Abstract
Genes involved in circadian regulation, such as circadian locomotor output cycles kaput [CLOCK], cryptochrome [CRY1], and period [PER], have been associated with sleep outcomes in prior animal and human research. However, it is unclear whether polymorphisms in these genes are associated with the sleep disturbances commonly experienced by adults living with human immunodeficiency virus/acquired immunodeficiency syndrome (HIV/AIDS). Thus, the purpose of this study was to describe polymorphisms in selected circadian genes that are associated with sleep duration or disruption as well as the sleep-wake rhythm strength and phase timing among adults living with HIV/AIDS. A convenience sample of 289 adults with HIV/AIDS was recruited from HIV clinics and community sites in the San Francisco Bay Area. A wrist actigraph was worn for 72 hours on weekdays to estimate sleep duration or total sleep time (TST), sleep disruption or percentage of wake after sleep onset (WASO), and several circadian rhythm parameters: mesor, amplitude, the ratio of mesor to amplitude (circadian quotient), and 24-hour autocorrelation. Circadian phase measures included clock time for peak activity (acrophase) from actigraphy movement data, and bed time and final wake time from actigraphy and self-report. Genotyping was conducted for polymorphisms in 5 candidate genes involved in circadian regulation: CLOCK, CRY1, PER1, PER2, and PER3. Demographic and clinical variables were evaluated as potential covariates. Interactions between genotype and HIV variables (i.e., viral load, years since HIV diagnosis) were also evaluated. Controlling for potentially confounding variables (e.g., race, gender, CD4+ T-cell count, waist circumference, medication use, smoking, depressive symptoms), CLOCK was associated with WASO, 24-hour autocorrelation, and objectively-measured bed time; CRY1 was associated with circadian quotient; PER1 was associated with mesor and self-reported habitual wake time; PER2 was associated with TST, mesor, circadian quotient, 24-hour autocorrelation, and bed and wake times; PER3 was associated with amplitude, 24-hour autocorrelation, acrophase, and bed and wake times. Most of the observed associations involved a significant interaction between genotype and HIV. In this chronic illness population, polymorphisms in several circadian genes were associated with measures of sleep disruption and timing. These findings extend the evidence for an association between genetic variability in circadian regulation and sleep outcomes to include the sleep-wake patterns experienced by adults living with HIV/AIDS. These results provide direction for future intervention research related to circadian sleep-wake behavior patterns.
Keywords: circadian, genetic, HIV, autocorrelation, mesor, acrophase, actigraphy, chronotype
Introduction
Sleep disturbance is a common symptom in chronic illness populations, and it is estimated that up to 75% of adults living with human immunodeficiency virus or acquired immunodeficiency syndrome (HIV/AIDS) experience sleep problems (Rubinstein & Selwyn, 1998). The most common type of sleep problem in HIV disease is difficulty with sleep maintenance (Phillips et al., 2005; Lee et al., 2012; Gamaldo et al., 2013b), although short sleep duration (Lee et al., 2012; Gamaldo et al., 2013a) and disturbed sleep-wake rhythm (Taibi et al., 2013) are also reported. Sleep problems can be influenced by mood disorders, lifestyle factors, and HIV disease processes and treatments. In addition, the inability to work may influence one’s regular daytime schedule of activity and affect circadian rhythms as well as sleep. Not surprisingly, sleep patterns and disturbances have also been associated with genes involved in circadian regulation, such as circadian locomotor output cycles kaput [CLOCK], cryptochrome [CRY1], and period [PER1, PER2, PER3] (Allebrandt et al., 2010; Ojeda et al., 2013; Zhang et al., 2013; Hida et al., 2014; Parsons et al., 2014). However, there are no published genetic association studies evaluating the extent to which such circadian genes might account for phenotypic sleep problems commonly experienced by adults living with HIV. Therefore, the purpose of this study was to describe associations between polymorphisms in selected candidate genes related to circadian regulation (i.e., CLOCK, CRY1, PER1, PER2, PER3) and phenotypes that include sleep duration, maintenance, and circadian sleep-wake phase timing and rhythm strength in a sample of adults living with HIV.
Circadian rhythms are under control of the circadian oscillator found in the anterior hypothalamus (Takahashi et al., 2008). The circadian oscillator is considered the master circadian clock and regulates the 24-hour cycle through interacting positive and negative feedback loops (Ko & Takahashi, 2006). These feedback loops act at the levels of molecular clock gene transcription and protein degradation (Dunlap, 1999; Lincoln et al., 2003; Ko & Takahashi, 2006). The core clock genes that drive circadian-related feedback loops include CLOCK, CRY1, PER1, PER2, and PER3 (Osland et al., 2011). CLOCK acts as a positive regulator, while CRY1, PER1, PER2, and PER3 act as negative regulators (Gekakis et al., 1998; Shearman et al., 2000; Ko & Takahashi, 2006). Circadian rhythms in the human body are driven by these clock genes, and we hypothesize that polymorphisms of CLOCK, CRY1, PER1, PER2, and PER3 will be associated with sleep-wake phenotypes in adults with HIV/AIDS. Circadian rhythm phenotype can also be influenced by employment, with fixed work schedules reinforcing 24-hour entrainment. Thus, unemployment and disability in a sample of adults with HIV infection may more readily reveal underlying circadian preferences.
Materials and Methods
Participants and Setting
The Symptom and Genetic Study was a longitudinal study aimed at identifying biomarkers of symptom experience among HIV-infected adults (Lee et al., 2009). This analysis focuses on potential circadian-related genetic markers of insomnia related to poor sleep maintenance and duration, as well as circadian rhythm and timing. The Committee on Human Research at the University of California at San Francisco (UCSF) approved the study protocol. Participants were recruited using flyers posted at local HIV clinics and community sites. Participants provided written informed consent and signed a Health Insurance Portability and Accountability Act release to access their protected medical information for this research. Study visits were conducted at a clinic visit in the UCSF Clinical Research Center.
Eligible participants were English-speaking adults at least 18 years of age in whom HIV had been diagnosed at least 30 days before enrollment. To specifically address HIV-related symptom experience, potential participants were excluded if they currently used illicit drugs (as determined by self-report or by positive urine drug testing); worked nights (i.e., at least 4 hours between midnight and 06:00 a.m.); reported having bipolar disorder, schizophrenia, or dementia; or were pregnant within the prior 3 months. Participants were not excluded for insomnia, but were excluded for other diagnosed sleep disorders, such as apnea or narcolepsy.
Measures
Demographic, clinical, and laboratory characteristics
A demographic questionnaire was used to collect information about the participant’s age, gender, race/ethnicity, and employment status. Health history (e.g., time since HIV diagnosis, prior AIDS diagnosis) and current medication regimen were obtained by self-report. Medications were categorized as antiretroviral therapy (ART), sleep medication, anxiolytic, antidepressant, neuroleptic, opiate, antiemetic, or anti-histamine based on the potential for such medications to affect sleep. Lifestyle factors likely to exacerbate sleep disturbance (smoking and daily consumption of caffeine and alcohol) were assessed using a 3-day diary. Trained research staff obtained measures of body mass index (BMI, weight in kilograms divided by squared height in meters) and waist circumference during the clinic office visit. CD4+ T-cell count and HIV viral load values were obtained from the most recent laboratory report in the patient’s medical record and were usually obtained from morning blood draws. Because circadian genes have been widely associated with mood disorders (Bunney et al., 2015), the Center for Epidemiological Studies – Depression Scale (CES-D) (Radloff, 1977) was used to assess depressive symptomatology as a potentially confounding variable. The CES-D has well-established concurrent and construct validity, and the Cronbach’s alpha coefficient in this study was .88. A cutpoint of 16 is used to identify individuals at risk for clinical depression.
Gene selection and genotyping
Five candidate genes related to circadian regulation were selected for analysis. Genomic DNA was extracted from peripheral blood mononuclear cells and maintained by the UCSF Genomic Markers of Symptoms Tissue Bank (Aouizerat et al., 2009) using the PUREGene DNA Isolation System (Invitrogen, Carlsbad, CA). Of the 350 participants enrolled in the study, DNA could be isolated from 348 samples.
