engrafted mice were sacrificed, and HSCs (LSK48-150+) were purified from bone marrow and transplanted intravenously into secondary Rag2−/−/IL2Rγc−/− mice (lower panel).
(F) After 12–14 weeks the presence of B (CD19+) and T (CD4+/CD8+) cells in peripheral blood, bone marrow and spleen was analyzed by flow cytometry.
(G) Analysis for the presence of B and T cells in peripheral blood after 3 months and in spleen and bone marrow after 8 months of secondary transplantation.
PS34CD45− Cells Generate B and T-lymphocytes and Engraft Immunodeficient Mice upon Notch Activation
We next asked if PS34CD45− cells could generate B and T lymphocytes after co-culture. PS34CD45− cells were co-cultured with OP9 (to generate B-cells) or OP9-DL1 (to generate T-cells) in the presence of IL-7, Flt3L and SCF. B and T cell precursors expand 3,000–30,000 fold after 2 weeks in culture (Figure S6A). 15 days after initiation of co-cultures on OP-9, CD19+B220+ B-lymphoid cells emerged (Figure S6B) that generate B-cell colonies (Figure S6B and S6C). When co-cultured with OP9-DL1 PS34CD45− cells and HSPCs produced immature CD25+CD44+ cells (day 15) and a population of CD4+CD8+ double positive cells (day 25) after reduction of cytokine concentrations (Figure S6D and S6E). To induce maturation we transplanted B and T cell precursors at day 15 into Rag2−/−/IL2Rγc−/− mice. 4 weeks later we detected single positive CD4+ and CD8+ T cells and CD19+B220+ B cells (Figure S6F and S6G). Single positive CD4 and CD8 T cells generated after maturation in vivo express the T-cell co-receptor CD3 (Figure S6H). Mice were sacrificed 4 weeks after transplantation and CD4+ and CD8+ T cells were isolated from blood, spleen and bone marrow for functional activity assays. After activation in culture, both PS34CD45− and HSPCs derived T cells secrete the Th1/Th2 cytokines IL-5, IL-4, INF-γ and TNF-α (Figure S6I). These data confirm that the PS34 phenotype marks HPs with the capacity to generate hematopoietic progenitors of myeloid and lymphoid lineages as well as mature cells that include macrophages, granulocytes, erythrocytes, megakaryocytes and B- and T- lymphocytes.
We next asked if PS34CD45− cells give rise to transplantable HSCs. We first performed transplantation experiments into congenic SJL mice. PS34CD45−cKit−, PS34CD45−cKit+ and PS34CD45+ cells were transplanted into sub lethally irradiated mice. In agreement with the lack of hematopoietic clonogenic activity in vitro, PS34CD45− or PS34CD45+ cells do not engraft adult SJL mice (Figure S7A). This result contrasts with whole placenta tissue transplantation at E11.5 and E12.5 where engraftment is robust (Figure S7A and S7B). We did not detect engraftment of whole placenta tissue at E10.5 (0.3 or 1 embryo equivalents per transplanted mouse) or PS34CD45− engraftment into more permissive Rag2−/−/IL2Rγc mice (not shown). We next asked whether an in vitro co-culture step with OP-9 would confer engraftment potential to the emergent hematopoietic progenitors (Figure S7B). We isolated PS34CD45− cells at E10.5, E11.5 and E12.5 and performed co-cultures with OP-9 for 3 or 4 days. We did not detect engraftment after OP-9 co-culture with cytokines (Figure S7B). This result suggests that hematopoietic progenitors may be pushed to differentiate in the culture conditions employed and activation of a critical signaling pathway(s) may be required to maintain self-renewal of in vitro generated HSPCs. Notch signaling is essential for the specification of HSC and definitive hematopoiesis (Kumano et al., 2003; Marcelo et al., 2013). During development, close contact between Notch signal-emitting cells and precursors is essential for successful HSC specification and maturation (Clements et al., 2011; Kobayashi