Abstract
Alcoholism is associated with dysregulation in the neural circuitry that mediates motivated and goal-directed behaviors. The dopaminergic connection between the ventral tegmental area (VTA) and the nucleus accumbens is viewed as a critical component of the neurocircuitry mediating alcohol’s rewarding and behavioral effects. We sought to determine the effects of binge alcohol drinking on global gene expression in VTA dopaminergic (DA) neurons. Alcohol-preferring C57BL/6J × FVB/NJ F1 hybrid female mice were exposed to a modified drinking in the dark (DID) procedure for 3 weeks, while control animals had access to water only. Global gene expression of laser-captured tyrosine hydroxylase - positive VTA DA neurons was measured using microarrays. 644 transcripts were differentially expressed between the drinking and non-drinking mice and 930 transcripts correlated with alcohol intake during the last two days of drinking in the alcohol group. Bioinformatics analysis of alcohol-responsive genes identified molecular pathways and networks perturbed in DA neurons by alcohol consumption, which included neuroimmune and epigenetic functions, alcohol metabolism and brain disorders. The majority of genes with high and specific expression in DA neurons were down regulated by or negatively correlated with alcohol consumption, suggesting a decreased activity of DA neurons in high drinking animals. These changes in the dopaminergic transcriptome provide a foundation for alcohol-induced neuroadaptations that may play a crucial role in the transition to addiction.
Keywords: alcohol consumption, ventral tegmental area, dopaminergic neurons, neuroadaptation, laser capture microdissection, gene expression, microarrays
Introduction
Alcohol (ethanol) abuse is one of the leading causes of death and disability, claiming millions of lives worldwide. Alcohol is frequently comorbid with other substances of abuse and psychiatric disorders (Wang et al., 2011), which emphasizes its particularly damaging effects on the nervous system. Alcohol is capable of altering brain function by causing gene expression changes in neurons and glia (Pignataro et al., 2009; Repunte-Canonigo et al., 2007; Mulligan et al., 2006; Mulligan et al., 2011; Ponomarev et al., 2012), which may ultimately control alcohol-induced neuroadaptations and behavioral modifications. The ventral tegmental area (VTA) is a key brain region involved in regulation of drinking behavior and rewarding effects of alcohol (Deehan et al., 2013; Nimitvilai et al., 2013; Hwa et al., 2013). VTA dopaminergic (DA) neurons form the first segment of the mesolimbic system, whose activation causes increases in dopamine levels in the nucleus accumbens, forming the crux of the rewarding effects of drugs of abuse (Watabe-Uchida et al., 2012; Sesack and Grace, 2010). Acute alcohol activates the mesolimbic pathway by increasing DA neuron firing (Brodie, 1999; Robinson et al., 2009; Morikawa and Morrisett, 2010). Several studies reported reduced firing rates in adult midbrain DA neurons shortly after the cessation of chronic alcohol administration (Mulholland et al., ACER, 2009 for review). It is hypothesized that repeated alcohol causes neuroadaptations in the DA neurons associated with a decrease in alcohol-induced firing and a subsequent increase in alcohol consumption (Hoffman and Tabakoff, 1996). Some evidence of such neuroadaptation is obtained by electrophysiological studies. For example, in vivo exposure to ethanol increased susceptibility to the induction of long-term potentiation of NMDA receptor-mediated transmission in VTA DA neurons (Bernier et al., 2011), while ethanol-exposed VTA slices showed an increase in GABA-mediated neurotransmission onto DA neurons (Theile et al., 2008). In addition, repeated cycles of alcohol exposure and withdrawal produced several alterations in the physiological properties of VTA DA neurons, including a reduction in functions of the small conductance calcium-dependent potassium (SK) channels (Hopf et al., 2007). However, molecular mechanisms underlying these neuroadaptations are not well understood.
In order to get an insight into these processes we used a combination of rapid antibody staining, laser capture microdissection (LCM) and microarrays to study global gene expression in tyrosine hydroxylase (TH)-positive DA neurons from the VTA of alcohol-drinking and non-drinking mice. We used a mouse model of binge drinking and identified multiple genes and molecular pathways regulated by alcohol intake in DA neurons. We hypothesized that these molecular changes underlie neuroadaptations to repeated exposure to alcohol in DA neurons, which may play an important role in early stages of addiction.
Materials and methods
Mice and drinking protocol
Female hybrid F1 mice were generated from reciprocal intercrosses of C57BL/6J (B6) × FVB/NJ (FVB) F1 and FVB/NJ × C57BL/6J F1 (maternal strain × paternal strain). No differences were found in ethanol intake using reciprocal crosses of these strains (Blednov et al., 2010). These mice consume high amounts of ethanol and can achieve elevated blood ethanol concentrations, which in our previous studies have been shown to reach up to 114 mg% after 9 hours of ethanol consumption (Blednov et al., 2005). The B6 and FVB breeders were procured from The Jackson Laboratory (Bar Harbor, ME) and mated at 8 weeks of age in the Texas Genetic Animal Core of the INIA (Integrated Neuroscience Initiative on Alcohol) at the University of Texas at Austin. We used a modified 2-bottle choice Drinking in the Dark (DID) paradigm. Mice (2–3 month old) were single-housed and were allowed to voluntarily consume 20% ethanol or water from two separate bottles (bottle positions were alternated daily) for 3 hours everyday, initiated at 3 hours into the dark cycle. Mice were allowed to drink for 20 days and sacrificed 24 hours after the beginning of the last drinking session. Minor increases were observed in body weight (7% increase compared to original weight) during the procedure. Brains were removed and snap frozen in liquid nitrogen. We used N=12 alcohol-drinking mice and N=11 littermate controls. This study is in compliance with animal research guidelines and was approved by the Institutional Review Board of the University of Texas at Austin.
Sectioning and Staining
12 µM thick serial frozen sections were cut through the Ventral Tegmental Area (VTA). Sections were collected at approximately 3.08 mm relative to bregma (Keith B. J Franklin & George Paxinos, mouse brain atlas) mounted on PALM polyethylene naphthalate membrane-coated slides (prod. no. 911724; Carl Zeiss, Microimaging Inc., Thornwood, NY, USA) and stored at −80°C till use. Tissue sections were thawed to room temperature for 15 seconds and were fixed with ice-cold acetone for 2 minutes, air dried and then blocked with normal goat serum (Vectastain Elite ABC Rabbit IgG kit; prod. no. PK-6101; Vector Laboratories, Burlingame, CA, USA) diluted in PBS containing 0.1% Triton X-100 (PBX-TX), for 2 minutes. The sections were subsequently incubated for 10 min with a rabbit anti-TH primary antiserum (Rabbit Polyclonal antibody- Invitrogen, Eugene, OR, USA) diluted in PBS-TX. The tissues were then sequentially incubated, for 3–5 min each, with biotinylated goat anti-rabbit antibodies and ABC reagent (Vectastain Elite ABC Rabbit IgG kit,) diluted in PBS. TH-immunoreactive (-ir) neurons were visualized by incubation with hydrogen peroxide (HRP) and (3,3’-diaminobenzidine (DAB) kit reagents (prod. no. SK-4100; Vector Laboratories) followed by dehydration using increasing concentrations of ethanol (70–100%). All reagents were prepared in diethyl pyrocarbonate-treated water, along with RNAse inhibitor (superase –In, Ambion, Austin, TX).