Genotyping was performed blinded to clinical status and included positive and negative controls. DNA samples were quantitated with a Nanodrop Spectrophotometer (ND-1000; Thermo Fisher Scientific, Waltham, MA) and normalized to a concentration of 50 ng/μL (diluted in 10 mM Tris/1 mM ethylenediaminetetraacetic acid [EDTA]). Samples were genotyped using the GoldenGate genotyping platform (Illumina, San Diego, CA) and processed according to the standard protocol using GenomeStudio (Illumina). Signal intensity profiles and resulting genotype call rates for each single nucleotide polymorphism (SNP) were visually inspected by two blinded reviewers. Disagreements were resolved by a third reviewer.
A combination of tagging SNPs and literature driven SNPs (i.e., SNPs reported as being associated with altered function) were selected for analysis. Tagging SNPs were required to be common (defined as having a minor allele frequency ≥ 0.05) in public databases (e.g., HapMap [http://www.hapmap.org]). In order to ensure robust genetic association analyses, quality-control filtering of SNPs was performed. All SNPs had call rates of > 95% and five SNPs were excluded with Hardy-Weinberg p-values of < 0.001. To maximize the power to detect genetic associations due to common genetic risk factors, SNPs with allele frequencies of < 5% (n = 1) or with fewer than three individuals homozygous for the rare allele (n = 3) were also excluded from analysis. In order to control for potential confounding due to population substructure (e.g., race/ethnicity), 106 ancestry informative marker (AIM) SNPs were genotyped. Nineteen SNPs among the 5 candidate genes (i.e., CLOCK, CRY1, PER1, PER2, PER3) passed all quality-control filters and were included in the genetic association analyses.
Actigraphy and sleep diary
Sleep and activity were estimated with a noninvasive battery-operated wrist actigraph microprocessor with a piezoelectric beam that detects movement and acceleration (Mini Motionlogger Actigraph model AAM-32, Ambulatory Monitoring, Inc. Ardsley, NY). Actigraphy provides continuous movement counts and data were sampled in 30-second epochs using zero-crossing mode. The actigraphy monitor was worn continuously on the nondominant wrist for 72 hours on three consecutive weekdays between Monday and Friday to control for potential weekend variability and to reduce subject burden in this chronic illness population. Sleep diaries were also completed each morning and evening of the actigraphy monitoring period for the purpose of cross-validating bed times and wake times. Wrist actigraphy has been validated with polysomnography measures of sleep and wake time for healthy and disturbed sleepers (Cole et al., 1992; Ancoli-Israel et al., 1997; Lichstein et al., 2006).
To reduce researcher scoring bias, actigraphy data were analyzed using an automatic sleep scoring program with the Cole-Kripke algorithm (Action4® Software Program, Ambulatory Monitoring Inc.). Bedtime and final wake times used in sleep scoring were determined by one of two approaches: (1) participant pressing the event marker on the actigraph to indicate “lights out” and “lights on” or (2) if no reliable event marker indication, the diary entry of clock time was used if it matched with a 50% change in movement during the same 10-min block of time on actigraphy. The Action 4 automatic scoring program was also used to conduct cosinor analysis and calculate lag correlations to estimate circadian rhythm strength and phase timing phenotypes. To obtain valid and reliable circadian rhythm estimates, we required a minimum of 48 hours of continuous actigraphy data with no more than 60 consecutive minutes of missing data on any given day and a minimum of either two peaks and a nadir or one peak and two nadirs; actigraphy recordings that did not meet these criteria (n = 28) were excluded from analyses.
Sleep disruption and duration phenotypes were estimated using two parameters: 1) wake after sleep onset (WASO) as a measure of disrupted sleep or poor sleep maintenance, and 2) total sleep time (TST) in minutes as a measure of sleep duration. WASO was standardized as a percentage of the person’s sleep period to control for varying sleep durations. The intraclass correlation coefficient across the 3 nights was 0.83 for WASO and 0.76 for TST. The 3-night means for WASO and TST were used for all analyses.
Circadian rhythm strength phenotypes were estimated from four parameters derived from the cosinor analysis and lag correlations: 1) middle estimated statistic of rhythm (mesor), which estimates the 24-hour adjusted mean level of activity, 2) amplitude, the difference between the mesor and peak (or nadir) activity, 3) circadian quotient, the ratio of amplitude to mesor, and 4) 24-hour autorcorrelation, an indicator of circadian rhythm strength.
Circadian phase timing phenotypes were estimated using three parameters: acrophase (time of peak of activity derived from the cosinor analysis) and bed times and wake times as estimates of sleep timing. The intraclass correlation coefficient across the 3 nights was 0.79 for bedtime, and 0.85 for wake time, and 3-night means were used for all analyses.
Self-reported habitual bed times, wake times, and chronotype
In addition to the 3-day sleep diary entries, habitual bed times and wake times were taken from the two items in the Pittsburgh Sleep Quality Index (PSQI) that ask about usual bed time and wake time during the prior month (Buysse et al., 1989). In cases where the participant reported a range for bed-time or final wake time, the range was averaged to obtain the estimates. The 19-item Horne-Ostberg Morningness-Eveningness Questionnaire (MEQ) was administered to all participants (Horne & Ostberg, 1976). While valid and reliable in most healthy populations, many of our participants had difficulty completing the entire 19-item instrument and thus, this measure was used to validate other measures of circadian phase, but was not included as a phenotype in this analysis.
Statistical Analysis
All statistical analyses were conducted using Stata (version 13, College Station, TX). Descriptive statistics were used to summarize demographic and clinical characteristics. Square root transformation was sufficient to normalize skewed distributions for WASO, wake time (PSQI), and CD4+ T-cell count, log transformation was sufficient to normalize acrophase, bed time (actigraphy and PSQI), and wake time (actigraphy) values, and square transformation was sufficient to normalize mesor and amplitude values. CD4+ T-cell count and HIV viral load were analyzed both as continuous variables and in clinically meaningful categories. Demographic and clinical associations with the sleep and rhythm parameters were evaluated using Spearman correlations, independent sample t-tests, or analysis of variance with Scheffé post hoc tests. Allele and genotype frequencies were determined by gene counting. Hardy-Weinberg equilibrium was assessed by the chi-square exact test. Measures of linkage disequilibrium (i.e., D′ and r2) were computed from participants’ genotypes with Haploview 4.1 (Barrett et al., 2005).
Unadjusted genetic associations with each sleep/rhythm parameter were determined using linear regression models. Three genetic models (i.e., additive, dominant, recessive) were tested, and the model that best fit the data by maximizing the significance of the p-value (barring trivial improvements of delta < 10%) was reported for each SNP. Genetic markers were further evaluated in adjusted linear regression models controlling for relevant covariates. Given evidence that sleep phenotypes differ by ancestry (Halder et al., 2015), all regression models controlled for genomic estimates of ancestry (described below) as well as self-reported race/ethnicity (i.e., White/Caucasian, Black/African American, other). In addition, all demographic and clinical variables associated with the sleep/rhythm parameters (p<.10) were evaluated as potential covariates. Covariates were retained if their significance was p<.05 prior to including genotype in the model. A model was fit for each genetic marker to estimate its unique contribution to the sleep/rhythm parameter when controlling for relevant demographic and clinical covariates. Given prior evidence that HIV interacts with CLOCK and PER3 (Konig et al., 2008), interactions between each genetic marker and measures of HIV exposure (i.e., HIV viral load, time since HIV diagnosis) were also evaluated.
Ancestry informative markers (AIMs) are used to minimize bias due to population substructure (Hoggart et al., 2003; Halder et al., 2008; Tian et al., 2008). Homogeneity in ancestry among participants was estimated by principal component analysis with orthogonal rotation (Price et al., 2006) using HelixTree software (GoldenHelix, Bozeman, MT). With 106 AIMs included in this analysis, principal components (PC) were sought that distinguished the major racial/ethnic groups in the sample (i.e., White/Caucasian, Black/African American, other) by visual inspection of scatterplots of orthogonal PCs (e.g., PC1 versus PC2, PC2 versus PC3). This procedure was repeated until no discernible clustering of participants by self-reported race/ethnicity was possible. The first three PCs for the AIMs were included as genomic estimates of ancestry in all adjusted regression models to allow for potential confounding due to genomic differences in ancestry.
