Abstract
Lamins are key structural components of the nuclear lamina, an intermediate filament meshwork that lies beneath the inner nuclear membrane. Lamins play a role in nuclear architecture, DNA replication, and gene expression. Mutations affecting A-type lamins have been associated with a variety of human diseases, including muscular dystrophy, cardiomyopathy, lipodystrophy, and progeria, but mutations in B-type lamins have never been identified in humans or in experimental animals. To investigate the in vivo function of lamin B1, the major B-type lamin, we generated mice with an insertional mutation in Lmnb1. The mutation resulted in the synthesis of a mutant lamin B1 protein lacking several key functional domains, including a portion of the rod domain, the nuclear localization signal, and the CAAX motif (the carboxyl-terminal signal for farnesylation). Homozygous Lmnb1 mutant mice survived embryonic development but died at birth with defects in lung and bone. Fibroblasts from mutant embryos grew under standard cell-culture conditions but displayed grossly misshapen nuclei, impaired differentiation, increased polyploidy, and premature senescence. Thus, the lamin B1 mutant mice provide evidence for a broad and nonredundant function of lamin B1 in mammalian development. These mutant mice and cell lines derived from them will be useful models for studying the role of the nuclear lamina in various cellular processes.
Keywords: nuclear envelope, lamins, knockout mice, gene trapping
The nuclear lamina, a protein meshwork that lines the inner nuclear membrane, is critical in fundamental cellular processes, including nuclear organization, chromatin segregation, DNA replication, and gene transcription (1–5). The principal protein components of the lamina are lamins, which are members of the intermediate filament protein family. Like other intermediate filament proteins, lamins possess an amino-terminal head domain and a highly conserved central α-rod domain for polymerization and oligomerization (6). Lamins are, however, distinguished from other intermediate filament proteins by a nuclear localization motif. In addition, prelamin A and lamins B1 and B2 contain a carboxyl-terminal CAAX motif that triggers a series of posttranslational modifications (farnesylation, endoproteolytic trimming of the last three amino acid residues, and methylation of the newly exposed farnesylcysteine) (6). Aside from their structural role in the formation of the nuclear lamina, lamins A and C are found in the nucleoplasm adjacent to sites of DNA synthesis and RNA processing, suggesting that these proteins could influence both DNA replication and gene expression (2, 7, 8).
In vertebrates, lamins are classified as A or B type, based on sequence homology, expression pattern, biochemical properties, and localization during mitosis. The A-type lamins, lamins A and C, are synthesized from alternatively spliced transcripts of LMNA and are expressed in most differentiated cells (9). Somatic cells also express two B-type lamins, lamin B1 and lamin B2, which are encoded by LMNB1 and LMNB2, respectively. Although the A- and B-type lamins interact with an overlapping set of other nuclear envelope proteins, they exhibit distinct expression patterns and different assembly properties, suggesting independent functions. For example, B-type lamins are expressed throughout development, whereas A-type lamins are expressed only after commitment of cells to a particular differentiation pathway (10, 11). Also, A- and B-type lamins exhibit distinct solubility properties during mitosis because of temporal differences in the association with chromatin, with the association of B-type lamins preceding that of the A-type lamins (12).
Mutations in LMNA produce an intriguingly diverse spectrum of diseases including muscular dystrophies (Emery–Dreifuss muscular dystrophy, limb-girdle muscular dystrophy type 1B), neuropathy (Charcot-Marie-Tooth disease type 2), dilated cardiomyopathy with conduction system disease, familial partial lipodystrophy (s.c. fat loss and diabetes), mandibuloacral dysplasia (skeletal malformations and lipodystrophy), atypical Werner's syndrome, and Hutchinson–Gilford progeria syndrome (precocious aging syndromes) (13–17). Many of the phenotypes associated with LMNA mutations in humans have been observed in mice harboring Lmna mutations. Mice with null mutations in the Lmna gene develop muscular dystrophy (18), peripheral neuropathy (19), and cardiomyopathy (20). Gene-targeted mice with mutations leading to multiple Lmna mRNA splicing abnormalities exhibit aging-like phenotypes akin to those observed in humans with progeria, such as reduced lifespan, osteolytic lesions of bones, dental abnormalities, and cells with misshapen nuclei (21). A deficiency in Zmpste24 (a metalloproteinase of the endoplasmic reticulum) prevents the processing of prelamin A to mature lamin A, resulting in the accumulation of prelamin A within cells (22, 23). Zmpste24-deficient mice exhibit multiple phenotypes reminiscent of progeria, including hair loss, dental abnormalities, and osteolysis of the clavicle and other bones.
Although dozens of disease-causing LMNA mutations have been catalogued, defects in the B-type lamins have never been identified. The absence of human disease suggests that the loss of one of the B-type lamins is either inconsequential or causes death early in development. “Knockdown” experiments with small interfering RNAs (siRNAs) favor the latter possibility, because reduced lamin B1 expression caused cultured cells to stop growing and undergo apoptosis, whereas reduced expression of the A-type lamins had no appreciable effect on cell growth (24).
To define the importance of lamin B1 in mammals, we generated Lmnb1 mutant mice. In view of the aforementioned siRNA experiments (24), we predicted that the mutant mice would die early in embryonic development. To our surprise, however, the mice survived until birth, albeit with bone and lung abnormalities. Lamin B1-deficient fibroblasts grew under standard culture conditions but exhibited misshapen nuclei and growth and differentiation abnormalities. Thus, lamin B1 is required for normal embryonic development and postnatal survival, but the lamin B1 mutation did not lead to lethality at the cellular level.