Laser capture micro-dissection and microarray analysis
TH quick immunolabeling procedure was utilized for identifying the DA neurons. A P.A.L.M. UV-A microlaser (Carl Zeiss Microimaging, Inc., Thornwood, NY, USA) was utilized to dissect and isolate individual TH positive-ir neurons exhibiting a visible intact nucleus and complete labeling of the cytoplasm (Briski et al., 2009). Laser dissected neurons were ejected from the object plane directly into a 0.5 ml P.A.L.M Adhesive Cap (prod. no. 415101-4400-255; Carl Zeiss Microimaging, Inc.). From each brain sample, about 300 TH-ir positive neurons (approximately 100 neurons from each of rostral, middle and caudal VTA) were collected into a single adhesive cap (Fig. 1a, b). After catapulting the neurons, a small portion of intact VTA tissue (whole VTA) was collected from the same sample into another separate adhesive cap for quality control. Total RNA was extracted according to the manufacturer’s protocol (RNAqueous micro kit, Ambion, Austin, TX), however, the volumes of ethanol, wash buffers and total elution buffer were limited to 25µl, 75µl, and 10µl respectively. 1µl of Rnase inhibitor (20units/µl) (superase In, Ambion, Austin, TX) was added to the elution buffer and the total RNA was concentrated by reducing the elution buffer volume to 3 µl using a vacuum centrifuge. Resulting final RNA sample was amplified with TargetAmp two-Round Aminoallyl–aRNA Amplification Kit-3.0 (Epicentre Biotechnologies, Madison, WI) according to the manufacturer’s protocol. Amplified biotinylated aminoallyl RNA was eluted with 40 ul of Rnase free water followed by analyzing the quality and estimating the average size of the product using Agilent Bioanalyzer 2100 (Agilent, Palo Alto, CA). RNA Integrity Number (RIN) values of whole VTA samples were above 6.0. Samples from 11 control and 12 alcohol-treated mice as well as 1 control sample from whole VTA were hybridized to the Illumina® (San Diego, CA, USA) MouseRef-6 BeadChip array and results were imported into BeadStudio (Illumina®) for analysis. The quality of the Illumina bead summary data was assessed using the Bioconductor packages Lumi and arrayQualityMetrics.
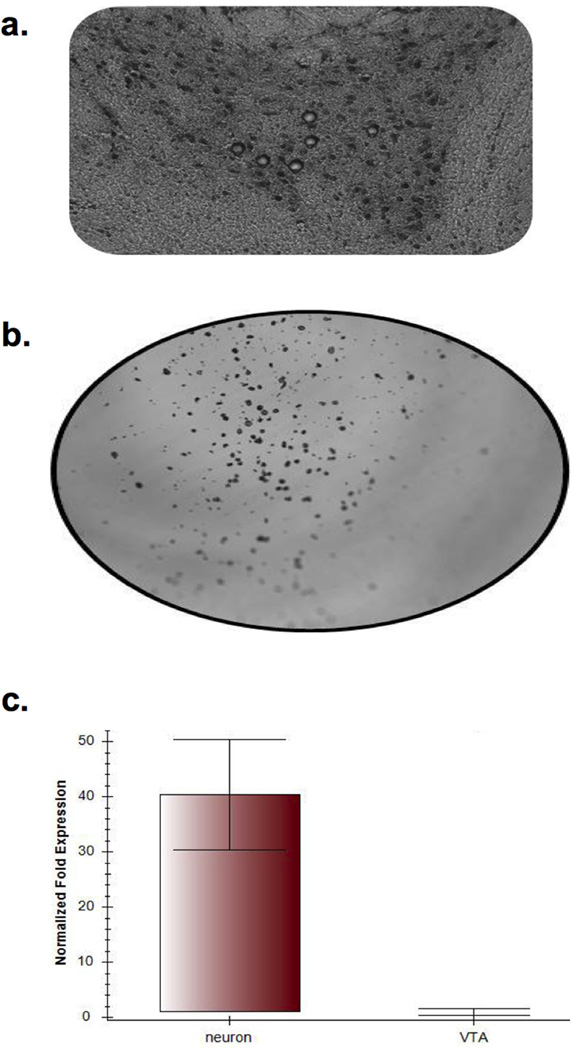
Figure 1. Laser-capture microdissection and validation of tyrosine hydroxylase enrichment in VTA DA neurons.
Frozen 12µm coronal brain sections of the VTA were fixed in acetone and stained with an antibody against tyrosine hydroxylase (TH) to label DA neurons (see methods for details). (a) A section of VTA stained for TH-positive neurons, with several neurons being removed using laser-assisted microdissection. (b). Laser captured neurons are catapulted onto P.A.L.M Adhesive Cap for collection (c). Results of RT-PCR to measure Th abundance using RNA extracted from 300 neurons or whole VTA (n=3 samples per group). A 40-fold enrichment was obtained in captured neurons vs. whole VTA tissue (two-tailed t-test p<0.01).
Statistical analysis
Behavior
Data for ethanol preference was averaged for three periods: days 1–6, 7–12 and 13–20 and compared to 0.5 no preference level using a one-sample t-test. The Bonferroni correction was applied to adjust for multiple tests.
Microarrays
Data preprocessing, including variance stabilization and quantile normalization (for all 24 samples) were carried out using the Lumi package. Transcripts with detection p-value < 0.05 in at least 75% of samples were considered detected. One sample from the alcohol group was excluded as an outlier based on clustering in Lumi and several individual transcript values were excluded based on Grubbs’ test for outlier detection within each group. Alcohol drinking and non-drinking mice were compared and differentially expressed genes (DEGs) at a nominal p<0.05 were determined using the Bioconductor package limma (Smyth, 2005). In addition, we correlated 6 behavioral variables (preference or intake during all 20 days, last 8 days or last 2 days of the procedure) with expression values of each detected gene within the ethanol group (11 subjects) using Pearson’s correlation (nominal p<0.05) and generated 6 lists of genes correlated with either alcohol intake or preference. Numbers of DEGs and correlated genes in each list were compared to the 5% chance level using a Chi square test and the Chi square p-values were adjusted using the Bonferroni correction. False discovery rate (FDR) for each gene list was estimated using the q-value approach (Storey, 2002). Fold enrichment of genes expressed in DA neurons was calculated by averaging the gene expression values for the 22 (11 alcohol and 11 control) neuronal samples divided by values obtained from the whole VTA, and genes with greater than 2-fold difference were considered as DA-enriched. Due to experimental restrictions, we have maximized the number of control and alcohol-treated neuronal samples with only 1 whole VTA sample profiled. Variability of gene expression between cell types is multi-fold greater, than those induced by perturbations within the same cell type and, therefore, a single whole VTA sample should be sufficient to estimate the enrichment in DA neurons.
RT-PCR
We validated TH expression in DA neurons in a separate experiment using RT-PCR following manufacturer’s instructions (3 neuronal samples vs. 3 samples from whole VTA; Th assay: Mm00447557_m1, Gapdh assay: Mm99999915_g1 used as endogenous control, Life technologies, Grand Island, NY). A t-test was used to analyze the data.