et al., 2014). We hypothesized that providing a Notch signal in vitro would confer engraftment potential to PS34CD45− cells. To test this we isolated PS34CD45− cells from E12.5 placentas and performed 4 day co-cultures on gelatin-coated dishes, OP-9 or OP-9DL1 cells, which express the Notch ligand Delta1 (Figure 7D, S7C and S7D). Twelve weeks after transplantation into Rag2−/−/IL2Rγc mice we detected a large population of B and T cells in peripheral blood only when PS34CD45− cells were co-cultured with OP9-DL1 (Figure 7E upper panel, and 7F). The populations of B and T cells are maintained in the peripheral blood for up to 8 months after transplantation (Figure 7E). We next asked if PS34CD45− derived cells reestablish an HSPC compartment in the bone marrow and maintain self-renewal. To address this we sacrificed transplanted mice at 14 weeks and isolated purified HSCs (LSKCD48−CD150+) from bone marrow and transplanted secondary Rag2−/−/IL2Rγc mice. We first confirmed that both the spleen and bone marrow of transplanted mice contained populations of B and T cells at 14 weeks (Figure 7F, lower panel). At 8 months after secondary transplantation we could still detect B and T lymphocytes in peripheral blood, bone marrow and spleen of these animals (Figure 7E and 7G). These results demonstrate that the PS34CD45− phenotype marks early precursors of bona fide HSCs and that Notch signaling is crucial for a productive EHT in vitro. In summary, based on the phenotype and gene expression data from our previous in vitro programming studies (Pereira et al., 2013) we have identified and characterized in detail HSC precursor cells.
Discussion
In developmental hematopoiesis there is no surface marker definition for the precursor cells to HE. CD34 is not expressed (or at low levels) in LT-HSC from bone marrow and fetal liver (Gekas et al., 2005; Osawa et al., 1996; Sanchez et al., 1996) but is highly expressed in HSCs from the placenta, AGM (Gekas et al., 2005; Kumano et al., 2003; Sanchez et al., 1996) and in repopulating cells from E9.5 yolk sacs (Yoder et al., 1997). CD34 is also expressed in the HE derived from human PSCs (Nakajima-Takagi et al., 2013; Sturgeon et al., 2014). The Ly6a gene that encodes the Sca1 antigen is expressed before HSC emergence in the dorsal aorta and is a marker of definitive but not primitive HE (Chen et al., 2011; Ling et al., 2004). Placental and embryonic HPs are also marked by the expression of the Prom1 gene. In humans, Prom1 is a marker for endothelial progenitor cells and HSCs. Indeed, it has been shown that a CD34+PROM1+ population from human umbilical cord blood can give rise to endothelial and hematopoietic cells (Wu et al., 2007). In the mouse however, Prom1 is not expressed in LT-HSCs isolated from bone marrow (Arndt et al., 2013). We showed that Prom1 expression rapidly declines during EHT allowing the exclusion of specified, mature HSPCs. These three markers showed peak expression in PS34CD45−cKit− cells where we also detected other genes previously implicated in HE or hemogenic programming (Sox17, Scl, Podxl, Kitl, cFos, Gata2) (Clarke et al., 2013; Pereira et al., 2013). Runx1 and Gfi1b, critical genes during EHT (Lancrin et al., 2012; Swiers et al., 2013), were only detected at low levels implying that PS34CD45− cells represent early stage HPs. Furthermore, along with the expression of endothelial markers we detected the expression of CD49f, an antigen identified in induced HPs (Pereira et al., 2013) and human HSCs (Notta et al., 2011). CD49f is expressed in both PS34 and emergent HSPCs. Taken together, these data highlight the relevance of the phenotype defined by Prom1, Sca1 and CD34 for the prospective isolation of early definitive HPs in vivo.