Results
Sample Characteristics
A convenience sample of 350 adults with HIV was enrolled in the study, and 61 participants were excluded prior to analysis: 31 screened positive for illicit drugs, 2 were unable to submit a urine or blood sample, and 28 had incomplete or invalid actigraphy data. Sample characteristics for the 289 participants included in the analysis are presented in Table 1. The sample was ethnically diverse and predominantly male, reflecting the local population of adults with HIV. Participants had been living with HIV for an average of 12.1 ± 6.9 years; AIDS had been diagnosed in 51%; 29% with a current AIDS diagnosis had a CD4+ T-cell count below 200 cells/mm3. Most were unemployed and receiving medical disability assistance (84%), 71% were currently receiving ART, and study participants were taking a daily average of 5.9 ± 4.0 different medications (median 6, range 0–20).
Table 1.
Sleep disruption (WASO) and duration (TST) by demographic and clinical characteristics (n = 289)
| N | Total mean ± SD or n (%) | WASO% rho or mean ± SD | TST mins rho or mean ± SD | Statistics (when p<.10) | |
|---|---|---|---|---|---|
| Age (years), range 22–77 | 289 | 44.9 ± 8.4 | rho = .028 | rho = −.047 | |
| Gender | 289 | ||||
| Male | 193 (67%) | 21.0 ± 15.5 | 369 ± 100 | ||
| Female | 73 (25%) | 19.0 ± 12.6 | 384 ± 97 | ||
| Transgender | 23 (8%) | 25.1 ± 14.4 | 349 ± 96 | ||
| Race | 289 | WASO: F(2,286) = 11.4, p < .001a | |||
| Caucasian | 118 (41%) | 16.1 ± 12.0 | 407 ± 93 | TST: F(2,286) = 15.4, p < .001a | |
| African American | 110 (38%) | 24.6 ± 14.4 | 339 ± 92 | ||
| Other | 61 (21%) | 23.1 ± 17.8 | 358 ± 99 | ||
| Employment | 289 | WASO: t(287) = 3.15, p = .002 | |||
| Employed or in school | 46 (16%) | 15.3 ± 12.9 | 395 ± 85 | TST: t(287) = 1.79, p = .075 | |
| Unemployed or on disability | 243 (84%) | 21.9 ± 14.9 | 366 ± 101 | ||
| CD4+ T-cell count (cells/mm3) | 276 | ||||
| Mean ± SD | 453 ± 267 | rho = −.166 | rho = .183 | WASO: p = .006; TST: p = .002 | |
| < 200 | 47 (17%) | 23.5 ± 15.2 | 350 ± 93 | ||
| ≥200 | 229 (83%) | 20.2 ± 14.6 | 374 ± 99 | ||
| Viral load (log10 copies/mL) | 270 | ||||
| Mean ± SD | 2.64 ± 1.20 | rho = .102 | rho = −.107 | WASO: p = .096; TST: p = .078 | |
| Undetectable | 137 (51%) | 18.7 ± 13.0 | 382 ± 93 | WASO: t(268) = 1.87, p = .062 | |
| Detectable | 133 (49%) | 22.5 ± 16.0 | 361 ± 103 | TST: t(268) = 1.78, p = .077 | |
| Antiretroviral therapy (ART) | 289 | ||||
| Not on treatment | 85 (29%) | 21.9 ± 13.3 | 367 ± 96 | ||
| On treatment | 204 (71%) | 20.4 ± 15.3 | 373 ± 100 | ||
| Depression | 287 | TST: t(285) = 2.24, p = .026 | |||
| No (CESD<16) | 147 (51%) | 21.6 ± 14.8 | 359 ± 91 | ||
| Yes (CESD≥16) | 140 (49%) | 19.9 ± 14.7 | 385 ± 104 | ||
| Neuroleptic medication use | 288 | WASO: t(286) = 1.74, p = .083 | |||
| No | 261 (91%) | 21.3 ± 15.1 | 367 ± 99 | TST: t(286) = 2.15, p = .033 | |
| Yes | 27 (9%) | 15.5 ± 9.5 | 410 ± 93 | ||
| Opiate medication use | 288 | WASO: t(286) = 3.40, p < .001 | |||
| No | 210 (73%) | 19.1 ± 14.3 | 374 ± 95 | ||
| Yes | 78 (27%) | 25.2 ± 15.2 | 364 ± 108 | ||
| Antiemetic medication use | 288 | WASO: t(286) = 2.67, p = .008 | |||
| No | 277 (96%) | 20.3 ± 14.5 | 374 ± 97 | TST: t(286) = 2.24, p = .026 | |
| Yes | 11 (4%) | 32.6 ± 15.9 | 306 ± 121 | ||
| Smoker? | 288 | WASO: t(286) = 1.71, p = .088 | |||
| No | 125 (43%) | 19.1 ± 13.8 | 386 ± 91 | TST: t(286) = 2.21, p = .028 | |
| Yes | 163 (57%) | 22.1 ± 15.4 | 360 ± 103 | ||
| Body mass index | 289 | ||||
| Male | 193 | 26.0 ± 4.8 | rho = .137 | rho = −.252 | WASO: p = .058; TST: p < .001 |
| Female | 73 | 29.0 ± 6.3 | rho = .114 | rho = .023 | |
| Transgender | 23 | 28.9 ± 6.8 | rho = −.079 | rho = .163 | |
| Waist circumference (cm) | 289 | ||||
| Male | 193 | 93.7 ± 12.4 | rho = .121 | rho = −.204 | M: WASO: p = .093; TST: p = .004 |
| Female | 73 | 93.1 ± 14.1 | rho = .215 | rho = −.079 | F: WASO: p = .067 |
| Transgender | 23 | 95.6 ± 13.4 | rho = −.026 | rho = .157 |
WASO and CD4+ T-cell count analyses were conducted with square root-transformed values. WASO and TST were unrelated to use of sleep medication, antidepressants, or anxiolytics, or to consumption of alcohol or caffeine (data not shown). CESD, Center for Epidemiological Studies – Depression Scale; SD, standard deviation; TST, total sleep time; WASO, wake after sleep onset. Bolded statistics are significant (p<.05).
Post-hoc analyses indicated that Caucasians had significantly less WASO and more TST than African-Americans and other races.
Sleep duration (TST) and disruption (WASO)
The sample generally had short sleep duration and poor sleep maintenance. Almost half (45%, n = 130) of the sample averaged less than 6 hours of sleep at night, and 35% (n = 101) had mean WASO values of more than 25% of their sleep period. As shown in Table 1, WASO and TST were both associated with race, and higher WASO was also associated with unemployment. Of the clinical variables, lower CD4+ T-cell count and antiemetic medication use were both associated with higher WASO and lower TST; depressive symptoms and neuroleptic medication were associated with more TST, and opiate medication was associated with more WASO. Consumption of alcohol or caffeine was unrelated to WASO and TST, but smoking was associated with shorter sleep duration (TST). Higher BMI and larger waist circumference were associated with shorter TST among men, but this pattern was not evident among women and was weaker among transgender adults.
Circadian rhythm strength
All circadian rhythm strength parameters were obtained from the wrist actigraphy analyses. The mean mesor value was 68.6 ± 14.5, mean amplitude was 45.6 ± 10.6, and mean circadian quotient was 0.68 ± 0.15. The mean 24-hour autocorrelation was 0.38 ± 0.16. As shown in Table 2, these rhythm parameters differed by gender, race, employment status, CD4+ T-cell count, viral load, smoking status, depressive symptoms, as well as use of sleep, opiate, and antidepressant medication. In general, stronger circadian rhythms were observed for women, Caucasians, and participants who were employed, did not smoke, were not depressed, and did not take sleep, opiate or antidepressant medication. Stronger rhythms were also associated with higher CD4+ T-cell counts and lower viral load. Among men, higher BMI and larger waist circumference were associated with lower circadian quotients.
Table 2.