Materials and Methods
An Insertional Mutation in Lmnb1. Mouse embryonic stem (ES) cells containing a gene-trap insertion in Lmnb1 (cell line XA130) were obtained from BayGenomics (San Francisco) (25). Insertion of the gene-trap vector into Lmnb1 was verified by direct sequencing of cDNA obtained by 5′ rapid amplification of cDNA ends (26). Chimeric mice were generated by blastocyst microinjection and crossed with C57BL/6J mice to create mice carrying the mutant Lmnb1 allele (Lmnb1+/Δ). Offspring were genotyped by PCR of genomic DNA with primers specific for the wild-type Lmnb1 allele (Lmnb1 forward, 5′-TCCGTGTCGTGTGGTAGGAGG-3′; Lmnb1 reverse, 5′-GCAGGAGGGTTGGGAAAGCC-3′) and for the mutant allele carrying the gene-trap insertion (Lmnb1 forward, as above; vector reverse, 5′-CACTCCAACCTCCGCAAACTC-3′).
Cell Culture and Western Blots. Primary mouse embryonic fibroblasts (MEFs) were prepared from embryos harvested 14.5 days post coitus (dpc) (27). Cells were maintained at 37°Cin5%CO2 with DMEM containing 10% FBS, penicillin/streptomycin, and 2 mM glutamine. Two days after reaching confluence, adipocyte differentiation was induced by adding insulin, dexamethasone, isomethylbutylxanthine, and the peroxisome proliferator activated receptor γ ligand, rosiglitazone (BRL 49653; a gift from Todd Leff, Wayne State University, Detroit) (28). Karyotype analysis was performed after treating cells for 5 h with 0.1 μg/ml colcemid (Sigma) (27). For Western blot analysis, cellular protein extracts (10 μg) were electrophoresed on 10% SDS/polyacrylamide gels and blotted to nitrocellulose for detection by chemiluminescence (Amersham Pharmacia). Primary antibodies were directed against lamin A/C (n-18; Santa Cruz Biotechnology), lamin-associated protein 2 (LAP2; clone 27; BD Biosciences Clontech), and lamin B (C-20; Santa Cruz Biotechnology). The latter appears to be lamin B1-specific, based on our Western blot and immunofluorescence studies.
RT-PCR. Gene expression levels in MEFs and embryos were assessed by RT-PCR. cDNA was prepared and analyzed as described (29). Primers were as follows: Lmnb1-3′ forward, TCAGGGAGAGGAGGTTGCTC-3′; Lmnb1-3′ reverse, TCTGCACTGTATACAGGACTC-3′; Lmnb1-5′ forward, 5′-GGGCGTCAGATTGAGTATGAG-3′; Lmnb1-5′ reverse, 5′-TTAGAGAGCTGTGAGGAGAGG-3′; Lmna forward, 5′-GACTTGGTGTGGAAGGCGCAGA-3′; Lmna reverse, 5′-GCTTCGAGTGACTGTGACACTGGA-3′; Lmnc forward, same as Lmna forward; Lmnc reverse, 5′-GCAGGGCTACCCTTGTGGGCT-3′; Hprt forward, CACAGCACTAGAACACCTGC-3′; and Hprt reverse, 5′-GCTGGTGAAAAGGACCTCT-3′.
Embryo Staining and Histology. Whole embryos or tissues were fixed in 10% formalin, dehydrated, cleared, embedded in paraffin, sectioned at 4 μm, and stained with hematoxylin/eosin. For lung histology, embryos were dissected in PBS to prevent inflation of lung tissue. β-galactosidase (β-gal) activity was detected in freshly isolated embryos by fixation in 0.2% glutaraldehyde and incubation with a β-gal staining solution (30). Staining of skeleton was performed with Alizarin red and Alcian blue (31).
Immunofluorescence Microscopy. Primary cells at passage 4–5 were grown on collagen-coated coverslips, fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS, and blocked with 10% goat or donkey serum for 30 min. Cells were incubated for 60 min with antibodies against lamin B, lamin A/C, LAP2, β-gal (MAb1802; Chemicon), or nuclear pore complex NPC (MAb414; BabCO, Richmond, CA). After washing, cells were stained first with Cy3-conjugated secondary antibodies (Jackson ImmunoResearch) and then with SYTOX Green (Molecular Probes) to visualize DNA. Images were obtained with an LSM 510 confocal laser scanning system attached to an Axiovert inverted microscope (Zeiss).
Results
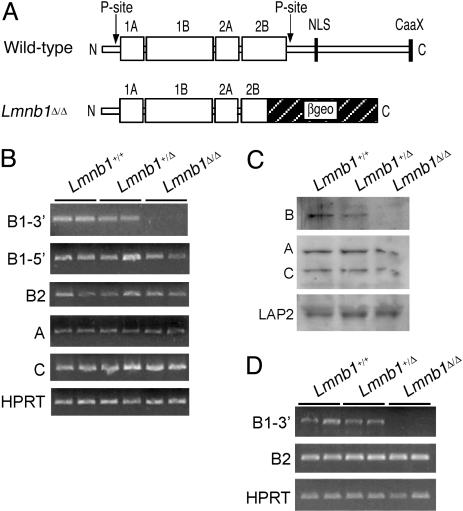
Lmnb1 Mutation Is Lethal at Birth. To produce Lmnb1 mutant mice, we used an ES cell line carrying an insertional mutation in Lmnb1 (BayGenomics cell line XA130). The insertion, located in intron 5 of Lmnb1 (Lmnb1 contains 11 exons and 10 introns), results in an mRNA fusion transcript containing the first five Lmnb1 exons joined in-frame to a βgeo reporter gene. The mutant lamin B1 protein contains the amino-terminal head domain and a truncated α-helical central rod domain fused to βgeo, but lacks the carboxyl-terminal 273 aa of lamin B1 (Fig. 1A). This deletion eliminates one of two known phosphorylation sites, the nuclear localization signal, and the carboxyl-terminal CAAX motif. We refer to the mutant Lmnb1 allele as Lmnb1Δ.
Fig. 1.