In the E10.5–E11.5 AGM region two types of HSC precursors that sequentially develop into HSCs have been described: type I pre-HSCs (VE-Caderin+CD45−CD41low) and type II pre-HSCs (VE-cadherin+CD45+) (Rybtsov et al., 2011; Taoudi et al., 2008). In the E9.5 mouse embryo, an earlier HSC precursor (pro-HSC, VEcadherin+CD45−CD41lowCD43−) that is SCF dependent has also been described (Rybtsov et al., 2014). A Runx1 23GFP reporter in combination with VE-Cadherin was used to isolate HE cells (Swiers et al., 2013). As suggested by the genome-wide gene expression data and the activity of the 23GFP reporter, PS34 cells represent an earlier precursor, where hematopoietic markers such as CD45, CD41 and Runx1 are not highly expressed. Indeed, Prom1+ cells are embedded in the endothelial layer and cells with hematopoietic morphology expressing Prom1 were rarely detected by immunofluorescence. Placental PS34 cells are derived from embryonic tissue and located in the vascular labyrinth at the maternal-fetal interface, in agreement with previous reports on placental HSCs (Gekas et al., 2005; Ottersbach and Dzierzak, 2005; Rhodes et al., 2008). It will be interesting to investigate the maternal and fetal factors (Rossant and Cross, 2001) that impact the specification and maturation of HPs into HSCs. Defining such factors may facilitate the efficient maturation of hemogenic cells in vitro.
We show that a co-culture step with OP9 supplemented with cytokines allows the specification of HPs into definitive HSPCs with myeloid and lymphoid clonogenic activity. The activation of the Notch pathway (by co-culture with OP9-DL1) endows serial transplantation capacity in Rag2/IL2Rγc mutant mice, highlighting the role of Notch and a permissive environment for precursor cell maturation (Hadland et al., 2015). We show that the transcripts for the adhesion molecule F11r (also know as Jam1a) (Kobayashi et al., 2014) and the TFs Foxc2 (Jang et al., 2015) and Hes1 (Bertrand et al., 2010b) are up-regulated both in placental PS34 cells and programmed hemogenic cells. Interestingly, in zebrafish Notch-dependent Gata2b expression within the hemogenic cell compartment is required to initiate Runx1 expression (Butko et al., 2015) supporting the notion that Gata2 functions in HPs before Runx1 (Ditadi et al., 2015). The ability to isolate HPs in large quantities from developing placentas and subsequently mature them in vitro into functional HSPCs provides an opportunity to dissect EHT and stem cell generation. Using this system it will be interesting to address the impact of inflammatory signals (Li et al., 2014), Wnt ligands (Ruiz-Herguido et al., 2012) and other substrates as well as soluble factors on HP maturation and the establishment of stem cell self-renewal.
Our PS34 dataset and integrative analysis between programmed precursors and in vivo precursor cells provides a stringent signature for HP cells. By integrating the two datasets we eliminate concerns of contamination with other cell-types (ex vivo isolation) and also aberrant gene activation from TF overexpression. Our data show that both programmed precursors from fibroblasts and placental PS34CD45−cKit− cells are strikingly similar and correlate with 23GFP+ cells from AGM. This generated signature can be used to test the relevance of specific pathways (such as integrin signaling, endothelial to mesenchymal transition, angiogenesis, cytoskeletal regulation by Rho GTPases, Netrin signaling, and others) for HP specification. We provide our full dataset and analysis in Table S2. It will be interesting to assess the role of individual or combination of genes enriched in HP clusters during EHT and stem cell generation (for example Aldh7a1, Cxcl12, Kitl, Sema3c, Par6b, Wnt7a, Vcam1). HP lineage divergence from endothelium may occur before extensive formation of intra-aortic clusters (Rybtsov et al., 2011; Swiers et al., 2013). The enrichment of angiogenesis, migration and adhesion-related genes in PS34CD45− cells also suggests that HP cells do not represent a cohort of mature endothelium but more likely a different lineage with endothelial features that is committed to the hematopoietic route (Ditadi et al., 2015). This result is also supported by the recent implication of cell migration as a feature of HPs in zebrafish (Kobayashi et al., 2014) and Runx1 targets in mice (Lie et al., 2014).
In summary, we used information generated during direct programming into hemogenic cells (Pereira et al., 2013) to provide insights into the specification of HSCs in the placenta and AGM. Direct cell reprogramming is therefore not only a methodology for generating specific cell types but also can provide the minimal TF network to kick start the specification of cell identity and cellular markers and phenotypes to clarify developmental specification. This approach can be complementary to traditional developmental approaches especially in scenarios where genetic studies might fail. For example, when genes functionally compensate each other or due to the complexity of embryonic tissue and spatial-temporal constraints. Previous studies of AP-1 in HSC specification illustrate the advantage of this approach. Deletion of cFos did not severely impair placental HSCs (Ottersbach and Dzierzak, 2005). However cFos overexpression was absolutely required for hemogenesis from fibroblasts (Pereira et al., 2013) or the homologous gene FOSB, from human endothelial cells (Sandler et al., 2014). In light of the recent implication of inflammatory pathways as major players during HSC specification (Li et al., 2014), it would be interesting to investigate the link between AP-1 activation, NF-kappaB and inflammation (Linnemann et al., 2011; Natoli, 2010).