Circadian rhythm strength parameters by demographic and clinical characteristics
| Mesor rho or mean ± SD n=278 | Amplitude rho or mean ± SD n=278 | Circadian Quotient rho or mean ± SD; n=278 | Auto-correlation rho or mean ± SD; n=278 | Statistics (when p < .10) | |
|---|---|---|---|---|---|
| Age (years) | rho = .034 | rho = −.010 | rho = −.022 | rho = .030 | |
| Gender | AMP: F(2,275) = 3.78, p = .024; F>M | ||||
| Male | 67.7 ± 14.7 | 44.4 ± 11.0 | 0.67 ± 0.15 | 0.38 ± 0.15 | CQ: F(2,275) = 3.36, p = .036 |
| Female | 69.0 ± 14.6 | 48.4 ± 9.8 | 0.72 ± 0.16 | 0.40 ± 0.15 | |
| Transgender | 74.5 ± 11.4 | 47.1 ± 8.4 | 0.61 ± 0.13 | 0.39 ± 0.15 | |
| Race | MESOR: F(2,275)=5.06, p = .007a | ||||
| Caucasian | 66.6 ± 12.8 | 47.7 ± 10.6 | 0.73 ± 0.14 | 0.41 ± 0.16 | AMP: F(2,275) = 5.24, p = .006 b |
| African American | 72.0 ± 15.4 | 44.9 ± 10.0 | 0.64 ± 0.15 | 0.38 ± 0.14 | CQ: F(2,275) = 10.3, p < .001c |
| Other | 66.0 ± 15.3 | 42.6 ± 11.1 | 0.66 ± 0.15 | 0.35 ± 0.16 | AUTO: F(2,275) = 3.32, p = .038 d |
| Employment | CQ: t(276) = 2.15, p = .032 | ||||
| Employed or in school | 66.4 ± 15.0 | 47.4 ± 11.3 | 0.72 ± 0.14 | 0.40 ± 0.16 | |
| Unemployed or on disability | 69.0 ± 14.5 | 45.2 ± 10.5 | 0.67 ± 0.16 | 0.38 ± 0.15 | |
| CD4+ T-cell count (cells/mm3) | n=265 | n=265 | n=265 | n=265 | AMP: p = .045; CQ: p = .001 |
| rho = −.114 | rho = .123 | rho = .197 | rho = .047 | AMP: t(263) = 1.78, p = .077 | |
| < 200 | 71.0 ± 15.4 | 43.7 ± 8.7 | 0.64 ± 0.15 | 0.39 ± 0.13 | CQ: t(263) = 2.08, p = .039 |
| ≥200 | 68.3 ± 13.9 | 46.2 ± 10.7 | 0.69 ± 0.15 | 0.39 ± 0.15 | |
| Viral load (log10 copies/mL) | n=259 | n=259 | n=262 | n=261 | |
| rho = .018 | rho = −.152 | rho = −.117 | rho = −.105 | AMP: p = .014; CQ: p = .084; | |
| Undetectable | 68.6 ± 13.3 | 46.8 ± 10.8 | 0.69 ± 0.16 | 0.40 ± 0.15 | AUTO: p = .093 |
| Detectable | 69.0 ± 15.0 | 44.9 ±10.1 | 0.67 ± 0.15 | 0.38 ± 0.15 | |
| Antiretroviral therapy | |||||
| Not on treatment | 69.6 ± 13.1 | 45.8 ± 9.8 | 0.67 ± 0.14 | 0.37 ± 0.16 | |
| On treatment | 68.1 ± 15.1 | 45.5 ± 11.0 | 0.69 ± 0.16 | 0.39 ± 0.15 | |
| Sleep medication use | AUTO: t(276) = 1.98, p = .048 | ||||
| No (n=240) | 68.6 ± 14.5 | 45.9 ± 10.6 | 0.68 ± 0.15 | 0.39 ± 0.15 | |
| Yes (n=49) | 68.1 ± 14.8 | 44.2 ± 10.6 | 0.66 ± 0.15 | 0.34 (0.16 | |
| Antidepressant medication use | MESOR: t(276) = 2.73, p = .007 | ||||
| No (n=166) | 70.5 ± 13.2 | 47.4 ± 10.2 | 0.69 ± 0.15 | 0.40 ± 0.15 | AMP: t(276) = 3.43, p < .001 |
| Yes (n=112) | 65.7 ± 16.0 | 43.0 ± 10.7 | 0.67 ± 0.16 | 0.36 ± 0.16 | AUTO: t(276) = 2.21, p = .028 |
| Depression | MESOR: t(275) = 2.68, p = .010 | ||||
| No (CESD<16) | 70.7 ± 13.4 | 46.8 ± 10.0 | 0.68 ± 0.15 | 0.41 ± 0.15 | AMP: t(275) = 1.85, p = .093 |
| Yes (CESD≥16) | 66.1 ± 15.2 | 44.4 ± 11.2 | 0.69 ± 0.16 | 0.36 ± 0.15 | AUTO: t(275) = 2.78, p = .006 |
| Opiate medication use | AMP: t(276) = 1.74, p= .083 | ||||
| No | 68.7 ± 14.9 | 46.2 ± 11.0 | 0.69 ± 0.15 | 0.40 ± 0.15 | AUTO: t(276) = 3.10, p = .002 |
| Yes | 68.0 ± 13.8 | 44.1 ± 9.5 | 0.67 ± 0.15 | 0.34 ± 0.15 | |
| Smoker? | AMP: t(276) = 3.70, p < .001 | ||||
| No | 67.9 ± 14.2 | 48.0 ± 11.2 | 0.72 ± 0.14 | 0.43 ± 0.16 | CQ: t(276) = 3.34, p < .001 |
| Yes | 69.0 ± 14.8 | 43.8 ± 9.9 | 0.66 ± 0.16 | 0.36 ± 0.15 | AUTO: t(276) = 3.67, p < .001 |
| Alcohol consumption | AMP: t(276) = 1.99, p = .048 | ||||
| None in last 3 days (n=243) | 68.9 ± 14.6 | 46.1 ± 10.4 | 0.69 ± 0.15 | 0.39 ± 0.15 | AUTO: t(276) = 1.68, p = .093 |
| Any in last 3 days (n=35) | 66.3 ± 13.9 | 42.1 ± 11.4 | 0.65 ± 0.16 | 0.35 ± 0.17 | |
| Body mass index | |||||
| Male (M) | rho = .091 | rho = −.087 | rho = −.187 | rho = −.019 | CQ: p = .010 |
| Female (F) | rho = .110 | rho = .095 | rho = .067 | rho = .142 | |
| Transgender (T) | rho = −.316 | rho = −.086 | rho = .120 | rho = −.050 | |
| Waist circumference (cm) | |||||
| Male | rho = .052 | rho = −.086 | rho = −.170 | rho = −.021 | CQ: p = .020 |
| Female | rho = .151 | rho = .084 | rho = −.005 | rho = .138 | |
| Transgender | rho = −.330 | rho = −.070 | rho = .163 | rho = −.047 | |
Mesor and amplitude analyses were conducted with square-transformed values and CD4+ T-cell count analyses were conducted with square root-transformed values. Circadian rhythm parameters were unrelated to use of anxiolytics, neuroleptics, or anti-emetics, or to consumption of caffeine (data not shown). AMP, amplitude; AUTO, 24-hour autocorrelation; CESD, Center for Epidemiological Studies – Depression Scale; CQ, circadian quotient; SD, standard deviation. Bolded statistics are significant (p<.05).
Post-hoc testing indicated that African-Americans had higher mesor than Caucasians and other races.
Post-hoc testing indicated that Caucasian had higher amplitude than other races.
Post-hoc testing indicated that Caucasians had higher circadian quotients than African-Americans and other races.
Despite the significant omnibus test, post-hoc testing indicated no significant racial differences in 24-hour autocorrelation.
Circadian phase timing
Measures of circadian phase were obtained from both the wrist actigraphy and self-report questionnaires. The sample had a mean acrophase of 14:53 ± 1:31, a mean bed time of 23:00 ± 1:29 by actigraphy, and 22:36 ±1:38 by PSQI self-report. Mean wake time was 7:07 ± 1:28 by actigraphy and 7:02 ± 1:43 by PSQI self-report. For the subset of 214 participants who also completed the entire MEQ, the correlations between the MEQ and our five measures of circadian phase ranged from 0.41 to 0.52 (all p<.001). As shown in Table 3, measures of circadian phase differed by race, employment, AIDS diagnosis, ART, smoking status, and use of opiate medication or alcohol. In general, later circadian phase timing was observed among Caucasians and participants who were employed, diagnosed with AIDS, taking ART, consumed alcohol, and did not smoke or take opiate medication. Among men, higher BMI and larger waist circumference were associated with earlier acrophase and wake time. Depressive symptoms were associated with slightly later wake times, but only by self-report.
Table 3.