Structure and expression of lamin B1 in gene-trap mice. (A) Structure of lamin B1 in wild-type and Lmnb1Δ/Δ mice. The amino-terminal half of lamin B1 consists primarily of the rod domain (1A, 1B, 2A, and 2B) and flanking phosphorylation sites (P-site). The C-terminal half contains the nuclear localization signal (NLS) and the CAAX motif. In Lmnb1Δ/Δ cells, a portion of the rod domain and the entire carboxyl-terminal domain are replaced by βgeo. (B) RT-PCR analysis of mRNA expression levels for lamins B1 (5′ and 3′ regions), B2, A, and C in Lmnb1+/+, Lmnb1+/Δ, and Lmnb1Δ/Δ MEFs from two embryos of each genotype. Hprt (hypoxanthine phosphoribosyl transferase) was used as a normalization control. (C) Immunoblot analysis of lamins and LAP2 in protein extracts from Lmnb1+/+, Lmnb1+/Δ, and Lmnb1Δ/Δ MEFs. (D) RT-PCR analysis of mRNA expression levels for lamins B1 and B2 in 7.5 dpc whole embryos.
ES cells carrying the Lmnb1 mutation were injected into mouse blastocysts, and chimeric offspring were bred to produce Lmnb1+/Δ mice, which were fertile and phenotypically indistinguishable from wild-type mice. However, no live Lmnb1Δ/Δ mice were identified from 140 offspring of heterozygote intercrosses. To determine when Lmnb1Δ/Δ embryos died, we followed their fate during embryonic development. The percentage of Lmnb1+/+, Lmnb1+/Δ, and Lmnb1Δ/Δ mice conformed to the expected Mendelian ratios through embryonic day 18.5, suggesting that Lmnb1Δ/Δ animals might survive until birth (Table 1). Indeed, upon further scrutiny, we observed that Lmnb1Δ/Δ pups were born alive but died within a few minutes and were cannibalized by the mother.
Table 1. Mendelian inheritance of the mutant Lmnb1 allele (Δ) in intercrosses of heterozygous mutant mice.
| Age of embryos, dpc | Lmnb1+/+ | Lmnb1+/Δ | Lmnb1Δ/Δ |
|---|---|---|---|
| 14.5 | 13 | 21 | 12 |
| 16.5 | 4 | 11 | 4 |
| 18.5 | 14 | 33 | 14 |
Lamin expression in mouse embryonic fibroblasts (MEFs) was examined by RT-PCR and Western blot analysis. In Lmnb1+/+ and Lmnb1+/Δ cells, the Lmnb1 mRNA was easily detected with primers against both 5′ and 3′ portions of the transcript. Only the 5′ Lmnb1 sequences were detected in the Lmnb1Δ/Δ cells (Fig. 1B). Expression of lamins B2, A, and C was not altered in Lmnb1Δ/Δ cells (Fig. 1B). The loss of lamin B1 expression in Lmnb1Δ/Δ cells was confirmed by Western blot analysis of fibroblast extracts (Fig. 1C). The lamin B1–βgeo fusion protein was not detected with an antibody raised against the C-terminal domain of B-type lamins. This antibody appears to be specific for lamin B1 because it did not detect the lamin B2 expressed by the Lmnb1Δ/Δ cells. Lamins A and C, as well as lamin-associated protein 2 (LAP2), were expressed at normal levels in Lmnb1Δ/Δ fibroblasts. Lamin B expression was also assessed in 7.5-dpc whole embryos by RT-PCR (Fig. 1D). As expected, the 3′ portion of lamin B1 was not detected, but lamin B2 was expressed at comparable levels in wild-type, Lmnb1+/Δ, and Lmnb1Δ/Δ embryos.
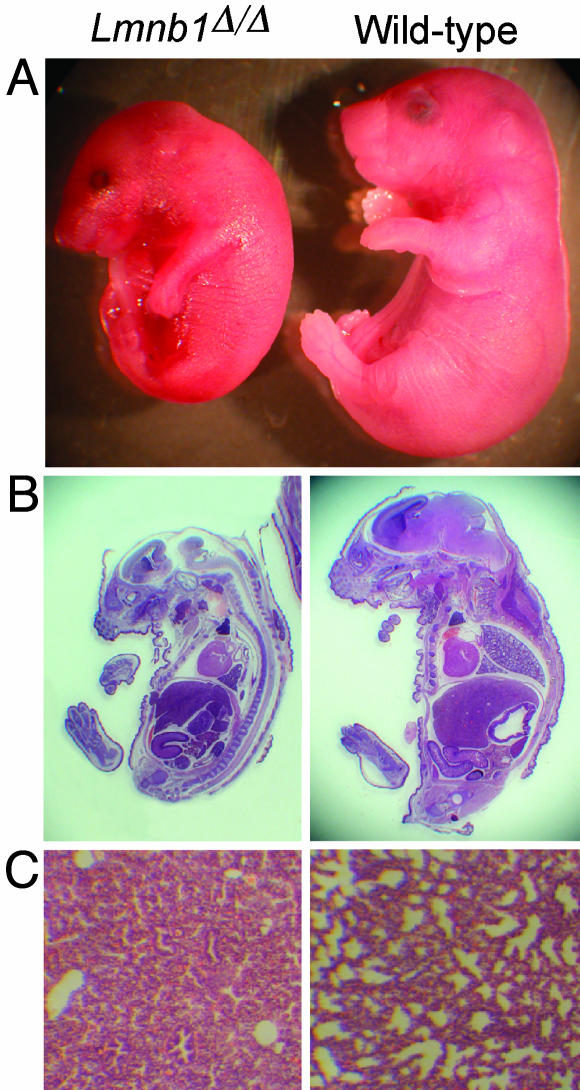
Lmnb1 Deficiency Results in Abnormal Lung Development and Bone Ossification. The Lmnb1Δ/Δ embryos were smaller than wild-type mice (609 ± 67 mg vs. 1,083 ± 166 mg at 18.5 dpc; P < 0.0001) and had an abnormally rounded posture with craniofacial dysmorphology (Fig. 2A). Staining of Lmnb1Δ/Δ and Lmnb1+/Δ embryos for β-gal expression indicated that Lmnb1 is expressed throughout embryonic development and in nearly all tissue types (data not shown). Most of the internal organs in Lmnb1Δ/Δ mice appeared grossly normal by routine histology (Fig. 2B). Lmnb1Δ/Δ embryos harvested at 18.5 dpc had a heartbeat, suggesting a functional circulatory system. Therefore, we suspected that the perinatal mortality in Lmnb1Δ/Δ mice might have been caused by respiratory failure. In support of that possibility, 9 of 11 Lmnb1Δ/Δ embryos examined had abnormal lung tissue with fewer alveoli than wild-type lungs and abnormally thick mesenchymal tissue (Fig. 2C). In two Lmnb1Δ/Δ embryos, the lung had no detectable alveoli.