Collectively, our results show that Prom1, Sca1 and CD34 mark HP cells. These precursors can be matured to serially engrafting HSCs in vivo. Our results support the view that HP lineage divergence is an early developmental event and underscore the requirement of Notch activation for their maturation into functional HSCs. These studies provide an in vitro platform for the molecular dissection of definitive EHT and HSC emergence. The derived gene expression platform will work both ways, suggesting new ways to improve in vitro hematopoietic reprogramming. Taken together our studies suggest that both processes will provide insights into the expansion of HSCs from both reprogrammed and donor/patient-derived HSCs for clinical transplant and regenerative medicine.
Experimental procedures
Mice and Placental Cell Isolation
C57BL/6 pregnant mice (The Jackson Laboratory) were used to dissect placentas between embryonic day 10.5 (E10.5) and E12.5. The day of copulation plug was regarded as E0.5. Double transgenic (designated 34/H2BGFP) placentas were derived from crosses of individual transgenic CD34tTA (Dan Tenen, Harvard) and TetO-H2BGFP (The Jackson Laboratory) mouse lines that had been backcrossed to C57BL/6J over 12 generations (Qiu et al., 2014). B10;B6-Rag2tm1Fwa Il2rgtm1Wjl (Rag2−/−/IL2Rγc) mice were purchased from Taconic (model 4111). Congenic B6.SJL-Ptprca Pepcb/BoyJ (SJL) mice were purchased from The Jackson Laboratory, bred, and maintained in house. Placentas were dissected and separated from the umbilical cord and maternal decidua. Tissues were kept on ice, washed in PBS, dissociated mechanically through a 18G needle, and treated with 0.2% collagenase type I (Sigma) in PBS with 20% FBS (Benchmark) for 1.5 hr at 37°C, followed by passages through 20G and 25G needles. Single cell suspensions were filtered through 70 μm cell strainers (BD Falcon). Placentas from the same litter were combined for cell isolation. Animal experiments and procedures were approved by the Institutional Animal Care and Use Committee and conducted in accordance with the Animal Welfare Act.
mRNAseq Library Preparation, Sequencing and Analysis
Cells isolated by FACS were lysed in Trizol (Ambion) and total RNA extracted by ethanol precipitation. RNA from each sample was used for library preparation with the TruSeq RNA Sample Preparation Kit v2 (Illumina). A common adapter was used for all samples and barcode sequences present in the reverse primer were introduced by 12–26 cycles of amplification. Each library was quantified by PCR using a library quantification kit for Illumina sequencing platforms (KAPA) and equimolar amounts of each barcoded library were mixed and single-end sequenced on an Illumina HiSeq 2500 Sequencing System. For each sample ~9–25 M 100-nt reads were obtained, pre-processed and aligned to the mouse genome (Mus musculus mm9 assembly) as previously described (Pereira et al., 2013). PS34CD45− and HSPCs were sorted and collected. Single-Cell cDNA synthesis was performed following the manufacturers instruction using the C1 Single-Cell Auto Prep System (Fluidigm). Additional analytical details are provided in Supplemental Experimental Procedures.