Circadian rhythm timing parameters by demographic and clinical characteristics
| Bed time | Wake Time | Statistics (when p<.10) | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| Acrophase rho or mean ± SD n=278 | Actigraphy rho or mean ± SD n=289 | PSQI rho or mean ± SD n=284 | Actigraphy rho or mean ± SD n=289 | PSQI rho or mean ± SD n=277 | ||
| Age (years) | rho = −.082 | rho = .022 | rho = .016 | rho = −.024 | rho = −.017 | |
| Gender | ACRO: F(2,275) = 2.99, p = .052 | |||||
| Male | 15:02 ± 1:34 | 23:05 ± 1:30 | 22:42 ± 1:34 | 7:11 ± 1:35 | 7:01 ± 1:41 | |
| Female | 14:30 ± 1:16 | 22:45 ± 1:26 | 22:16 ± 1:30 | 6:58 ± 1:08 | 6:56 ± 1:40 | |
| Transgender | 14:53 ± 1:47 | 23:06 ± 1:34 | 22:40 ± 2:22 | 7:13 ± 1:23 | 7:26 ± 2:05 | |
| Race | ACRO: F(2,275)=5.09, p=.007a | |||||
| Caucasian | 15:13 ± 1:43 | 23:09 ± 1:36 | 22:52 ± 1:41 | 7:31 ± 1:42 | 7:26 ± 1:48 | BED-P: F(2,281)=4.86, p=.008a |
| African American | 14:33 ± 1.15 | 22:49 ± 1:14 | 22:13 ± 1:21 | 6:41 ± 1:05 | 6:32 ± 1:29 | WAKE-A: F(2,286)=8.11, p<.001a |
| Other | 14:51 ± 1:25 | 23:02 ± 1:39 | 22:43 ± 1:51 | 7:09 ± 1:22 | 7:05 ± 1:43 | WAKE-P: F(2,274)=7.75, p<.001a |
| Employment | ACRO: t(276) = 1.80, p = .073 | |||||
| Employed or in school | 15:16 ± 1:36 | 23:27 ± 1:30 | 22:57 ± 1:24 | 7:34 ± 1:34 | 7:30 ± 1:58 | BED-A: t(287) = 2.26, p = .024 |
| Unemployed/ disabled | 14:49 ± 1:30 | 22:55 ± 1:28 | 22:31 ±1:40 | 7:03 ± 1:26 | 6:56 ± 1:39 | BED-P: t(282) = 1.73, p = .084 |
| WAKE-A: t(287) = 2.15, p =.032 | ||||||
| WAKE-P: t(275) = 1.97, p =.050 | ||||||
| CD4+ T-cell count (cells/mm3) | (n=265) | |||||
| rho = −.065 | rho = −.075 | rho = −.041 | rho = −.022 | rho = .000 | ||
| < 200 | 14:57 ± 1:01 | 23:05 ± 1:29 | 22:30 ± 1:34 | 7:01 ± 1:06 | 6:57 ± 1:28 | |
| ≥200 | 14:49 ± 1:35 | 22:58 ± 1:28 | 22:36 ± 1:38 | 7:06 ± 1:30 | 7:01 ± 1:45 | |
| Viral load (log10copies/mL) | (n=259) | |||||
| rho = −.004 | rho = −.013 | rho = .037 | rho = −.015 | rho = −.053 | ||
| Undetectable | 14:57 ± 1:39 | 23:03 ± 1:34 | 22:34 ± 1:38 | 7:11 ± 1:36 | 7:08 ± 1:47 | |
| Detectable | 14:47 ± 1:19 | 22:57 ± 1:22 | 22:39 ± 1:36 | 7:03 ± 1:16 | 6:55 ± 1:36 | |
| AIDS diagnosis | ACRO: t(276) = 1.72, p = .087 | |||||
| No | 14:44 ± 1:26 | 22:45 ± 1:20 | 22:23 ± 1:28 | 7:00 ± 1:20 | 6:54 ± 1:30 | BED-A: t(287) = 2.76, p = .006 |
| Yes | 15:03 ± 1:35 | 23:14 ± 1:36 | 22:47 ± 1:45 | 7:15 ± 1:35 | 7:09 ± 1:53 | BED-P: t(282) = 1.99, p = .048 |
| Antiretroviral therapy | ACRO: t(276) = 2.12, p = .035 | |||||
| Not on treatment | 14:36 ± 1:29 | 22:45 ± 1:23 | 22:30 ± 1:24 | 6:59 ± 1:22 | 6:55 ± 1:41 | BED-A: t(287) = 1.78, p = .076 |
| On treatment | 15:01 ± 1:32 | 23:06 ± 1:31 | 22:38 ± 1:43 | 7:11 ± 1:30 | 7:04 ± 1:43 | |
| Depression | WAKE-A: t(285) = 2.07, p =.040 | |||||
| No (CESD<16) | 14:50 ± 1:37 | 23:02 ± 1:28 | 22:35 ± 1:31 | 6:58 ± 1:33 | 6:58 ± 1:38 | |
| Yes (CESD≥16) | 14:57 ± 1:25 | 22:56 ± 1:29 | 22:35 ± 1:44 | 7:17 ± 1:21 | 7:06 ± 1:48 | |
| Opiate medication | ACRO: t(276) = 1.73, p = .085 | |||||
| No | 14:59 ± 1:30 | 23:06 ±1:22 | 22:39 ± 1:31 | 7:07 ± 1:28 | 7:03 ± 1:36 | BED-A: t(286) = 2.27, p = .024 |
| Yes | 14:39 ± 1:35 | 22:41 ± 1:45 | 22:26 ± 1:55 | 7:09 ± 1:27 | 6:57 ± 2:00 | |
| Smoker? | ACRO: t(276) = 2.92, p = .004 | |||||
| No | 15:12 ± 1:34 | 23:12 ± 1:30 | 22:52 ± 1:36 | 7:28 ± 1:30 | 7:24 ± 1:45 | BED-A: t(286) = 1.92, p = .056 |
| Yes | 14:40 ± 1:27 | 22:51 ± 1:29 | 22:22 ± 1:37 | 6:51 ± 1:23 | 6:44 ± 1:38 | BED-P: t(282) = 2.64, p = .008 |
| WAKE-A: t(282)=2.64, p <.001 | ||||||
| WAKE-P: t(275)=3.22, p<.001 | ||||||
| Alcohol consumption | ACRO: t(276) = 2.00, p = .047 | |||||
| None in last 3 days | 14:49 ± 1:30 | 22:56 ± 1:28 | 22:36 ± 1:36 | 7:04 ± 1:26 | 6:58 ± 1:38 | BED-A: t(286) = 1.85, p = .065 |
| Any in last 3 days | 15:23 ± 1:38 | 23:26 ± 1:33 | 22:32 ± 1:49 | 7:29 ± 1:39 | 7:24 ± 2:11 | |
| Body mass index | ||||||
| Male | rho = −.133 | rho = −.049 | rho = −.008 | rho = −.225 | rho = −.178 | Males: ACRO: p = .070; |
| Female | rho = −.044 | rho = −.180 | rho = .127 | rho = −.147 | rho = −.192 | WAKE-A: p =.002; |
| Transgender | rho = .026 | rho = −.294 | rho = −.148 | rho = −.166 | rho = .002 | WAKE-P: p = .014 |
| Waist circumference (cm) | Males: ACRO: p =.022; | |||||
| Male | rho = −.167 | rho = −.070 | rho = .001 | rho = −.208 | rho = .171 | WAKE-A: p =.004; |
| Female | rho = −.068 | rho = −.119 | rho = .100 | rho = −.163 | rho = −.210 | WAKE-P: p = .018 |
| Transgender | rho = .255 | rho = −.106 | rho = .047 | rho = .059 | rho = .192 | Females: WAKE-P: p = .091 |
Analyses were based on log-transformed acrophase, bedtime (actigraphy and PSQI), and wake time (actigraphy) values and square root-transformed wake time (PSQI) and CD4+ T-cell count values. None of the five circadian timing parameters were associated with use of sleep medications, antidepressants, anxiolytics, neuroleptics, opiates, or anti-emetics (data not shown). ACRO, acrophase; BED-A, bedtime based on actigraphy; BED-P, habitual bedtime based on self-report PSQI; CESD, Center for Epidemiological Studies – Depression Scale; PSQI, Pittsburgh Sleep Quality Index; SD, standard deviation. WAKE-A, wake time based on actigraphy; WAKE-P, habitual waketime based on self-report PSQI. Bolded statistics are significant (p<.05).
Post-hoc analyses of race indicated that Caucasians had significantly later circadian timing than African-Americans.