Fig. 2.
Appearance of Lmnb1Δ/Δ embryos. (A) Reduced size and abnormal shape of Lmnb1Δ/Δ embryos at 18.5 dpc. (B) Sagittal sections of embryos stained with hematoxylin/eosin. (C) Lung sections obtained before inflation, stained with hematoxylin/eosin.
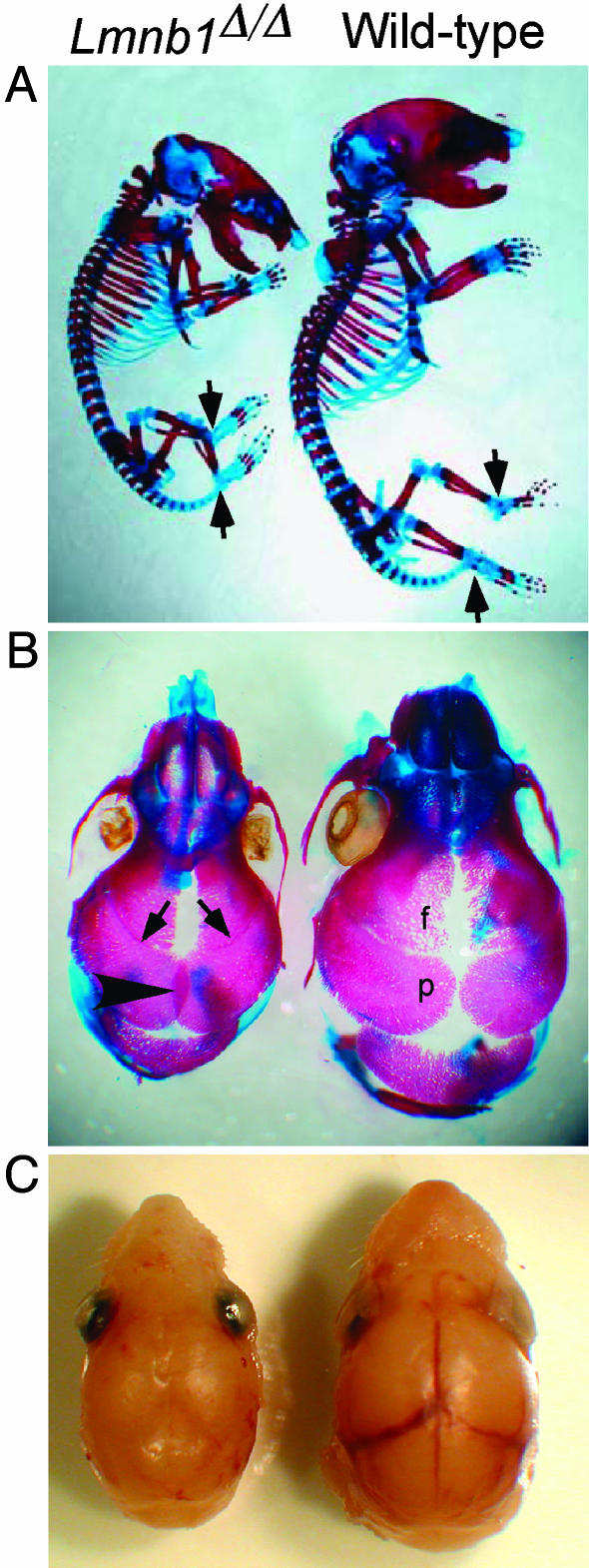
The small size and abnormal curvature of the spine in Lmnb1Δ/Δ embryos suggested that lamin B1 might have a role in skeletal development. Alcian blue and Alizarin red staining of Lmnb1Δ/Δ embryos revealed distortion of the thoracic vertebral column (Fig. 3A). Also, there was reduced Alizarin red staining of the top cervical vertebrae, as well as the metacarpal and metatarsal bones, phalanges, talus, and calcaneus, suggesting retarded or impaired ossification. The lengths of the long bones (humerus, radius, ulna, femur, tibia, and fibula) were reduced in proportion to the decreased size of Lmnb1Δ/Δ embryos. Of note, however, the degree of ossification, as judged by Alizarin red staining, was reduced in the mutant embryos (Table 2).
Fig. 3.
Abnormal skeleton and skull shape in Lmnb1Δ/Δ mice. (A) Alizarin red (bone) and Alcian blue (cartilage) staining of Lmnb1Δ/Δ skeleton (Left) shows a flattened skull, an abnormal vertebral column, and reduced ossification in phalanges, talus, and calcaneus (arrows). (B) Dorsal view of cranial vault. Alizarin red stain shows closure of the coronal sutures between parietal and frontal bones (arrows) and overlapping of the parietal (p) bones at the sagittal suture (arrowhead) in the Lmnb1Δ/Δ embryo (Left). (C) Removal of scalp reveals obscured view of underlying blood vessels due to suture closure in the Lmnb1Δ/Δ embryo (Left).