Cell Culture, Hemogenic Assays and Colony-Forming Assays
For hemogenic assays placental-derived PS34CD45− cells were cultured in gelatin-coated dishes. For cobblestone forming assays PS34CD45− cells were plated on irradiated AFT024 feeder cells. Cultures were maintained in Myelocult Media (M5300; Stem Cell Technologies) supplemented with Hydrocortisone (10−6 M; Stem Cell Technologies) and 100 ngml−1 SCF, 100 ngml−1 Flt3L, 20 ngml−1 IL-3, 10 ngml−1 TPO and 20 ngml−1 IL-6 (R&D). To generate clonogenic progenitors PS34CD45− cells were co-cultured with live OP9 cells for 4 days without cytokines or in the presence of 100 ngml−1 SCF, 100 ngml−1 Flt3L, 20 ngml−1 IL-3, 10 ngml−1 TPO and 20 ngml−1 IL-6. PS34CD45− cells were also co-cultured on gelatin, OP-9, and OP-9-DL1 cells for 4 days in the presence of 50 ngml−1 SCF, 20 ngml−1 Flt3L and 50 ngml−1 IL-3 and then transplanted. Experimental details for immunofluorescence, FACS sorting, cell culture, transplantation assays and other assays are provided in Supplemental Experimental Procedures.
Statistical Analysis
Comparisons among groups were performed by one-way ANOVA followed by Tukey-Kramer multiple comparison test with GraphPad Prism 5 software. P-values are shown when relevant (*p<0.05; **p<0.01, ***p<0.001).
Supplementary Material
Highlights.
Direct Reprogramming informs identification of Hemogenic Precursors (HP) in vivo
HP cells display a Prominin1, Sca1, CD34 phenotype that is also CD45 negative
Gene expression signatures were established at the population and single cell level
HP cells can be matured into bona fide hematopoietic stem cells
Acknowledgments
We thank N. Speck (University of Pennsylvania) for the OP9 cell lines. We thank the members of the Lemischka and Moore laboratories for useful discussions and Y. Liu for laboratory management. We would like to thank V. Gouon-Evans, O. Goldman, S. Ghaffari, G. Camprecios and P. Rimmele for assistance. We thank J. Kovacic and A. Nomura-Kitabayashi for the R26StopYFP mice and advice with lineage tracing and H. Schöler for critical reading of the manuscript. We would also like to thank the Mount Sinai Pluripotent Stem Cell Core and S. L. D’Souza for help with materials and protocols and the Mount Sinai Genomics, Flow Cytometry and Mouse facilities. This study was made possible in part by funds granted by the Charles H. Revson Foundation (C.F.P., senior fellow in biomedical science) and by NIH 1R01HL119404 (K.A.M. and I.R.L.). The statements made and views expressed are solely the responsibility of the authors.
Footnotes
Accession numbers
mRNAseq data are deposited in the Gene Expression Omnibus database under accession number GSE54574.
The authors have no conflicts of interest.
Author Contributions
C.F.P., B.C, A.G., J.B., X.N. and C.S. conducted placental experiments; G.S., E.A. and M.F.T.R.B. conducted AGM experiments; B.C., D.P. and C.F.P analyzed mRNA-Seq data; C.F.P., I.R.L. and K.A.M. designed the experiments and wrote the paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arndt K, Grinenko T, Mende N, Reichert D, Portz M, Ripich T, Carmeliet P, Corbeil D, Waskow C. CD133 is a modifier of hematopoietic progenitor frequencies but is dispensable for the maintenance of mouse hematopoietic stem cells. Proc Natl Acad Sci U S A. 2013;110:5582–5587. doi: 10.1073/pnas.1215438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DY, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010a;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Cisson JL, Stachura DL, Traver D. Notch signaling distinguishes 2 waves of definitive hematopoiesis in the zebrafish embryo. Blood. 2010b;115:2777–2783. doi: 10.1182/blood-2009-09-244590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset JC, Clapes T, Klaus A, Papazian N, Onderwater J, Mommaas-Kienhuis M, Cupedo T, Robin C. Progressive maturation towards hematopoietic stem cells in the mouse embryo aorta. Blood. 2014 doi: 10.1182/blood-2014-07-588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Butko E, Distel M, Pouget C, Weijts B, Kobayashi I, Ng K, Mosimann C, Poulain FE, McPherson A, Ni CW, et al. Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development. 2015;142:1050–1061. doi: 10.1242/dev.119180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Li Y, De Obaldia ME, Yang Q, Yzaguirre AD, Yamada-Inagawa T, Vink CS, Bhandoola A, Dzierzak E, Speck NA. Erythroid/myeloid progenitors and hematopoietic stem cells originate from distinct populations of endothelial cells. Cell Stem Cell. 2011;9:541–552. doi: 10.1016/j.stem.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RL, Yzaguirre AD, Yashiro-Ohtani Y, Bondue A, Blanpain C, Pear WS, Speck NA, Keller G. The expression of Sox17 identifies and regulates haemogenic endothelium. Nat Cell Biol. 2013;15:502–510. doi: 10.1038/ncb2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements WK, Kim AD, Ong KG, Moore JC, Lawson ND, Traver D. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature. 2011;474:220–224. doi: 10.1038/nature10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditadi A, Sturgeon CM, Tober J, Awong G, Kennedy M, Yzaguirre AD, Azzola L, Ng ES, Stanley EG, French DL, et al. Human definitive haemogenic endothelium and arterial vascular endothelium represent distinct lineages. Nat Cell Biol. 2015;17:580–591. doi: 10.1038/ncb3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doulatov S, Daley GQ. Development. A stem cell perspective on cellular engineering. Science. 2013;342:700–702. doi: 10.1126/science.1238363. [DOI] [PubMed] [Google Scholar]
- Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nature immunology. 2008;9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilken HM, Nishikawa S, Schroeder T. Continuous single-cell imaging of blood generation from haemogenic endothelium. Nature. 2009;457:896–900. doi: 10.1038/nature07760. [DOI] [PubMed] [Google Scholar]
- Gekas C, Dieterlen-Lievre F, Orkin SH, Mikkola HK. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8:365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Hadland BK, Varnum-Finney B, Poulos MG, Moon RT, Butler JM, Rafii S, Bernstein ID. Endothelium and NOTCH specify and amplify aorta-gonad-mesonephros-derived hematopoietic stem cells. The Journal of clinical investigation. 2015;125:2032–2045. doi: 10.1172/JCI80137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Nakano Y, Tanaka M, Tamura K, Sekiguchi T, Minehata K, Copeland NG, Jenkins NA, Okabe M, Kogo H, et al. Identification of podocalyxin-like protein 1 as a novel cell surface marker for hemangioblasts in the murine aorta-gonad-mesonephros region. Immunity. 1999;11:567–578. doi: 10.1016/s1074-7613(00)80132-6. [DOI] [PubMed] [Google Scholar]
- Jang IH, Lu YF, Zhao L, Wenzel PL, Kume T, Datta SM, Arora N, Guiu J, Lagha M, Kim PG, et al. Notch1 acts via Foxc2 to promote definitive hematopoiesis via effects on hemogenic endothelium. Blood. 2015;125:1418–1426. doi: 10.1182/blood-2014-04-568170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieusseian A, Brunet de la Grange P, Burlen-Defranoux O, Godin I, Cumano A. Immature hematopoietic stem cells undergo maturation in the fetal liver. Development. 2012;139:3521–3530. doi: 10.1242/dev.079210. [DOI] [PubMed] [Google Scholar]
- Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- Kobayashi I, Kobayashi-Sun J, Kim AD, Pouget C, Fujita N, Suda T, Traver D. Jam1a-Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature. 2014;512:319–323. doi: 10.1038/nature13623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K, Chiba S, Kunisato A, Sata M, Saito T, Nakagami-Yamaguchi E, Yamaguchi T, Masuda S, Shimizu K, Takahashi T, et al. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 2003;18:699–711. doi: 10.1016/s1074-7613(03)00117-1. [DOI] [PubMed] [Google Scholar]
- Lancrin C, Mazan M, Stefanska M, Patel R, Lichtinger M, Costa G, Vargel O, Wilson NK, Moroy T, Bonifer C, et al. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood. 2012;120:314–322. doi: 10.1182/blood-2011-10-386094. [DOI] [PubMed] [Google Scholar]