Genetic Associations with Sleep Disruption (WASO) and Duration (TST)
Of the 19 SNPs examined, 4 SNPs (i.e., PER1 rs2253820, PER2 rs10198215 and rs10462023, PER3 rs2640908) in 3 of the 5 candidate genes were significantly associated with WASO in unadjusted analyses, and 5 SNPs (i.e., CLOCK rs11932595, PER1 rs2253820, and PER2 rs10198215, rs2304674, and rs10462023) in 3 of the 5 candidate genes were significantly associated with TST in unadjusted analyses (Supplemental Table 1). To better estimate the magnitude of the genetic associations when adjusting for relevant covariates, multiple linear regression models were fit predicting WASO, and separate models were fit predicting TST. Genomic estimates of ancestry and self-reported race/ethnicity were forced into all models. All WASO models were also adjusted for gender, the interaction of gender and race/ethnicity, CD4+ T-cell count, waist circumference, and use of opiate or antiemetic medication. All TST models were also adjusted for gender, use of antiemetic or neuroleptic medication, smoking status, and waist circumference. One of the TST models (i.e., PER2 rs10198215) was also adjusted for viral load and its significant interaction with genotype.
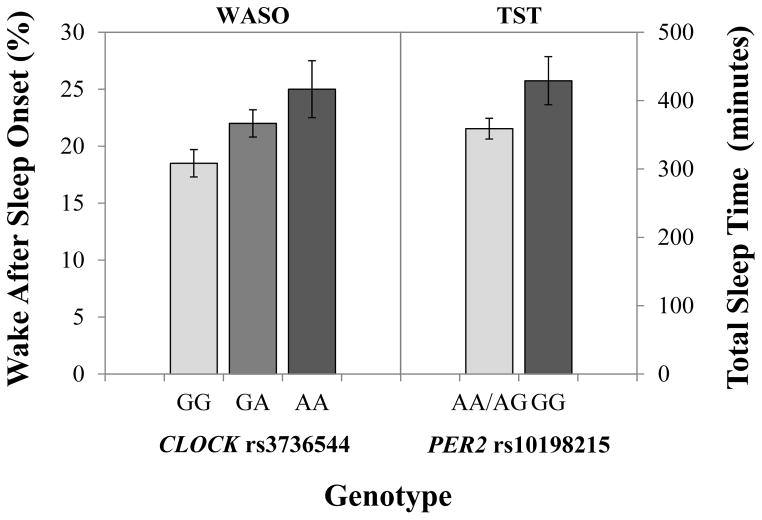
Of the 4 SNPs associated with WASO and the 5 SNPs associated with TST in unadjusted analyses (Supplemental Table 1), only one (PER2 rs10198215) remained significant in adjusted analyses (Table 4). PER2 rs10198215 and its interaction with viral load were significantly associated with TST after adjusting for relevant covariates (Table 5). The overall model explained 20.6% of the variance in TST, with the genotype and its interaction with viral load accounting for 3.1% of the variance (Table 5). Adjusted differences in TST by this PER2 polymorphism are illustrated in Figure 1.
Table 4.
Patterns of adjusted associations between genotypes, their interactions with HIV, and sleep/circadian rhythm measures
| GENE | SNP | SLEEP DISRUPTION AND DURATION (SNP-HIV interaction) | CIRCADIAN RHYTHM STRENGTH (SNP-HIV interaction) | CIRCADIAN RHYTHM PHASE (SNP-HIV interaction) |
|---|---|---|---|---|
| CLOCK | rs1801260 | BED-A (x YRS) a | ||
| rs3736544 | WASO (none) | |||
| rs6849474 | AUTO (x YRS) | |||
| rs11932595 | ||||
| rs11735267 | WASO (none)b | |||
| rs2070062 | BED-A (x YRS) | |||
| rs6850524 | ||||
|
| ||||
| CRY1 | rs10746075 | CQ (x YRS) | ||
|
| ||||
| PER1 | rs2253820 | MESOR (x YRS) | WAKE-P (x VL) | |
| rs885747 | MESOR (x YRS) | |||
|
| ||||
| PER2 | rs10198215 | TST (x VL)c | MESOR (x VL)c | |
| CQ (x VL, YRS)c,d | ||||
| rs2304674 | AUTO (x YRS) | |||
| rs10462023 | BED-A (none) | |||
| WAKE-A (x VL, YRS)d | ||||
| WAKE-P (none) | ||||
|
| ||||
| PER3 | rs228729 | AMP (x VL) | WAKE-P (x YRS)c | |
| AUTO (x VL) | ||||
| rs228682 | WAKE-A (none) c | |||
| rs1012477 | AMP (x VL) | WAKE-P (x VL) | ||
| rs707465 | ACRO (none)c | |||
| WAKE-A (none) c | ||||
| WAKE-P (none)c | ||||
| rs4908482 | AMP (x VL) | |||
| rs2640908 | AUTO (none)c | BED-A (x YRS) | ||
| WAKE-A (YRS) c | ||||
Interactions (x) between genotype and HIV are listed in parentheses: none, no significant interaction; VL, interaction with viral load (log-transformed); YRS, interaction with years since HIV diagnosis.
ACRO, acrophase; AMP, amplitude; AUTO, 24-hour autocorrelation; BED-A, bed time by actigraphy; BED-P, habitual bed time by self-report PSQI; CLOCK, circadian locomotor output cycles kaput gene; CRY, cryptochrome gene; CQ, circadian quotient (amplitude/mesor); PER, period circadian clock gene; PSQI, Pittsburgh Sleep Quality Index; SNP, single nucleotide polymorphism; TST, total sleep time; WAKE-A, wake time by actigraphy; WAKE-P, habitual wake time by self-report PSQI; WASO, wake after sleep onset.
CLOCK rs1801260 was in complete disequilibrium with CLOCK rs2070062.
CLOCK rs11735267 was in complete disequilibrium with CLOCK rs3736544.
This association was also significant in unadjusted analyses.
The genotype interacted with both viral load and years since HIV diagnosis in separate models.
Table 5.
Significant adjusted associations between genotype and sleep-wake rhythm measures
| GENE | SNP / *interaction | Phenotype | Model | n | β | t | p | ΔR2 | R2 | Full models |
|---|---|---|---|---|---|---|---|---|---|---|
| CLOCK | rs3736544a | WASO | A | 273 | .157 | 2.85 | .005 | .024 | .247 | F(16,256) = 5.26, p<.001 |
|
| ||||||||||
| rs6849474 | AUTO | R | 275 | −.351 | 3.03 | .003 | .030 | .161 | F(14,260) = 3.57, p<.001 | |
| YRS*rs6849474 | .300 | 2.59 | .010 | |||||||
|
| ||||||||||
| rs2070062b | BED-A | D | 286 | −.165 | 1.39 | .165 | .020 | .074 | F(10,275) = 2.19, p=.019 | |
| YRS*rs2070062 | .269 | 2.12 | .035 | |||||||
|
| ||||||||||
| CRY1 | rs10746075 | CQ | R | 276 | .182 | 1.59 | .113 | .015 | .170 | F(12,263) = 4.48, p<.001 |
| YRS *rs10746075 | −.246 | 2.10 | .037 | |||||||
|
| ||||||||||
| PER1 | rs885747 | MESOR | R | 276 | .195 | 1.60 | .110 | .018 | .083 | F(9,266) = 2.66, p=.006 |
| YRS*rs885747 | −.261 | 2.13 | .034 | |||||||
|
| ||||||||||
| rs2253820 | MESOR | D | 276 | −.155 | 1.24 | .216 | .018 | .082 | F(9,266) = 2.64, p=.006 | |
| YRS*rs2253820 | .263 | 1.98 | .049 | |||||||
|
| ||||||||||
| rs2253820 | WAKE-P | R | 257 | .324 | 1.94 | .053 | .020 | .113 | F(9,247) = 3.51, p<.001 | |
| VL*rs2253820 | −.385 | 2.30 | .022 | |||||||
|
| ||||||||||
| PER2 | rs10198215 | TST | R | 267 | .298 | 2.21 | .028 | .031 | .206 | F(14,252) = 4.66, p<.001 |
| VL*rs10198215 | −.382 | 2.81 | .005 | |||||||
|
| ||||||||||
| rs10198215 | MESOR | R | 257 | −.271 | 1.88 | .062 | .038 | .102 | F(9,247) = 3.12, p=.001 | |
| VL*rs10198215 | .416 | 2.87 | .004 | |||||||
|
| ||||||||||
| rs10198215 | CQ | R | 257 | .232 | 1.64 | .103 | .026 | .172 | F(12,244) = 4.22, p<.001 | |
| VL*rs10198215 | −.339 | 2.38 | .018 | |||||||
|
| ||||||||||
| rs10198215 | CQ | R | 276 | −.282 | 2.48 | .014 | .020 | .175 | F(12,263) = 4.64, p<.001 | |
| YRS*rs10198215 | .257 | 2.29 | .023 | |||||||
|
| ||||||||||
| rs2304674 | AUTO | D | 275 | .263 | 2.17 | .031 | .016 | .148 | F(13,262) = 2.97, p<.001 | |
| YRS*rs2304674 | −.294 | 2.10 | .037 | |||||||
|
| ||||||||||
| rs10462023 | BED-A | A | 286 | −.131 | 2.01 | .045 | .014 | .067 | F(8,277) = 2.50, p=.012 | |
|
| ||||||||||
| rs10462023c | WAKE-A | R | 267 | −.315 | 2.11 | .036 | .018 | .139 | F(10,256) = 4.11, p<.001 | |
| VL*rs10462023 | .327 | 2.23 | .027 | |||||||
|