Table 2. Bone measurement and percent ossification in wild-type vs. Lmnb1Δ/Δ mice.
| Humerus | Radius | Ulna | Femur | Tibia | Fibula | |
|---|---|---|---|---|---|---|
| WT (n = 10) | ||||||
| Length, mm | 4.0 ± 0.3 | 3.2 ± 0.3 | 4.2 ± 0.3 | 3.7 ± 0.3 | 3.9 ± 0.3 | 3.7 ± 0.3 |
| Ossification, % | 58.8 ± 4.9 | 70.9 ± 2.5 | 67.8 ± 4.1 | 54.7 ± 3.6 | 61.9 ± 6.3 | 61.5 ± 7.1 |
| Lmnb1Δ/Δ (n = 8) | ||||||
| Length, mm | 3.7 ± 0.2* | 2.9 ± 0.2** | 3.8 ± 0.2** | 3.3 ± 0.3** | 3.3 ± 0.3*** | 3.2 ± 0.3** |
| Ossification, % | 52.8 ± 4.0* | 60.5 ± 5.3*** | 60.9 ± 3.9** | 44.7 ± 6.0*** | 51.8 ± 7.6** | 52.3 ± 7.8* |
, P < 0.05; **, P < 0.01; ***, P < 0.001.
We also observed abnormal development of bones forming the roof of the skull as evidenced by the flattened appearance of the skull (Fig. 3A). Hypermineralization was also visible in the nasal bones as increased Alizarin red staining (Fig. 3B). Additional abnormalities were evident in the appearance of cranial sutures, fibrous joints that allow the calvarial bones to expand as the brain enlarges during development. Normally, all but one of the cranial sutures remain open throughout the life span of the animal (32). Wild-type embryos showed patent coronal and sagittal sutures, but in Lmnb1Δ/Δ mice, the coronal suture was narrowed (Fig. 3B, arrows) and the sagittal suture was obscured by bone overlap, suggesting bone overgrowth (Fig. 3B, arrowhead). The absence of normal cranial sutures was also evident from the lack of visible blood vessels underneath sutures in Lmnb1Δ/Δ mice, which were readily detected in wild-type embryos (Fig. 3C).
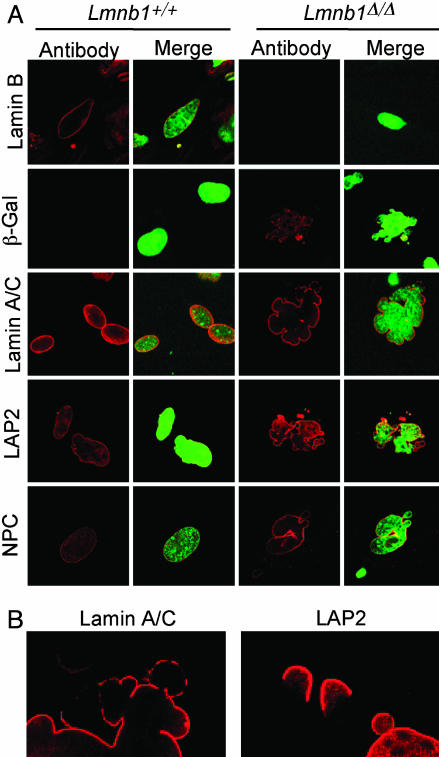
Nuclear Abnormalities in Primary Lmnb1Δ/Δ Cells. To assess the effect of lamin B1 mutation at the cellular level, we characterized nuclear structure and protein localization in primary MEFs from Lmnb1+/+ and Lmnb1Δ/Δ embryos. As expected, lamin B1 in Lmnb1+/+ MEFs was localized to the inner surface of the nuclear envelope and was undetectable in Lmnb1Δ/Δ cells (Fig. 4A). The mutant cells also exhibited striking nuclear dysmorphology; analysis of 260 cells of each genotype in two independent experiments showed multiple large blebs in 38–39% of Lmnb1Δ/Δ nuclei but in only 2–8% of wild-type nuclei (Fig. 4A and data not shown).
Fig. 4.
Abnormal nuclear architecture in Lmnb1Δ/Δ MEFs. (A) Confocal micrographs of MEFs from Lmnb1+/+ or Lmnb1Δ/Δ embryos stained with antibodies for nuclear proteins (Left) and merged with DNA stain for nuclei (Right). A- and B-type lamins, LAP2, and nuclear pore complex (NPC) proteins were detected with specific antibodies; the lamin B1-βgeo fusion in cells was detected with an antibody against β-gal. (B) Higher magnification of lamin A/C and LAP2 staining showing nonuniform distribution in Lmnb1Δ/Δ cells.
The insertional mutation results in production of a lamin B1–βgeo fusion protein. We could therefore detect the product of the Lmnb1Δ/Δ allele by immunofluorescence with an antibody against β-gal. The lamin B1–βgeo fusion protein, which was present only in the mutant cells, was detected in both the nucleoplasm and at the nuclear membrane (Fig. 4A). The nuclear localization was somewhat unexpected because the fusion protein lacks the nuclear localization signal.
Although the Lmnb1Δ/Δ mutation did not alter the level of expression of lamin A/C or LAP2 (see Fig. 1), it did affect their localization. Both were distributed unevenly along the periphery of the nucleus, with noticeably reduced staining of some blebs of the nucleus (Fig. 4B). In addition, there was less DNA in some blebs, especially in those with reduced staining for lamin A/C and LAP2, possibly reflecting an altered lamina–chromatin interaction (see merged images in Fig. 4A Right). In contrast, the distribution of nuclear pore complex proteins, which span both the inner and outer nuclear membranes, appeared normal in Lmnb1Δ/Δ cells. The examination of Lmnb1+/Δ fibroblasts gave the same results as wild-type cells, indicating that the mutant protein does not act in a dominant manner (data not shown).