Bioinformatics analysis
Two gene lists: DEGs and genes correlated with the last two days of alcohol intake (Alcohol-Correlated Genes; ACGs) were used as input for Ingenuity Pathway Analysis (IPA) (www.ingenuity.com) software for carrying out network and canonical pathway analyses using all detected genes (~10,000) from our experiment as the reference (background) set and default settings for other parameters. In addition, these two gene lists were subjected to an over-representation analysis for biological pathways using Enrichr (Chen et al., BMC Bioinf., 2013; http://amp.pharm.mssm.edu/Enrichr). The Enrichr-based pathway analysis includes several well-curated databases including: KEGG, Wiki pathways, Reactome, Biocarta and Panther). Over-representation (enrichment) p-value for each pathway was calculated based on the Fisher’s exact test. Because Enrichr does not use user-uploaded backgrounds, we compared our results with 3 random lists of 600 to 900 genes from our data set to determine if any of the over-represented functional groups in DEG/ACG lists were detected by chance. Those molecular pathways overlapping between DEG/ACG lists and randomly generated lists were excluded. To nominate candidate genes for future studies we used a systems approach based on convergent validity for each candidate. We considered all genes with nominal p-value < 0.05 and discussed those that are either enriched in dopamine neurons, belong to an over-represented molecular pathway, or play functional roles in neurons in general. This approach combines statistical and biological significance to reach an adequate balance between false positives and false negatives. It may result in higher rates of false positives, compared to strict statistical approaches, but, most importantly, it can contribute to the discovery of novel functional relationships, leading to biological hypotheses.
Results
Genes enriched in dopamine neurons
To validate our approach of profiling individual neuronal populations, we determined DA-enriched genes, i.e., genes with higher expression in laser-captured neuronal samples, compared to the sample from whole VTA tissue. Out of 9,971 transcripts with measurable expression (detection p<0.05), 439 were highly expressed and showed at least 2-fold enrichment in DA neuronal samples (Supplementary File 1). Several dopamine-specific markers were among the DA-enriched genes, including tyrosine hydroxylase (Th) and dopamine transporter (Slc6a3), while markers for GABAergic neurons (Gad1, Gad2) and glia (Mobp, Mbp, Slc1a2, Mag, Mal) were enriched in the whole VTA sample. RT-PCR validation of Th expression showed a ~40 fold enrichment in neuronal samples (Fig. 1c), which, given a ~4 fold microarray-based enrichment of Th, suggests that microarrays provide a rather conservative estimate of changes in highly expressed genes. This may be due to a relatively limited dynamic range of detection and saturation of signal in microarrays compared to RT-PCR.
Ethanol consumption produces global changes in gene expression in DA neurons
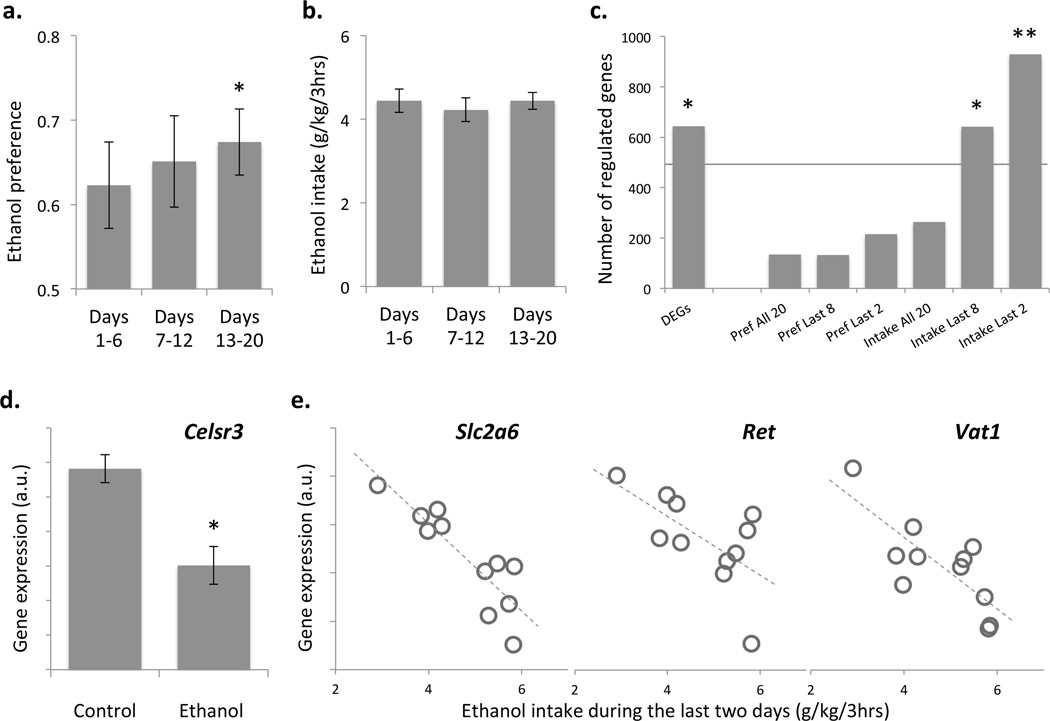
Mice from the alcohol group showed stable alcohol intake and a slight progressive increase in preference for the ethanol solution during the 20-day exposure (Fig. 2a and 2b). Alcohol consumption resulted in moderate changes in global gene expression, as 644 genes (DEGs) were significantly different in expression between the drinking and non-drinking mice (nominal p<0.05). 260 DEGs were down regulated while 384 were up regulated in the alcohol group. Correlation analysis of gene expression and alcohol consumption within the alcohol group showed that consumption during the last 2 days of the procedure was the best predictor of changes in global gene expression (Fig. 2c), as 930 genes (ACGs) were correlated with this phenotype (nominal p<0.05), of which 376 genes correlated negatively, while 554 genes correlated positively (Supplementary File 1). The numbers of DEGs and ACGs were significantly greater than those expected by chance (adjusted χ2 p<0.01), indicating marked effects of moderate doses of ethanol on the transcriptome of DA neurons (Fig. 2c). The greater number of ACGs, compared to DEGs, suggests that differences in alcohol intake during the last 2 days in the alcohol group contribute more to differences in gene expression, than the 20-day history of alcohol exposure. This variability in the alcohol group might have also contributed to fewer statistically significant changes between alcohol and control groups. Several DA-enriched genes are shown as examples to be either differentially expressed between ethanol and control groups (Fig. 2d) or correlated with the last two days of intake (Fig. 2e). In contrast to the general trend of more up-regulation in DEGs and more genes positively correlated with drinking, the majority of ethanol-regulated genes enriched in DA neurons were either down-regulated by ethanol (16 out of 19) or negatively correlated with ethanol consumption (44 out of 44) (Supplemental File 1).
Figure 2. Drinking behavior and global gene expression.
Female B6 × FVB F1 hybrid mice were subjected to a 2-bottle choice DID paradigm (20% ethanol, 3 hours daily, 20 days). Data for alcohol preference (a) and intake (b) were averaged for three periods: days 1–6, 7–12 and 13–20. Asterisk indicates a significant difference from 0.5 no preference level (one sample t-test adjusted p<0.01). (c). Numbers of genes (transcripts) differentially expressed between alcohol and control groups (DEGs) or correlated with either preference (Pref) or intake in the alcohol group averaged during three time periods (all 20 days, last 8 days or last 2 days) of the procedure. Horizontal line at ~500 gene level represents the 5% chance. Numbers of regulated genes in three gene lists were significantly greater than chance (adjusted χ2 p values: * = p<0.01; ** = p<0.0001). (d). DA-enriched differentially expressed gene, cadherin, Celsr3 (* = p<0.01). (e). Three DA-enriched genes significantly correlated with last two days of ethanol intake (glucose transporter, Slc2a6, R=−0.87; proto-oncogene, Ret, R=−0.63; vesicle amine transporter, Vat1, R=−0.78; all p<0.05).