| ||||||||||
| rs10462023 | WAKE-P | R | 275 | −.133 | 2.21 | .028 | .016 | .106 | F(7,267) = 4.53, p<.001 | |
|
| ||||||||||
| PER3 | rs228729 | AMP | D | 257 | −.382 | 2.67 | .008 | .032 | .205 | F(12,244) = 5.24, p<.001 |
| VL*rs228729 | .471 | 3.10 | .002 | |||||||
|
| ||||||||||
| rs228729 | AUTO | D | 256 | −.381 | 2.57 | .011 | .031 | .156 | F(14,241) = 3.19, p<.001 | |
| VL*rs228729 | .391 | 2.47 | .014 | |||||||
|
| ||||||||||
| rs228729 | WAKE-P | D | 275 | −.178 | 1.51 | .132 | .036 | .126 | F(9,265) = 4.24, p<.001 | |
| YRS*rs228729 | .370 | 2.72 | .007 | |||||||
|
| ||||||||||
| rs228682 | WAKE-A | D | 286 | .113 | 1.98 | .049 | .012 | .136 | F(8,277) = 5.47, p<.001 | |
|
| ||||||||||
| rs1012477 | AMP | D | 257 | .342 | 2.32 | .021 | .018 | .191 | F(12,244) = 4.80, p<.001 | |
| VL*rs1012477 | −.333 | 2.18 | .030 | |||||||
|
| ||||||||||
| rs1012477 | WAKE-P | D | 257 | .312 | 2.09 | .038 | .019 | .112 | F(9,247) = 3.45, p<.001 | |
| VL*rs1012477 | −.344 | 2.25 | .025 | |||||||
|
| ||||||||||
| rs707465 | ACRO | R | 276 | −.134 | 2.20 | .029 | .016 | .094 | F(8,267) = 3.45, p<.001 | |
|
| ||||||||||
| rs707465 | WAKE-A | R | 286 | −.163 | 2.82 | .005 | .025 | .149 | F(8,277) = 6.04, p<.001 | |
|
| ||||||||||
| rs707465 | WAKE-P | R | 275 | −.127 | 2.11 | .036 | .015 | .105 | F(7,267) = 4.46, p<.001 | |
|
| ||||||||||
| rs4908482 | AMP | D | 257 | .292 | 2.05 | .042 | .015 | .189 | F(12,244) = 4.72, p<.001 | |
| VL*rs4908482 | −.344 | 2.13 | .034 | |||||||
|
| ||||||||||
| rs2640908 | AUTO | R | 275 | −.120 | 2.05 | .041 | .014 | .125 | F(12,262) = 3.71, p<.001 | |
|
| ||||||||||
| rs2640908 | BED-A | R | 286 | .203 | 1.90 | .059 | .023 | .077 | F(10,275) = 2.28, p=.014 | |
| YRS*rs2640908 | −.265 | 2.48 | .014 | |||||||
|
| ||||||||||
| rs2640908c | WAKE-A | R | 286 | .103 | 1.01 | .314 | .033 | .158 | F(10,275) = 5.14, p<.001 | |
| YRS*rs2640908 | −.260 | 2.54 | .012 | |||||||
All models adjusted for genomic estimates of ancestry and self-reported race. In addition, WASO (square root-transformed) models also adjusted for self-reported gender, interaction of self-reported race and gender, CD4 T-cell count (square root-transformed), waist circumference, and use of opiate or anti-emetic medication. TST models adjusted for self-reported gender. Mesor (square-transformed) models adjusted for use of anti-depressant medication, amplitude (square-transformed) models adjusted for gender, viral load (log-transformed), use of anti-depressant medication and smoking status. Circadian quotient models adjusted for gender, employment, and smoking status, and autocorrelation models adjusted for gender, use of anti-depressant or opiate medication, depressive symptoms, and smoking status,. Acrophase (log-transformed) models also adjusted for smoking and alcohol use. Actigraphy bed time (log-transformed) models also adjusted for employment and AIDS status, and actigraphy wake time (log-transformed) models also adjusted for smoking status and waist circumference, Habitual self-reported bedtime (log-transformed) and self-reported wake time (square root-transformed) models both adjusted for smoking status. A, additive model; CLOCK, circadian locomotor output cycles kaput gene; D, dominant model; VL, viral load (interaction with SNP); PER2, period 2 gene; PER3, period 3 gene; R, recessive model; R2, proportion of variance in sleep outcome explained by the full model; ΔR2, proportion of variance in sleep outcome accounted for by genotype when adjusting for covariates; SNP, single nucleotide polymorphism; YRS, years since HIV diagnosis (interaction with SNP).
CLOCK rs11735267 was also associated with WASO, but was not included in the table because it is in complete disequilibrium with CLOCK rs3736544.
CLOCK rs1801260 was also associated with bed time by actigraphy, but was not included in the table because it is in complete disequilibrium with CLOCK rs2070062.
The interaction between PER2 rs10462023 and years since HIV diagnosis was also associated with wake time by actigraphy, but since the interaction with viral load was stronger, it is the one reported.
PER3 rs2640908 was also associated with wake time by actigraphy (p=.042) when the interaction with years since HIV diagnosis was excluded, but since the model with the interaction term was stronger, it is the one reported.
Figure 1.
Adjusted mean wake after sleep onset (WASO) and total sleep time (TST) by genotype. Each dose of the CLOCK rs3736544 rare allele (A) was associated with an increase in wake after sleep onset (p=.005). Carriers of two doses of the PER2 rs10198215 rare allele (GG) had significantly longer TST than carriers of the common allele (A; p=.028). The interaction between PER2 rs10198215 and viral load resulted in minimal differences in TST and is not shown.
In addition, two CLOCK SNPs (rs3736544 and rs11735267) that were not associated with WASO in bivariate analyses were significantly associated with WASO after adjusting for relevant covariates (Table 4). Due to the complete collinearity, or shared variance, between these SNPs (i.e., linkage disequilibrium r2 = 1.0), we selected rs3736544 as a surrogate for rs11735267, and the model for rs11735267 is not shown (Table 5). The overall model explained 24.7% of the variance in WASO, with genotype accounting for 2.4% of the variance. Adjusted differences in WASO by CLOCK genotype are illustrated in Figure 1.
Genetic Associations with Circadian Rhythm Strength
Of the 19 SNPs examined for the candidate genes in unadjusted analyses, three PER2 SNPs (i.e., rs10198215, rs2304674, rs10462023) were associated with mesor and circadian quotient, one PER3 SNP (rs2640908) was associated with 24-hour autocorrelation, and none were associated with amplitude (Supplemental Table 2). All multivariate models adjusted for Genomic estimates of ancestry and self-reported race/ethnicity, and an interaction between genotype and either viral load or years since HIV diagnosis was included in the model when significant (Table 4). In addition, mesor models also adjusted for anti-depressant use, and amplitude models adjusted for gender, viral load, anti-depressant use, and smoking status. Circadian quotient models adjusted for gender, employment status, and smoking status, and 24-hour autocorrelation models adjusted for gender, smoking status, use of anti-depressant or opiate medication, and depressive symptoms (i.e., CES-D scores).