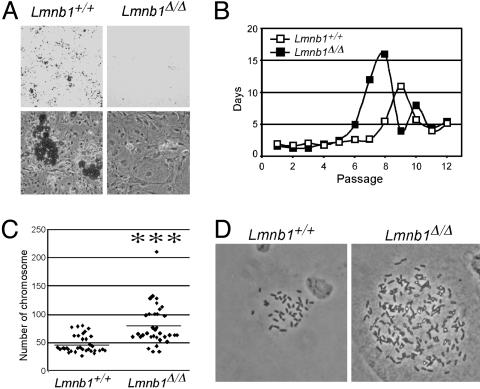
We next asked whether the disruption in nuclear structure in Lmnb1Δ/Δ cells affected cell differentiation or proliferative capacity. Lmnb1+/+ and Lmnb1Δ/Δ MEFs were grown to confluence, and induced to differentiate into adipocytes 2 days later. Differentiation into mature adipocytes was monitored by staining for triglyceride accumulation with Oil Red O. Wild-type MEFs accumulated substantial amounts of triglyceride after 6 days of differentiation (Fig. 5A). In contrast, Lmnb1Δ/Δ cells failed to accumulate lipid, suggesting impaired differentiation.
Fig. 5.
Cell proliferation and differentiation defects in Lmnb1Δ/Δ MEFs. (A) Lmnb1+/+ and Lmnb1Δ/Δ MEFs were grown to confluence and, 2 days later, induced to differentiate into adipocytes. Six days later, cells were stained with Oil Red O to detect lipid accumulation. Representative cultures are shown at ×6 (Upper) and ×200 (Lower) magnification. (B) MEFs were continuously passaged at 95% confluence and replating at 30% confluent density. Passage number was plotted against the number of days between passages, averaged for three independent MEF isolates of each genotype. Passage 1 denotes the initial isolation of MEFs. (C) Analysis of chromosome number in metaphase spreads of Lmnb1Δ/Δ and Lmnb1+/+ MEFs at passage 4. Scatterplot points show the distribution of chromosome numbers; horizontal lines show mean values. ***, P < 0.00001. Photomicrographs at right show representative diploid Lmnb1+/+ and polyploid Lmnb1Δ/Δ cells (magnification, ×800).
To further characterize the defect in lamin B1 mutant cells, we evaluated the proliferative capacity of freshly prepared MEFs. We recorded the number of days required for wild-type and Lmnb1Δ/Δ fibroblasts to reach 95% confluence at each passage and determined the number of passages until cells entered a senescent phase (33). The passage times for the two cell types were similar through passage 5. Wild-type cells entered a proliferative crisis, as indicated by the increase in passage time, at passage 8 (Fig. 5B). In contrast, Lmnb1Δ/Δ cells reached the proliferative crisis two passages earlier, indicating reduced replicative potential in the mutant cell population. Similar results were obtained with primary fibroblasts from two embryos of each genotype. No differences were observed between Lmnb1+/+ and Lmnb1+/Δ cells (data not shown). Despite the more rapid entry of Lmnb1Δ/Δ fibroblasts into a senescent phase, those cells could nevertheless be immortalized by continued passaging.
We performed karyotype analysis to determine whether alterations in chromosome number are associated with the premature senescence observed in Lmnb1Δ/Δ cells. Passage 4 MEFs from two independent wild-type and Lmnb1Δ/Δ embryos were treated with colcemid to arrest the cell cycle at metaphase, and chromosomes were counted. The average chromosome number in wild-type cells was ≈2n (47 ± 15), whereas >80% of Lmnb1Δ/Δ cells were polyploid, with a significantly increased number of chromosomes (80 ± 35) (Fig. 5C). Furthermore, one-third of the Lmnb1Δ/Δ cells had accumulated >100 chromosomes. Such an increase was never observed in wild-type cells.
Discussion
We investigated Lmnb1 mutant mice to define the role of lamin B1 in mammalian development and nuclear architecture. In contrast to lamin A/C-deficient mice, which survive for 4–6 weeks after birth (18), Lmnb1Δ/Δ embryos exhibited impaired growth and died shortly after birth. The retarded or abnormal lung development may have led to respiratory failure and perinatal lethality, although we could not exclude the possibility that abnormalities in other organs also contributed to their demise. In addition, there was a striking reduction in long bone ossification and an abnormal cranial structure. The apparent closure of coronal sutures and the parietal bone overgrowth at the sagittal suture are analogous to craniosynostosis that has been characterized in mouse strains with altered expression of genes involved in neural development and cellular growth (34–36). At the cellular level, the lamin B1 mutation disrupted the organization of the lamina and the structure of the nuclear envelope, producing grossly misshapen nuclei and abnormal chromatin distribution. Lmnb1Δ/Δ fibroblasts exhibited premature senescence, impaired adipocyte differentiation, and an increased occurrence of polyploidy.
The more severe phenotype in lamin B1-mutant mice compared with lamin A/C-deficient mice might reflect the fact that Lmnb1 expression exhibits a broader tissue distribution than Lmna. All mammalian cells express at least one form of the B-type lamins, and the expression is constitutive during development; expression is detectable in ES cells as well as stem cells for the immune, hematopoeitic, and neuroendocrine systems (10, 11, 37, 38). In contrast, A-type lamins cannot be detected in any cell type before day 7 or 8 of embryonic development (10, 11, 37). Interestingly, Rober et al. (11) found that lamin A/C is present in many mouse tissues at birth, but is not detectable in the lung until after postnatal day 1, suggesting that B-type lamins may be particularly important in that tissue. The apparent respiratory failure in newborn Lmnb1Δ/Δ mice is consistent with this finding.