Gene networks and molecular pathways associated with ethanol consumption in DA neurons
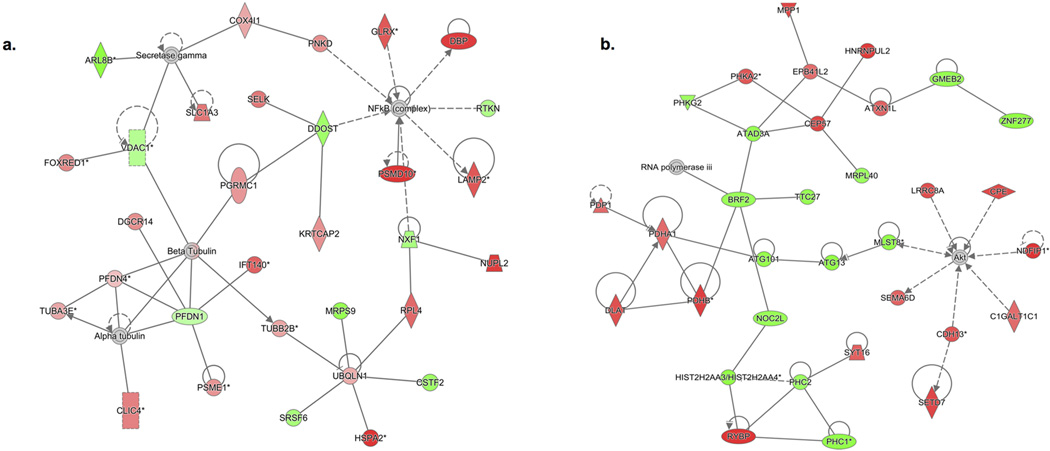
Bioinformatics analysis identified several gene networks and molecular pathways affected by ethanol consumption in DA neurons. Top IPA-based networks were: Neurological/Psychological Disorders and Lipid and Nucleic Acid Metabolism for DEGs and ACGs respectively (Fig. 3). Additional networks are shown in Supplemental Figure 1 (two for each gene list). Noticeably, two of the top six networks centered around the NF-kB signaling complex, a molecular pathway implicated in the neuroimmune system. Top molecular pathways overrepresented for DEGs and ACGs are shown in Tables 1 and 2 respectively and include ethanol degradation, mRNA splicing/processing, TCA cycle and chromatin organization.
Figure 3. Top gene networks identified using Ingenuity Pathway Analysis (IPA).
IPA generates networks based on the Ingenuity Knowledge Base that relies on known biological relationships among genes. Shown are top gene networks for DEGs (a): Neurological Disease, Psychological Disorders and Post Translational Modifications (IPA Score = 51) and ACGs (b): Lipid metabolism, Nucleic acid metabolism, Small Molecular Biochemistry (IPA Score = 51). Red represents molecules that are upregulated in alcohol group or positively correlated with alcohol intake, while green represents those that are downregulated or negatively correlated. Solid lines represent direct interactions while dashed lines represent indirect interactions between molecules.
Table 1. Molecular pathways and functional groups overrepresented in genes differentially expressed between alcohol drinking and non-drinking mice (DEGs).
Functional enrichment analysis was carried out using either IPA or Enrichr (see Methods for detail).
| Molecular Pathway/ Functional Group | Pathway Database |
Genes | P-value |
|---|---|---|---|
| Control of gene expression by vitamin d receptor | BioCarta | Smarce1, Smarcc2, Hdac1, Cops2, Actl6a | 0.00202 |
| Mechanism of protein import into the nucleus | BioCarta | Nutf2, Kpna2, Rangap1 | 0.00857 |
| Fc Epsilon RI Signaling | IPA | Gab1, Inpp5f, Map2k3, Mapk12, Nras, Ocrl, Ptpn11, Rac3, Raf1, Vav2 | 0.00130 |
| Glutathione Redox Reactions II | IPA | Glrx, Txndc12 | 0.00529 |
| Ethanol degradation | IPA | Acss1, Akr1a1, Aldh2, Hsd17b10 | 0.00725 |
| Butanoate metabolism | KEGG | Hmgcl, Echs1, Aldh2, Ilvbl, Ddhd1, Hsd17b10 | 0.00141 |
| Glycolysis and gluconeogenesis | KEGG | Ldha, Tpi1, Aldh2, Akr1a1, Aldoc, Dlat, Acss1 | 0.00149 |
| Propanoate metabolism | KEGG | Ldha, Echs1, Aldh2, Acadm, Acss1 | 0.00216 |
| Valine leucine and isoleucine degradation | KEGG | Hmgcl, Echs1, Aldh2, Acadm, Hsd17b10 | 0.00662 |
| Caprolactam degradation | KEGG | Echs1, Akr1a1, Hsd17b10 | 0.00767 |
| RNA Polymerase II Transcription | Reactome | Ercc3, Taf13, Cstf2, Snrpg, Gtf2h1, Srsf5, Taf9b, Srsf6, Polr2l, Nelfe | 0.00171 |
| Global Genomic NER (GG-NER) | Reactome | Ddb1, Lig1, Ercc3, Rpa1, Gtf2h1 | 0.00513 |
| Chromatin organization | Reactome | Smarce1, Smarcc2, Kdm2b, Hdac10, Hdac1, Kmt2c, Actl6a, Gps2, Elp2, Coprs, Kansl3, Hat1, Epc1, Kdm7a | 0.00520 |
| Processing of Capped Intron-Containing Pre-mRNA | Reactome | Nxf1, Sf3b2, Cstf2, Snrpg, Plrg1, Srsf5, Nupl2, Txnl4a, Prpf19, Srsf6, Polr2l | 0.00541 |
| Nucleotide Excision Repair | Reactome | Ddb1, Lig1, Ercc3, Rpa1, Gtf2h1, Polr2l | 0.00547 |
| mRNA Splicing | Reactome | Sf3b2, Cstf2, Snrpg, Plrg1, Srsf5, Txnl4a, Prpf19, Srsf6, Polr2l | 0.00900 |
| Mismatch repair | WikiPathways | Msh6, Lig1, Rpa1 | 0.00394 |
| Eukaryotic Transcription Initiation | WikiPathways | Ercc3, Taf13, Polr1a, Ilk, Gtf2h1 | 0.00803 |
P-values (P<0.01) are based on Fisher's exact test.
Table 2. Molecular pathways and functional groups overrepresented in genes correlated with alcohol intake during the last 2 days of the procedure (ACGs).
Functional enrichment analysis was carried out using either IPA or Enrichr (see Methods for detail).
| Molecular Pathway/ Functional Group | Pathway Database |
Genes | P-value |
|---|---|---|---|
| Phagosome maturation | IPA | Atp6v0a2, Atp6v0d1, Atp6v1b2, Atp6v1c1, Atp6v1h, Dync1i2, Nsf, Pik3c3, Rab5a, Rab7a, Tuba3e, Tubb, Vps37b, Vti1b, Ykt6 | 0.00436 |
| mRNA Splicing | Reactome | Ncbp1, Cstf3, Srsf1, Prpf19, Hnrnpm, Snrpd2, Hnrnph1, Pcbp2, Polr2d, Dhx38, Hnrnpc, Srsf7, Snrpa | 0.00293 |
| Processing of Capped Intron-Containing Pre-mRNA | Reactome | Ncbp1, Nup210, Cstf3, Srsf1, Prpf19, Hnrnpm, Snrpd2, Hnrnph1, Pcbp2, Polr2d, Dhx38, Hnrnpc, Rae1, Srsf7, Snrpa | 0.00346 |
| Regulation of pyruvate dehydrogenase (PDH) complex | Reactome | Pdp1, Pdha1, Dlat, Pdhb | 0.00642 |
| Insulin receptor recycling | Reactome | Atp6v1b2, Atp6v0a2, Atp6v1h, Atp6v0d1, Atp6v1c1 | 0.00974 |
| TCA Cycle | WikiPathways | Pdp1, Pdha1, Mdh1, Ogdh, Dlst, Dlat, Pdhb | 0.00041 |
P-values (P<0.01) are based on Fisher's exact test.