- Lancrin C, Sroczynska P, Stephenson C, Allen T, Kouskoff V, Lacaud G. The haemangioblast generates haematopoietic cells through a haemogenic endothelium stage. Nature. 2009;457:892–895. doi: 10.1038/nature07679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Esain V, Teng L, Xu J, Kwan W, Frost IM, Yzaguirre AD, Cai X, Cortes M, Maijenburg MW, et al. Inflammatory signaling regulates embryonic hematopoietic stem and progenitor cell production. Genes Dev. 2014;28:2597–2612. doi: 10.1101/gad.253302.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie ALM, Marinopoulou E, Li Y, Patel R, Stefanska M, Bonifer C, Miller C, Kouskoff V, Lacaud G. RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. Blood. 2014 doi: 10.1182/blood-2014-04-572958. [DOI] [PubMed] [Google Scholar]
- Ling KW, Ottersbach K, van Hamburg JP, Oziemlak A, Tsai FY, Orkin SH, Ploemacher R, Hendriks RW, Dzierzak E. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med. 2004;200:871–882. doi: 10.1084/jem.20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann AK, O’Geen H, Keles S, Farnham PJ, Bresnick EH. Genetic framework for GATA factor function in vascular biology. Proc Natl Acad Sci U S A. 2011;108:13641–13646. doi: 10.1073/pnas.1108440108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelo KL, Sills TM, Coskun S, Vasavada H, Sanglikar S, Goldie LC, Hirschi KK. Hemogenic endothelial cell specification requires c-Kit, Notch signaling, and p27-mediated cell-cycle control. Dev Cell. 2013;27:504–515. doi: 10.1016/j.devcel.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvinsky A, Rybtsov S, Taoudi S. Embryonic origin of the adult hematopoietic system: advances and questions. Development. 2011;138:1017–1031. doi: 10.1242/dev.040998. [DOI] [PubMed] [Google Scholar]
- Mikkola HK, Orkin SH. The journey of developing hematopoietic stem cells. Development. 2006;133:3733–3744. doi: 10.1242/dev.02568. [DOI] [PubMed] [Google Scholar]
- Nakajima-Takagi Y, Osawa M, Oshima M, Takagi H, Miyagi S, Endoh M, Endo TA, Takayama N, Eto K, Toyoda T, et al. Role of SOX17 in hematopoietic development from human embryonic stem cells. Blood. 2013;121:447–458. doi: 10.1182/blood-2012-05-431403. [DOI] [PubMed] [Google Scholar]
- Natoli G. NF-kappaB: no longer an island, but a piece of a continent. EMBO Rep. 2010;11:246–248. doi: 10.1038/embor.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobuhisa I, Osawa M, Uemura M, Kishikawa Y, Anani M, Harada K, Takagi H, Saito K, Kanai-Azuma M, Kanai Y, et al. Sox17-mediated maintenance of fetal intra-aortic hematopoietic cell clusters. Molecular and cellular biology. 2014;34:1976–1990. doi: 10.1128/MCB.01485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notta F, Doulatov S, Laurenti E, Poeppl A, Jurisica I, Dick JE. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Ottersbach K, Dzierzak E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell. 2005;8:377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, Hendry CE, Clark NR, Nomura-Kitabayashi A, Kovacic JC, Ma’ayan A, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira CF, Lemischka IR, Moore K. Reprogramming cell fates: insights from combinatorial approaches. Ann N Y Acad Sci. 2012;1266:7–17. doi: 10.1111/j.1749-6632.2012.06508.x. [DOI] [PubMed] [Google Scholar]
- Qiu J, Papatsenko D, Niu X, Schaniel C, Moore K. Divisional History and Hematopoietic Stem Cell Function during Homeostasis. Stem cell reports. 2014;2:473–490. doi: 10.1016/j.stemcr.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes KE, Gekas C, Wang Y, Lux CT, Francis CS, Chan DN, Conway S, Orkin SH, Yoder MC, Mikkola HK. The emergence of hematopoietic stem cells is initiated in the placental vasculature in the absence of circulation. Cell Stem Cell. 2008;2:252–263. doi: 10.1016/j.stem.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin C, Ottersbach K, Boisset JC, Oziemlak A, Dzierzak E. CD41 is developmentally regulated and differentially expressed on mouse hematopoietic stem cells. Blood. 2011;117:5088–5091. doi: 10.1182/blood-2011-01-329516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Placental development: lessons from mouse mutants. Nature reviews Genetics. 2001;2:538–548. doi: 10.1038/35080570. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herguido C, Guiu J, D’Altri T, Ingles-Esteve J, Dzierzak E, Espinosa L, Bigas A. Hematopoietic stem cell development requires transient Wnt/beta-catenin activity. J Exp Med. 2012;209:1457–1468. doi: 10.1084/jem.20120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybtsov S, Batsivari A, Bilotkach K, Paruzina D, Senserrich J, Nerushev O, Medvinsky A. Tracing the origin of the HSC hierarchy reveals an SCF-dependent, IL-3-independent CD43(−) embryonic precursor. Stem cell reports. 2014;3:489–501. doi: 10.1016/j.stemcr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybtsov S, Sobiesiak M, Taoudi S, Souilhol C, Senserrich J, Liakhovitskaia A, Ivanovs A, Frampton J, Zhao S, Medvinsky A. Hierarchical organization and early hematopoietic specification of the developing HSC lineage in the AGM region. J Exp Med. 2011;208:1305–1315. doi: 10.1084/jem.20102419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez MJ, Holmes A, Miles C, Dzierzak E. Characterization of the first definitive hematopoietic stem cells in the AGM and liver of the mouse embryo. Immunity. 1996;5:513–525. doi: 10.1016/s1074-7613(00)80267-8. [DOI] [PubMed] [Google Scholar]
- Sandler VM, Lis R, Liu Y, Kedem A, James D, Elemento O, Butler JM, Scandura JM, Rafii S. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 2014;511:312–318. doi: 10.1038/nature13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgeon CM, Ditadi A, Awong G, Kennedy M, Keller G. Wnt signaling controls the specification of definitive and primitive hematopoiesis from human pluripotent stem cells. Nat Biotechnol. 2014;32:554–561. doi: 10.1038/nbt.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiers G, Baumann C, O’Rourke J, Giannoulatou E, Taylor S, Joshi A, Moignard V, Pina C, Bee T, Kokkaliaris KD, et al. Early dynamic fate changes in haemogenic endothelium characterized at the single-cell level. Nature communications. 2013;4:2924. doi: 10.1038/ncomms3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoudi S, Gonneau C, Moore K, Sheridan JM, Blackburn CC, Taylor E, Medvinsky A. Extensive hematopoietic stem cell generation in the AGM region via maturation of VE-cadherin+CD45+ pre-definitive HSCs. Cell Stem Cell. 2008;3:99–108. doi: 10.1016/j.stem.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Wapinski OL, Vierbuchen T, Qu K, Lee QY, Chanda S, Fuentes DR, Giresi PG, Ng YH, Marro S, Neff NF, et al. Hierarchical mechanisms for direct reprogramming of fibroblasts to neurons. Cell. 2013;155:621–635. doi: 10.1016/j.cell.2013.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson RN, Pouget C, Gering M, Russell AJ, Davies SG, Kimelman D, Patient R. Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Dev Cell. 2009;16:909–916. doi: 10.1016/j.devcel.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Lensch MW, Wylie-Sears J, Daley GQ, Bischoff J. Hemogenic endothelial progenitor cells isolated from human umbilical cord blood. Stem Cells. 2007;25:2770–2776. doi: 10.1634/stemcells.2006-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Du Y, Deng H. Direct Lineage Reprogramming: Strategies, Mechanisms, and Applications. Cell Stem Cell. 2015;16:119–134. doi: 10.1016/j.stem.2015.01.013. [DOI] [PubMed] [Google Scholar]
- Yoder MC, Hiatt K, Dutt P, Mukherjee P, Bodine DM, Orlic D. Characterization of definitive lymphohematopoietic stem cells in the day 9 murine yolk sac. Immunity. 1997;7:335–344. doi: 10.1016/s1074-7613(00)80355-6. [DOI] [PubMed] [Google Scholar]
- Zhu L, Gibson P, Currle DS, Tong Y, Richardson RJ, Bayazitov IT, Poppleton H, Zakharenko S, Ellison DW, Gilbertson RJ. Prominin 1 marks intestinal stem cells that are susceptible to neoplastic transformation. Nature. 2009;457:603–607. doi: 10.1038/nature07589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zovein AC, Hofmann JJ, Lynch M, French WJ, Turlo KA, Yang Y, Becker MS, Zanetta L, Dejana E, Gasson JC, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 2008;3:625–636. doi: 10.1016/j.stem.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.