The fact that the Lmnb1Δ/Δ mice survived until birth, and that we were able to culture and even immortalize Lmnb1Δ/Δ fibroblasts, was surprising. From knockdown experiments with siRNAs (24), we had expected that the Lmnb1 mutation would cause death early in development, preventing us from culturing Lmnb1-deficient cells. Initially, we suspected that the relatively mild phenotype might have been caused by “intrinsic leakiness” of the insertional mutation. However, this was not the case. No full-length Lmnb1 transcripts were detected by RT-PCR, and no full-length lamin B1 protein was present on Western blots. Because we did detect normal lamin B2 mRNA levels in Lmnb1Δ/Δ embryos and primary cells, it is possible that this related B-type lamin could partially compensate for the loss of lamin B1 during development. Another possibility is that the Lmnb1Δ/Δ mutation represents a hypomorphic allele that produces a lamin B1–βgeo fusion protein that retains some biological activity. However, in vitro studies of lamin B1 mutants do not support the notion that the lamin B1 fusion protein could be functional. Deletion of the carboxyl-terminal half of lamin B1, similar to the deletion encoded by the Lmnb1Δ/Δ allele, leads to a complete loss of chromatin-binding regions of the protein (39). Moreover, our lamin B1 mutant protein lacks the carboxyl-terminal CAAX motif, and lamin B1 mutants lacking this domain interfere with the nuclear membrane targeting of A-type lamins (40). Nevertheless, we cannot completely exclude the possibility that the lamin B1–βgeo fusion protein retains some function, particularly because it localized to the nucleus. The nuclear localization was unexpected because the protein lacks both the nuclear localization signal and the CAAX sequence, two motifs that are important for the targeting of lamin B1 to the nuclear envelope. One possibility is that the fusion protein retains some capacity to dimerize with other lamina proteins (e.g., lamin B2).
Primary Lmnb1Δ/Δ MEFs exhibited grossly misshapen nuclei, reached a replicative crisis several passages before wild-type MEFs, and grew slowly. The early cell senescence and misshapen nuclei are reminiscent of findings reported for MEFs homozygous for a mutant Lmna allele characterized by multiple mRNA splicing abnormalities and a missense mutation (L530P) (21). In the case of the Lmnb1Δ/Δ MEFs, the early replicative crisis was accompanied by an accumulation of polyploid cells in the culture population, suggesting a possible role for lamin B1 in chromosome segregation during cell division. The inability of polyploid cells to continue to divide could contribute to the reduced DNA replication and premature senescence of Lmnb1Δ/Δ cells. Whether the Lmna mutant allele was also associated with karyotypic abnormalities is not known. MEFs from Lmnb1Δ/Δ embryos exhibited impaired adipocyte differentiation, an intriguing feature in light of the fact that certain LMNA mutations in humans cause a partial lipodystrophy phenotype. In the future, it will be interesting to determine whether early replicative crisis and impaired differentiation are general features of mutations that disrupt the nuclear lamina.
We were intrigued to identify skeletal abnormalities in the Lmnb1Δ/Δ mice. No bone abnormalities were reported in the lamin A/C-null mice. However, osteolysis and dental abnormalities are features of Hutchinson–Gilford progeria syndrome (41), which is caused by lamin A/C mutations. Also, Bergo et al. (22) observed nonhealing bone fractures, osteolysis, osteopenia, and dental abnormalities in mice lacking Zmpste24, an endoprotease that is required for the maturation of prelamin A to mature lamin A. At this point, the mechanism for bone abnormalities associated with the lamin abnormalities remains mysterious.
The lamin B1 mutant mice are a resource for investigators interested in the nuclear lamina and should spur interest in developing animal models with mutations in other nuclear envelope proteins. Moreover, the Lmnb1Δ/Δ MEFs will be useful to further elucidate the role of lamin B1 in maintaining nuclear structure and function. Experiments with those cells and MEFs from the lamin A/C-deficient mice (18) should facilitate an in-depth understanding of the gene expression changes that accompany structural abnormalities in the nuclear lamina.
Acknowledgments
We thank Ms. Merav Strauss and Ms. Lorraine Ponce for mouse colony management, and Dr. Lise Zakin for technical advice on mouse developmental studies. These studies were supported by National Heart, Lung, and Blood Institute Programs for Genomic Applications Grant U01 HL66621.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ES, embryonic stem; MEF, mouse embryonic fibroblast; β-gal, β-galactosidase; dpc, days post coitus.
References
- 1.Cohen, M., Lee, K. K., Wilson, K. L. & Gruenbaum, Y. (2001) Trends Biochem. Sci. 26, 41–47. [DOI] [PubMed] [Google Scholar]
- 2.Wilson, K. L., Zastrow, M. S. & Lee, K. K. (2001) Cell 104, 647–650. [PubMed] [Google Scholar]
- 3.Goldman, R. D., Gruenbaum, Y., Moir, R. D., Shumaker, D. K. & Spann, T. P. (2002) Genes Dev. 16, 533–547. [DOI] [PubMed] [Google Scholar]
- 4.Burke, B. & Stewart, C. L. (2002) Nat. Rev. Mol. Cell. Biol. 3, 575–585. [DOI] [PubMed] [Google Scholar]
- 5.Hutchison, C. J. (2002) Nat. Rev. Mol. Cell. Biol. 3, 848–858. [DOI] [PubMed] [Google Scholar]
- 6.Stuurman, N., Heins, S. & Aebi, U. (1998) J. Struct. Biol. 122, 42–66. [DOI] [PubMed] [Google Scholar]