Discussion
Dopaminergic neurons in the VTA play a critical role in mediating the rewarding and reinforcing effects of several drugs of abuse including ethanol (Nimitvilai et al., 2013; Ishikawa et al., 2013; Brodie et al., 1999, Koob et al., 1998). Repeated ethanol produces changes in DA neurons that may contribute to increased alcohol consumption and the development of alcohol dependence; however the molecular mechanisms of this neuroadaptation are not well understood. Here, we used a binge model of alcohol consumption and measured alcohol-induced changes in gene expression in DA neurons. We specifically targeted the 24 hr. time point, when mice anticipate the delivery of ethanol, thus enhancing the potential motivational value of the drug. We identified multiple genes that are either differentially expressed between the alcohol and control groups (DEGs) or correlated with ethanol consumption within the alcohol group (ACGs). Some of these changes may underlie initial neuroadaptation to alcohol that may play an important role in the transition to addiction. To nominate candidate genes for mechanistic studies, we used a systems approach that combines moderate statistical significance with relevant functional significance for each candidate. The following discussion is based on alcohol-related genes from three prioritized categories: a) genes with higher expression in DA neurons (DA-enriched), b) genes with known functions in neuronal physiology and c) genes as parts of overrepresented molecular pathways.
One surprising finding was that the majority of DA-enriched DEGs were down-regulated by alcohol and all DA-enriched ACGs were negatively correlated with alcohol consumption. Genes with high cell type – specific expression are thought to be important for cellular functions and we hypothesize that the alcohol-induced down-regulation of DA-enriched genes is associated with a decrease in activity of DA neuron and subsequent release of dopamine in target areas. These changes induced by repeated ethanol may represent a form of tolerance to the activating effects of initial administration of the drug. Low dopamine levels in the nucleus accumbens have been associated with increased alcohol preference and intake in drinking models of mice (George et al., 1995) and rats (McBride et al., 1995) and the increased consumption of alcohol continues until dopamine levels reach those of control animals (Weiss et al., 1996). Therefore, alcohol-induced molecular neuroadaptations may reinforce the addiction cycle by increasing the need to drink in order to normalize dopamine levels. DA-enriched genes that were both down-regulated by alcohol and negatively correlated with drinking included a cadherin, Celsr3 and a member of ATP–binding cassette subfamily, Abca7. Mice lacking Celsr3 show marked deficits in cortical and sub cortical connections particularly in the anterior commissure, internal capsule and corticospinal tract (Zhou et al., 2008) and Celsr3 mRNA is altered in rats as a result of binge alcohol consumption (Himes et al., 2008), while Abca7 has been implicated in psychiatric disorders and shown to cause memory deficits in knockout mice (Logge et al., 2012). A gene coding for another ATP-binding protein, Abca3, was among DA-enriched ACGs and was previously shown to be down-regulated in the amygdala of human alcoholics (Ponomarev et al., 2012). Chronic ethanol administration in mice can increase ATPase activity in brain and liver (Israel and Kuriyama, 1971); and decrease ATP concentrations in the brain (Rawat and Kuriyama 1972), which may alter levels of ATP binding proteins. Another ACG, Ret is a receptor tyrosine kinase highly expressed in VTA DA neurons (Trupp et al., 1997, Pascual et al., 2011) and an important mediator of behavioral adaptations to several drugs of abuse (Messer et al., 2000).
Several genes previously implicated in neuronal functions and mechanisms of drugs of abuse were also regulated in DA neurons after alcohol. These included cholinergic (Chrna3, Chrnb3), GABA-ergic (Gabrg2) and glutamatergic (Grik2) ionotropic receptor subunits, calcium (Cacna1g, Cacng2), potassium (Kcnk1, Kcnb1), chloride (Clic4) and sodium (Scn3b, Scn2a1) ion channel subunits and other genes coding for synaptic proteins (Vat1, Stx3, Stx5a, Ncam1, Cartpt, Crhr1, Gphn). For example, Cartpt is the precursor of cocaine- and amphetamine-regulated transcript protein, which is involved in mediating the behavioral effects of several drugs of abuse (Hurd et al., 1999; Rogge et al., 2008; Salinas et al., 2006) and is associated with alcoholism in a Korean population (Jung et al., 2004). The gamma 2 subunit of gamma-aminobutyric acid (GABA) A receptor is required for clustering of GABAA receptors and is down-regulated in postmortem brain from alcohol and cocaine addicts, compared to control cases (Enoch et al., 2012). The nicotinic acetylcholine receptor CHRNA5, A3, B4 gene cluster has been show to harbor polymorphisms that are predictive of age of initiation of tobacco use (Schlaepfer et al., 2008), while rare missense variants in CHRNA3 and CHRNB3 have been implicated in alcohol and cocaine abuse in a recent Collaborative Study on the Genetics of Alcoholism (COGA) study (Haller et al., 2014). In addition, chloride intracellular channel 4, Clic4, has been implicated in behavioral responses to alcohol in multiple species (Bhandari et al., 2012). Taken together, these molecular changes may provide a mechanistic foundation for ethanol-induced neuroadaptations in DA neurons.
Functional group analysis revealed several gene networks and molecular pathways overrepresented in the DEG and ACG lists. All genes in “ethanol degradation” and “glutathione redox reactions” categories were up-regulated in the alcohol group, implicating activation of detoxification pathways in response to the ethanol challenge in DA neurons. Aldehyde dehydrogenase 2 (Aldh2), one of the upregulated genes, is a mitochondrial enzyme involved in ethanol metabolism. The ALDH2*2 allele that encodes an inactive form of the enzyme is associated with low risk for alcoholism in several East Asian populations including Japanese (Higuchi, 1994), Koreans (Shen et al., 1997) and Han-Chinese (Chen et al., 1996). Interestingly, multiple lines of evidence suggest that brain-generated ethanol metabolites play an important role in the early development of alcohol reinforcement (Israel et al., 2015). Majority of genes in the TCA or Krebs cycle pathway were positively correlated with levels of alcohol intake, providing additional support for the involvement of energy metabolism in DA neurons in the regulation of alcohol consumption. Alcohol-induced neuroimmune response is proposed to be a critical factor in alcohol addiction (Crews and Vetreno, 2011; Osterndorff-Kahanek et al., 2013) and we detected several alcohol-responsive biological categories related to the immune function, including the nuclear factor kappa-light-chain enhancer of activated B cells (NF-kB) pathway. Chronic alcohol consumption can activate the NF-kB pathway and increase production of proinflammatory cytokines and chemokines that, coupled with decreased CREB signaling can cause hyperexcitability and neuronal damage (Vetreno and Crews, 2014).
“Chromatin organization” was one of the overrepresented functional groups among DEGs. There is growing evidence that chromatin (epigenetic) modifications play an important role in mediating the actions of drugs of abuse (for review, see Nestler, 2014; Andrzejewski et al., 2013) and alcohol can affect gene expression and downstream behavior via changes in chromatin landscape (Ponomarev et al., 2012; Farris et al., 2015; for review see Ponomarev, 2013; Zhou et al., 2014; Kyzar and Pandey, 2015). Alcohol consumption affected several enzymes involved in histone modifications, such as acetylation (Hat1), deacetylation (Hdac1, Hdac10), lysine-specific methylation (Kmt2c) and demethylation (Kdm2b, Kdm7a), as well as ATP-dependent chromatin remodeling (Smarce1, Smarcc2). A growing body of evidence suggests that targeting epigenetic modifications such as histone acetylation can result in alteration of drinking behavior. The histone deacetylase (HDAC) inhibitors Valproic acid and MS-275 have been shown to inhibit excessive alcohol intake in dependent rats (Simon O’Brien et al., 2015), while the HDAC inhibitor Trichostatin A (TSA) is capable of decreasing anxiety-like behavior and alcohol intake in alcohol preferring rats which is accompanied by a corresponding decrease in HDAC2 protein levels and increase in global histone acetylation in the amygdala (Sakharkar et al., 2014). In addition, methyl CpG binding protein 2, Mecp2, was positively correlated with drinking in our study. Repeated exposure to psychostimulants has been shown to increase levels of Mecp2 in the rat brain (Cassel et al., 2006) and Mecp2 phosphorylation (Deng et al., 2014), causing changes in gene expression and selectively modulating region-specific changes in plasticity accompanying drug exposure. DNA methylation and histone modifications may result in long-lasting changes in gene expression and we hypothesize that alcohol-induced changes in expression of genes involved in regulation of chromatin states can contribute to long-term neuroadaptations via epigenetic changes.
Several previous studies investigated the effects of alcohol consumption on gene expression in whole tissue VTA (Mulligan et al., 2011; McBride et al., 2013; Flatscher-Bader et al., 2008). To our knowledge, our study is the first to identify individual genes and molecular pathways regulated by and correlated with high alcohol intake in LCM-isolated DA neurons. Identification of cell type – specific alcohol-sensitive molecular targets is potentially very important because of the possibility to specifically affect activity of individual neurons and neuronal circuits, which may prove to be crucial for the development of new medications for drug addiction. For example, given the known role of VTA DA projections to the nucleus accumbens in drug reinforcement, possible pharmacotherapeutic agents acting on specific neuronal populations have the potential to decrease the motivational value of the drug or to inhibit conditioned responses to stimuli predicting drug availability. The ultimate goal of this research is to determine roles of individual neuronal populations in alcohol actions and to identify cell type - specific alcohol-sensitive genes and gene products as potential therapeutic targets for alcoholism.
Supplementary Material
Shown are top 2nd and 3rd networks for DEGs (a,b) and ACGs (c,d) (a). Cellular Assembly and Organization, Post-Translational Modification, Cancer (IPA Score = 51) (b). Cellular Assembly and Organization, Embryonic Development, Organismal Development (IPA Score = 43) (c). Connective Tissue Disorders, Dermatological Diseases and Conditions, Developmental Disorder (IPA Score = 41), d. Developmental Disorder, Hereditary Disorder, Metabolic Disease (IPA Score = 40). Red represents molecules that are upregulated in alcohol group or positively correlated with alcohol intake, while green represents those that are downregulated or negatively correlated. Solid lines represent direct interactions while dashed lines represent indirect interactions between molecules.
Highlighted in yellow are the genes that were 2 fold or more enriched in DA neurons vs. whole VTA tissue. Genes highlighted in bold represent an overlap between DEGs and ACGs.
Acknowledgments
This work was supported by NIH NIAAA grants AA017234 to IP, AA017838, AA013520 and AA020683 to RAH. The authors thank Elizabeth Osterndorff-Kahanek for technical assistance with RT-PCR.
Footnotes
The authors declare no conflict of interest.
References
- 1.Andrzejewski ME, McKee BL, Baldwin AE, Burns L, Hernandez P. The clinical relevance of neuroplasticity in corticostriatal networks during operant learning. Neurosci Biobehav Rev. 2013;37:2071–2080. doi: 10.1016/j.neubiorev.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernier BE, Whitaker LR, Morikawa H. Previous ethanol experience enhances synaptic plasticity of NMDA receptors in the ventral tegmental area. J Neurosci. 2011;31:5205–5212. doi: 10.1523/JNEUROSCI.5282-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhandari P, Hill JS, Farris SP, Costin B, Martin I, Chan CL, Alaimo JT, Bettinger JC, Davies AG, Miles MF, Grotewiel M. Chloride intracellular channels modulate acute ethanol behaviors in Drosophila, Caenorhabditis elegans and mice. Genes Brain Behav. 2012;11:387–397. doi: 10.1111/j.1601-183X.2012.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blednov YA, Metten P, Finn DA, Rhodes JS, Bergeson SE, Harris RA, Crabbe JC. Hybrid C57BL/6J × FVB/NJ mice drink more alcohol than do C57BL/6J mice. Alcohol Clin Exp Res. 2005;29:1949–1958. doi: 10.1097/01.alc.0000187605.91468.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blednov YA, Ozburn AR, Walker D, Ahmed S, Belknap JK, Harris RA. Hybrid mice as genetic models of high alcohol consumption. Behavior genetics. 2010;40:93–110. doi: 10.1007/s10519-009-9298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briski KP, Cherian AK, Genabai NK, Vavaiya KV. In situ coexpression of glucose and monocarboxylate transporter mRNAs in metabolic-sensitive caudal dorsal vagal complex catecholaminergic neurons: transcriptional reactivity to insulin-induced hypoglycemia and caudal hindbrain glucose or lactate repletion during insulin-induced hypoglycemia. Neuroscience. 2009;164:1152–1160. doi: 10.1016/j.neuroscience.2009.08.074. [DOI] [PubMed] [Google Scholar]
- 7.Brodie MS, Pesold C, Appel SB. Ethanol directly excites dopaminergic ventral tegmental area reward neurons. Alcohol Clin Exp Res. 1999;23:1848–1852. [PubMed] [Google Scholar]
- 8.Cassel S, Carouge D, Gensburger C, Anglard P, Burgun C, Dietrich JB, Aunis D, Zwiller J. Fluoxetine and cocaine induce the epigenetic factors MeCP2 and MBD1 in adult rat brain. Mol Pharmacol. 2006;70:487–492. doi: 10.1124/mol.106.022301. [DOI] [PubMed] [Google Scholar]
- 9.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen WJ, Loh EW, Hsu YP, Chen CC, Yu JM, Cheng AT. Alcohol-metabolising genes and alcoholism among Taiwanese Han men: independent effect of ADH2, ADH3 and ALDH2. Br J Psychiatry. 1996;168:762–767. doi: 10.1192/bjp.168.6.762. [DOI] [PubMed] [Google Scholar]