- 7.Jagatheesan, G., Thanumalayan, S., Muralikrishna, B., Rangaraj, N., Karande, A. A. & Parnaik, V. K. (1999) J. Cell Sci. 112, 4651–4661. [DOI] [PubMed] [Google Scholar]
- 8.Spann, T. P., Goldman, A. E., Wang, C., Huang, S. & Goldman, R. D. (2002) J. Cell Biol. 156, 603–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin, F. & Worman, H. J. (1993) J. Biol. Chem. 268, 16321–16326. [PubMed] [Google Scholar]
- 10.Stewart, C. & Burke, B. (1987) Cell 51, 383–392. [DOI] [PubMed] [Google Scholar]
- 11.Rober, R. A., Weber, K. & Osborn, M. (1989) Development (Cambridge, U.K.) 105, 365–378. [DOI] [PubMed] [Google Scholar]
- 12.Moir, R. D., Yoon, M., Khuon, S. & Goldman, R. D. (2000) J. Cell Biol. 151, 1155–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mounkes, L., Kozlov, S., Burke, B. & Stewart, C. L. (2003) Curr. Opin. Genet. Dev. 13, 223–230. [DOI] [PubMed] [Google Scholar]
- 14.Ostlund, C. & Worman, H. J. (2003) Muscle Nerve 27, 393–406. [DOI] [PubMed] [Google Scholar]
- 15.Eriksson, M., Brown, W. T., Gordon, L. B., Glynn, M. W., Singer, J., Scott, L., Erdos, M. R., Robbins, C. M., Moses, T. Y., Berglund, P., et al. (2003) Nature 423, 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Sandre-Giovannoli, A., Bernard, R., Cau, P., Navarro, C., Amiel, J., Boccaccio, I., Lyonnet, S., Stewart, C. L., Munnich, A., Le Merrer, M. & Levy, N. (2003) Science 300, 2055. [DOI] [PubMed] [Google Scholar]
- 17.Chen, L., Lee, L., Kudlow, B. A., Dos Santos, H. G., Sletvold, O., Shafeghati, Y., Botha, E. G., Garg, A., Hanson, N. B., Martin, G. M., et al. (2003) Lancet 362, 440–445. [DOI] [PubMed] [Google Scholar]
- 18.Sullivan, T., Escalante-Alcalde, D., Bhatt, H., Anver, M., Bhat, N., Nagashima, K., Stewart, C. L. & Burke, B. (1999) J. Cell Biol. 147, 913–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Sandre-Giovannoli, A., Chaouch, M., Kozlov, S., Vallat, J. M., Tazir, M., Kassouri, N., Szepetowski, P., Hammadouche, T., Vandenberghe, A., Stewart, C. L., et al. (2002) Am. J. Hum. Genet. 70, 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolova, V., Leimena, C., McMahon, A. C., Tan, J. C., Chandar, S., Jogia, D., Kesteven, S. H., Michalicek, J., Otway, R., Verheyen, F., et al. (2004) J. Clin. Invest. 113, 357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mounkes, L. C., Kozlov, S., Hernandez, L., Sullivan, T. & Stewart, C. L. (2003) Nature 423, 298–301. [DOI] [PubMed] [Google Scholar]
- 22.Bergo, M. O., Gavino, B., Ross, J., Schmidt, W. K., Hong, C., Kendall, L. V., Mohr, A., Meta, M., Genant, H., Jiang, Y., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 13049–13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pendas, A. M., Zhou, Z., Cadinanos, J., Freije, J. M., Wang, J., Hultenby, K., Astudillo, A., Wernerson, A., Rodriguez, F., Tryggvason, K. & Lopez-Otin, C. (2002) Nat. Genet. 31, 94–99. [DOI] [PubMed] [Google Scholar]
- 24.Harborth, J., Elbashir, S. M., Bechert, K., Tuschl, T. & Weber, K. (2001) J. Cell Sci. 114, 4557–4565. [DOI] [PubMed] [Google Scholar]
- 25.Stryke, D., Kawamoto, M., Huang, C. C., Johns, S. J., King, L. A., Harper, C. A., Meng, E. C., Lee, R. E., Yee, A., L'Italien, L., et al. (2003) Nucleic Acids Res. 31, 278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Townley, D. J., Avery, B. J., Rosen, B. & Skarnes, W. C. (1997) Genome Res. 7, 293–298. [DOI] [PubMed] [Google Scholar]
- 27.Freshney, R. I. (1984) Culture of Animal Cells: A Manual of Basic Technique (Liss, New York).
- 28.Alexander, D. L., Ganem, L. G., Fernandez-Salguero, P., Gonzalez, F. & Jefcoate, C. R. (1998) J. Cell Sci. 111, 3311–3322. [DOI] [PubMed] [Google Scholar]
- 29.Vergnes, L., Phan, J., Stolz, A. & Reue, K. (2003) J. Lipid Res. 44, 503–511. [DOI] [PubMed] [Google Scholar]
- 30.Hogan, B., Constantini, R. & Lacy, E. (1986) Manipulating the Mouse Embryo (Cold Spring Harbor Lab. Press, Plainview, NY).
- 31.Kaufman, M. H. (1992) The Atlas of Mouse Development (Academic, San Diego).
- 32.Warren, S. M. & Longaker, M. T. (2001) Yonsei Med. J. 42, 646–659. [DOI] [PubMed] [Google Scholar]
- 33.Todaro, G. J. & Green, H. (1963) J. Cell Biol. 17, 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, Y. H., Kundu, R., Wu, L., Luo, W., Ignelzi, M. A., Jr., Snead, M. L. & Maxson, R. E., Jr. (1995) Proc. Natl. Acad. Sci. USA 92, 6137–6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang, X., Kuroda, S., Carpenter, D., Nishimura, I., Soo, C., Moats, R., Iida, K., Wisner, E., Hu, F. Y., Miao, S., et al. (2002) J. Clin. Invest. 110, 861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eswarakumar, V. P., Monsonego-Ornan, E., Pines, M., Antonopoulou, I., Morriss-Kay, G. M. & Lonai, P. (2002) Development (Cambridge, U.K.) 129, 3783–3793. [DOI] [PubMed] [Google Scholar]
- 37.Rober, R. A., Sauter, H., Weber, K. & Osborn, M. (1990) J. Cell Sci. 95, 587–598. [DOI] [PubMed] [Google Scholar]
- 38.Broers, J. L., Machiels, B. M., Kuijpers, H. J., Smedts, F., van den Kieboom, R., Raymond, Y. & Ramaekers, F. C. (1997) Histochem. Cell Biol. 107, 505–517. [DOI] [PubMed] [Google Scholar]
- 39.Taniura, H., Glass, C. & Gerace, L. (1995) J. Cell Biol. 131, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izumi, M., Vaughan, O. A., Hutchison, C. J. & Gilbert, D. M. (2000) Mol. Biol. Cell 11, 4323–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackerman, J. & Gilbert-Barness, E. (2002) Pediatr. Pathol. Mol. Med. 21, 1–13. [DOI] [PubMed] [Google Scholar]