- 11.Crews FT, Vetreno RP. Addiction, adolescence, and innate immune gene induction. Front Psychiatry. 2011;2:19. doi: 10.3389/fpsyt.2011.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deehan GA, Jr, Hauser SR, Wilden JA, Truitt WA, Rodd ZA. Elucidating the biological basis for the reinforcing actions of alcohol in the mesolimbic dopamine system: the role of active metabolites of alcohol. Front Behav Neurosci. 2013;7:104. doi: 10.3389/fnbeh.2013.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng JV, Wan Y, Wang X, Cohen S, Wetsel WC, Greenberg ME, Kenny PJ, Calakos N, West AE. MeCP2 phosphorylation limits psychostimulant-induced behavioral and neuronal plasticity. J Neurosci. 2014;34:4519–4527. doi: 10.1523/JNEUROSCI.2821-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enoch MA, Zhou Z, Kimura M, Mash DC, Yuan Q, Goldman D. GABAergic gene expression in postmortem hippocampus from alcoholics and cocaine addicts; corresponding findings in alcohol-naive P and NP rats. PLoS One. 2012;7:e29369. doi: 10.1371/journal.pone.0029369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farris SP, Harris RA, Ponomarev I. Epigenetic modulation of brain gene networks for cocaine and alcohol abuse. Front Neurosci. 2015;9:176. doi: 10.3389/fnins.2015.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flatscher-Bader T, Zuvela N, Landis N, Wilce PA. Smoking and alcoholism target genes associated with plasticity and glutamate transmission in the human ventral tegmental area. Hum Mol Genet. 2008;17:38–51. doi: 10.1093/hmg/ddm283. [DOI] [PubMed] [Google Scholar]
- 17.George SR, Fan T, Ng GY, Jung SY, O'Dowd BF, Naranjo CA. Low endogenous dopamine function in brain predisposes to high alcohol preference and consumption: reversal by increasing synaptic dopamine. J Pharmacol Exp Ther. 1995;273:373–379. [PubMed] [Google Scholar]
- 18.Haller G, Kapoor M, Budde J, Xuei X, Edenberg H, Nurnberger J, Kramer J, Brooks A, Tischfield J, Almasy L, Agrawal A, Bucholz K, Rice J, Saccone N, Bierut L, Goate A. Rare missense variants in CHRNB3 and CHRNA3 are associated with risk of alcohol and cocaine dependence. Hum Mol Genet. 2014;23:810–819. doi: 10.1093/hmg/ddt463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higuchi S. Polymorphisms of ethanol metabolizing enzyme genes and alcoholism. Alcohol Alcohol Suppl. 1994;2:29–34. [PubMed] [Google Scholar]
- 20.Himes R, Wezeman FH, Callaci JJ. Identification of novel bone-specific molecular targets of binge alcohol and ibandronate by transcriptome analysis. Alcohol Clin Exp Res. 2008;32:1167–1180. doi: 10.1111/j.1530-0277.2008.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman PL, Tabakoff B. Alcohol dependence: a commentary on mechanisms. Alcohol Alcohol. 1996;31:333–340. doi: 10.1093/oxfordjournals.alcalc.a008159. [DOI] [PubMed] [Google Scholar]
- 22.Hopf FW, Martin M, Chen BT, Bowers MS, Mohamedi MM, Bonci A. Withdrawal from intermittent ethanol exposure increases probability of burst firing in VTA neurons in vitro. J Neurophysiol. 2007;98:2297–2310. doi: 10.1152/jn.00824.2007. [DOI] [PubMed] [Google Scholar]
- 23.Hurd YL, Svensson P, Ponten M. The role of dopamine, dynorphin, and CART systems in the ventral striatum and amygdala in cocaine abuse. Ann N Y Acad Sci. 1999;877:499–506. doi: 10.1111/j.1749-6632.1999.tb09285.x. [DOI] [PubMed] [Google Scholar]
- 24.Hwa LS, Debold JF, Miczek KA. Alcohol in excess: CRF(1) receptors in the rat and mouse VTA and DRN. Psychopharmacology (Berl) 2013;225:313–327. doi: 10.1007/s00213-012-2820-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishikawa M, Otaka M, Huang YH, Neumann PA, Winters BD, Grace AA, Schluter OM, Dong Y. Dopamine triggers heterosynaptic plasticity. J Neurosci. 2013;33:6759–6765. doi: 10.1523/JNEUROSCI.4694-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Israel MA, Kuriyama K. Effect of in vivo ethanol administration on adenosinetriphosphatase activity of subcellular fractions of mouse brain and liver. Life Sci II. 1971;10:591–599. doi: 10.1016/0024-3205(71)90196-2. [DOI] [PubMed] [Google Scholar]
- 27.Israel Y, Quintanilla ME, Karahanian E, Rivera-Meza M, Herrera-Marschitz M. The “first hit” toward alcohol reinforcement: role of ethanol metabolites. Alcohol Clin Exp Res. 2015;39:776–786. doi: 10.1111/acer.12709. [DOI] [PubMed] [Google Scholar]
- 28.Jung SK, Hong MS, Suh GJ, Jin SY, Lee HJ, Kim BS, Lim YJ, Kim MK, Park HK, Chung JH, Yim SV. Association between polymorphism in intron 1 of cocaine- and amphetamine-regulated transcript gene with alcoholism, but not with bipolar disorder and schizophrenia in Korean population. Neurosci Lett. 2004;365:54–57. doi: 10.1016/j.neulet.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 29.Koob GF, Roberts AJ, Schulteis G, Parsons LH, Heyser CJ, Hyytia P, Merlo-Pich E, Weiss F. Neurocircuitry targets in ethanol reward and dependence. Alcohol Clin Exp Res. 1998;22:3–9. [PubMed] [Google Scholar]
- 30.Kyzar EJ, Pandey SC. Molecular mechanisms of synaptic remodeling in alcoholism. Neurosci Lett. 2015 doi: 10.1016/j.neulet.2015.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Logge W, Cheng D, Chesworth R, Bhatia S, Garner B, Kim WS, Karl T. Role of Abca7 in mouse behaviours relevant to neurodegenerative diseases. PLoS One. 2012;7:e45959. doi: 10.1371/journal.pone.0045959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McBride WJ, Bodart B, Lumeng L, Li TK. Association between low contents of dopamine and serotonin in the nucleus accumbens and high alcohol preference. Alcohol Clin Exp Res. 1995;19:1420–1422. doi: 10.1111/j.1530-0277.1995.tb01001.x. [DOI] [PubMed] [Google Scholar]
- 33.McBride WJ, Kimpel MW, McClintick JN, Ding ZM, Hauser SR, Edenberg HJ, Bell RL, Rodd ZA. Changes in gene expression within the ventral tegmental area following repeated excessive binge-like alcohol drinking by alcohol-preferring (P) rats. Alcohol. 2013;47:367–380. doi: 10.1016/j.alcohol.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Messer CJ, Eisch AJ, Carlezon WA, Jr, Whisler K, Shen L, Wolf DH, Westphal H, Collins F, Russell DS, Nestler EJ. Role for GDNF in biochemical and behavioral adaptations to drugs of abuse. Neuron. 2000;26:247–257. doi: 10.1016/s0896-6273(00)81154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morikawa H, Morrisett RA. Ethanol action on dopaminergic neurons in the ventral tegmental area: interaction with intrinsic ion channels and neurotransmitter inputs. Int Rev Neurobiol. 2010;91:235–288. doi: 10.1016/S0074-7742(10)91008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mulholland PJ, Hopf FW, Bukiya AN, Martin GE, Liu J, Dopico AM, Bonci A, Treistman SN, Chandler LJ. Sizing up ethanol-induced plasticity: the role of small and large conductance calcium-activated potassium channels. Alcohol Clin Exp Res. 2009;33:1125–1135. doi: 10.1111/j.1530-0277.2009.00936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov YA, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci U S A. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulligan MK, Rhodes JS, Crabbe JC, Mayfield RD, Harris RA, Ponomarev I. Molecular profiles of drinking alcohol to intoxication in C57BL/6J mice. Alcohol Clin Exp Res. 2011;35:659–670. doi: 10.1111/j.1530-0277.2010.01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology. 2014;76 Pt B:259–268. doi: 10.1016/j.neuropharm.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nimitvilai S, Arora DS, You C, McElvain M, Brodie MS. Phorbol ester reduces ethanol excitation of dopaminergic neurons of the ventral tegmental area: involvement of protein kinase C theta. Front Integr Neurosci. 2013;7:96. doi: 10.3389/fnint.2013.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Osterndorff-Kahanek E, Ponomarev I, Blednov YA, Harris RA. Gene expression in brain and liver produced by three different regimens of alcohol consumption in mice: comparison with immune activation. PLoS One. 2013;8:e59870. doi: 10.1371/journal.pone.0059870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pascual A, Hidalgo-Figueroa M, Gomez-Diaz R, Lopez-Barneo J. GDNF and protection of adult central catecholaminergic neurons. J Mol Endocrinol. 2011;46:R83–R92. doi: 10.1530/JME-10-0125. [DOI] [PubMed] [Google Scholar]
- 43.Pignataro L, Varodayan FP, Tannenholz LE, Harrison NL. The regulation of neuronal gene expression by alcohol. Pharmacol Ther. 2009;124:324–335. doi: 10.1016/j.pharmthera.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ponomarev I. Epigenetic control of gene expression in the alcoholic brain. Alcohol Res. 2013;35:69–76. doi: 10.35946/arcr.v35.1.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–1897. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rawat AK, Kuriyama K. Ethanol oxidation: effect on the redox state of brain in mouse. Science. 1972;176:1133–1135. doi: 10.1126/science.176.4039.1133. [DOI] [PubMed] [Google Scholar]
- 47.Repunte-Canonigo V, Lutjens R, van der Stap LD, Sanna PP. Increased expression of protein kinase A inhibitor alpha (PKI-alpha) and decreased PKA-regulated genes in chronic intermittent alcohol exposure. Brain Res. 2007;1138:48–56. doi: 10.1016/j.brainres.2006.09.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robinson DL, Howard EC, McConnell S, Gonzales RA, Wightman RM. Disparity Between Tonic and Phasic Ethanol-Induced Dopamine Increases in the Nucleus Accumbens of Rats. Alcoholism-Clinical and Experimental Research. 2009;33:1187–1196. doi: 10.1111/j.1530-0277.2009.00942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogge G, Jones D, Hubert GW, Lin Y, Kuhar MJ. CART peptides: regulators of body weight, reward and other functions. Nat Rev Neurosci. 2008;9:747–758. doi: 10.1038/nrn2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakharkar AJ, Zhang H, Tang L, Baxstrom K, Shi G, Moonat S, Pandey SC. Effects of histone deacetylase inhibitors on amygdaloid histone acetylation and neuropeptide Y expression: a role in anxiety-like and alcohol-drinking behaviours. Int J Neuropsychopharmacol. 2014;17:1207–1220. doi: 10.1017/S1461145714000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salinas A, Wilde JD, Maldve RE. Ethanol enhancement of cocaine- and amphetamine-regulated transcript mRNA and peptide expression in the nucleus accumbens. J Neurochem. 2006;97:408–415. doi: 10.1111/j.1471-4159.2006.03745.x. [DOI] [PubMed] [Google Scholar]
- 52.Schlaepfer IR, Hoft NR, Collins AC, Corley RP, Hewitt JK, Hopfer CJ, Lessem JM, McQueen MB, Rhee SH, Ehringer MA. The CHRNA5/A3/B4 gene cluster variability as an important determinant of early alcohol and tobacco initiation in young adults. Biol Psychiatry. 2008;63:1039–1046. doi: 10.1016/j.biopsych.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sesack SR, Grace AA. Cortico-Basal Ganglia reward network: microcircuitry. Neuropsychopharmacology. 2010;35:27–47. doi: 10.1038/npp.2009.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen YC, Fan JH, Edenberg HJ, Li TK, Cui YH, Wang YF, Tian CH, Zhou CF, Zhou RL, Wang J, Zhao ZL, Xia GY. Polymorphism of ADH and ALDH genes among four ethnic groups in China and effects upon the risk for alcoholism. Alcohol Clin Exp Res. 1997;21:1272–1277. [PubMed] [Google Scholar]
- 55.Simon-O'Brien E, Alaux-Cantin S, Warnault V, Buttolo R, Naassila M, Vilpoux C. The histone deacetylase inhibitor sodium butyrate decreases excessive ethanol intake in dependent animals. Addict Biol. 2015;20:676–689. doi: 10.1111/adb.12161. [DOI] [PubMed] [Google Scholar]
- 56.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 57.Storey JD, et al. A direct approach to false discovery rates: Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2002;64(3):479–498. [Google Scholar]
- 58.Theile JW, Morikawa H, Gonzales RA, Morrisett RA. Ethanol enhances GABAergic transmission onto dopamine neurons in the ventral tegmental area of the rat. Alcohol Clin Exp Res. 2008;32:1040–1048. doi: 10.1111/j.1530-0277.2008.00665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trupp M, Belluardo N, Funakoshi H, Ibanez CF. Complementary and overlapping expression of glial cell line-derived neurotrophic factor (GDNF), c-ret proto-oncogene, and GDNF receptor-alpha indicates multiple mechanisms of trophic actions in the adult rat CNS. J Neurosci. 1997;17:3554–3567. doi: 10.1523/JNEUROSCI.17-10-03554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vetreno RP, Crews FT. Current hypotheses on the mechanisms of alcoholism. Handb Clin Neurol. 2014;125:477–497. doi: 10.1016/B978-0-444-62619-6.00027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang KS, Liu X, Zhang Q, Pan Y, Aragam N, Zeng M. A meta-analysis of two genome-wide association studies identifies 3 new loci for alcohol dependence. J Psychiatr Res. 2011;45:1419–1425. doi: 10.1016/j.jpsychires.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 62.Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 63.Weiss F, Parsons LH, Schulteis G, Hyytia P, Lorang MT, Bloom FE, Koob GF. Ethanol self-administration restores withdrawal-associated deficiencies in accumbal dopamine and 5-hydroxytryptamine release in dependent rats. J Neurosci. 1996;16:3474–3485. doi: 10.1523/JNEUROSCI.16-10-03474.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou L, Bar I, Achouri Y, Campbell K, De Backer O, Hebert JM, Jones K, Kessaris N, de Rouvroit CL, O'Leary D, Richardson WD, Goffinet AM, Tissir F. Early forebrain wiring: genetic dissection using conditional Celsr3 mutant mice. Science. 2008;320:946–949. doi: 10.1126/science.1155244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhou Z, Enoch MA, Goldman D. Gene expression in the addicted brain. Int Rev Neurobiol. 2014;116:251–273. doi: 10.1016/B978-0-12-801105-8.00010-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Shown are top 2nd and 3rd networks for DEGs (a,b) and ACGs (c,d) (a). Cellular Assembly and Organization, Post-Translational Modification, Cancer (IPA Score = 51) (b). Cellular Assembly and Organization, Embryonic Development, Organismal Development (IPA Score = 43) (c). Connective Tissue Disorders, Dermatological Diseases and Conditions, Developmental Disorder (IPA Score = 41), d. Developmental Disorder, Hereditary Disorder, Metabolic Disease (IPA Score = 40). Red represents molecules that are upregulated in alcohol group or positively correlated with alcohol intake, while green represents those that are downregulated or negatively correlated. Solid lines represent direct interactions while dashed lines represent indirect interactions between molecules.
Highlighted in yellow are the genes that were 2 fold or more enriched in DA neurons vs. whole VTA tissue. Genes highlighted in bold represent an overlap between DEGs and ACGs.