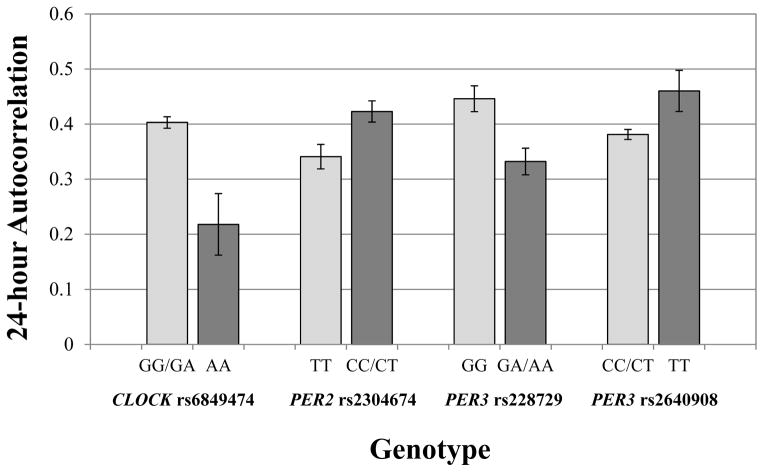
In adjusted analyses, 10 of the 19 SNPs across CLOCK, CRY1, PER1, PER2, and PER3 were associated with one or more of the circadian rhythm strength measures, for a total of 12 observed associations (Table 4). Five associations included an interaction between the polymorphism and viral load, five associations included an interaction with the number of years since HIV diagnosis, one included interactions with both HIV variables, and one association did not include an interaction with either HIV variable. Three of the adjusted associations were also significant in the unadjusted analyses, but 9 polymorphisms were only significantly associated with circadian rhythm strength after adjusting for other relevant covariates. As shown in Table 5, the overall models explained between 8.2% and 20.5% of the variance in the circadian rhythm strength measures, with genotype accounting for 1.5% to 3.8% of the variance. Adjusted differences in 24-hour autocorrelation rhythm strength by genotype are illustrated in Figure 2.
Figure 2.
Adjusted mean 24-hour autocorrelation by genotype. Carriers of two doses of the CLOCK rs6849474 rare allele (AA) had a weaker circadian sleep-wake rhythm than carriers of the common allele (G; p=.003). Carriers of the PER2 rs2304674 rare allele (C) had a stronger circadian sleep-wake rhythm than carriers of two doses of the common allele (TT; p=.031). Carriers of the PER3 rs228729 rare allele (A) had a weaker circadian sleep-wake rhythm than carriers of two doses of the common allele (GG; p =.011). Lastly, carriers of two doses of the PER3 rs2640908 rare allele (TT) had a stronger circadian sleep-wake rhythm than carriers of the common allele (C; p=.041). The genotype-HIV interactions had minimal impact on 24-hour autocorrelation and are not shown.
Genetic Associations with Circadian Phase Timing
Of the 19 SNPs examined for the candidate genes, 6 SNPs were associated with at least one of the five circadian phase timing parameters in unadjusted analyses (Supplemental Table 3). Acrophase and actigraphy bed time were associated with PER3 rs707465; habitual bed time was associated with CLOCK rs6850524 and PER2 rs10198215. Actigraphy wake time was associated with PER2 rs10198215 and four PER3 SNPs (i.e., rs228729, rs228682, rs707465, rs2640908) and habitual wake time was associated with three PER3 SNPs (i.e., rs228729, rs228682, rs707465).
As in prior adjusted analyses, genomic estimates of ancestry and self-reported race/ethnicity were forced into all multivariate models, and an interaction between genotype and either viral load or years since HIV diagnosis was included in the model when significant (Table 4). In addition, acrophase models adjusted for smoking status and use of alcohol, actigraphy bed time models adjusted for employment status and AIDS diagnosis, and actigraphy wake time models adjusted for smoking status and waist circumference. All habitual bed time and wake time models adjusted for smoking status.
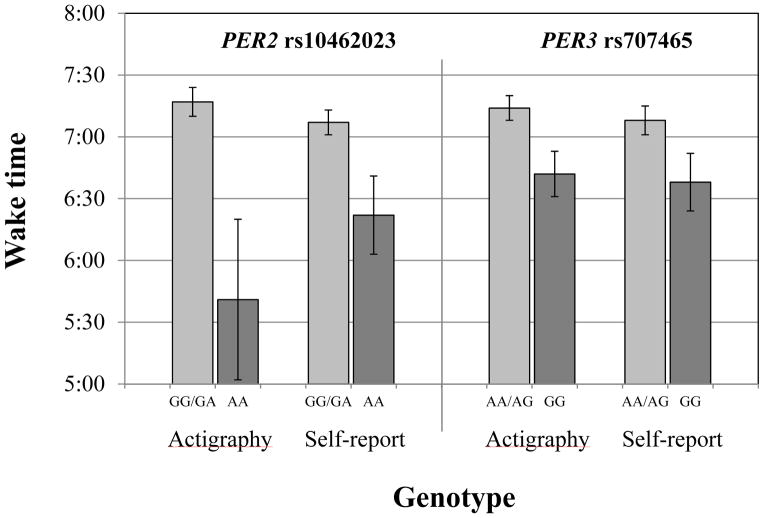
In adjusted analyses, 4 SNPs that were significant in bivariate analyses (i.e., PER3 rs228729, rs228682, rs707465 and rs2640908) remained associated with at least one of the circadian timing parameters after adjusting for relevant covariates (Table 4). In addition, 5 SNPs that were not significant in bivariate analyses (i.e., CLOCK rs1801260 and rs2070062, PER1 rs2253820, PER2 rs10462023, PER3 rs1012477) were significantly associated with at least one of the circadian phase parameters after adjusting for relevant covariates. Of the 16 observed genetic associations, 3 associations involved an interaction between the polymorphism and viral load, 6 associations included an interaction with the number of years since HIV diagnosis, and 7 associations did not include an interaction. Overall, the adjusted models explained between 6.7% and 15.8% of the variance in the circadian phase measures, with genotype accounting for 1.2% to 3.6% of the variance (Table 5). Adjusted differences in wake times by genotype are illustrated in Figure 3.
Figure 3.
Adjusted mean wake times (actigraphy and self-report) by PER2 rs10462023 and PER3 rs707465 genotype. Carriers of two doses of the PER2 rs10462023 rare allele (AA) or the PER3 rs707465 rare allele (G) had a significantly earlier wake time by both actigraphy and self-report than carriers of the common allele (G for PER2 rs10462023 and A for PER3 rs707465; all p<.05). The genotype-HIV interactions had minimal impact on wake time and are not shown.
Discussion
The results of this study indicate that polymorphisms in several circadian genes (i.e., CLOCK, CRY1, PER1, PER2, PER3) are associated with poor sleep maintenance and disturbed sleep-wake rhythms among adults living with HIV/AIDS. These findings are consistent with some prior studies documenting associations between circadian genes and sleep behavior in healthy populations, but to our knowledge, this is the first study to report such associations among adults with chronic illness where the majority are receiving unemployment disability compensation, and thus do not have fixed schedules related to employment. Mood disorders have been associated with circadian genes in prior studies, but controlling for depressive symptoms had minimal effect on the observed associations in this study.
CLOCK polymorphisms have been previously associated with short sleep duration (Allebrandt et al., 2010; Parsons et al., 2014), insomnia risk (Serretti et al., 2005; Ziv-Gal et al., 2013), and diurnal preference (Katzenberg et al., 1998), but other studies have yielded conflicting findings (Serretti et al., 2010; Barclay et al., 2011). The CLOCK gene, located on 4q12, encodes for a protein that is an important transcription factor for the molecular circadian clock and plays an important role in circadian rhythm regulation (Allebrandt et al., 2010; Ziv-Gal et al., 2013). In the present study, there were several unadjusted CLOCK associations with TST and circadian phase timing. After adjusting for genomic estimates of ancestry, self-reported race/ethnicity, and other relevant covariates, genetic variations in CLOCK were associated with poor sleep maintenance as indicated by high WASO (rs3736544), with circadian rhythm strength, as indicated by 24-hour autocorrelation (i.e., rs6849474 and its interaction with years since HIV diagnosis), and with circadian phase as indicated by actigraphy bed time (i.e, interaction between rs2070062 and the number of years since HIV diagnosis). CLOCK rs3736544 is a synonymous polymorphism, and it has been suggested that this SNP may affect biological function in the brain (Kishi et al., 2009). Our findings suggest CLOCK rs3736544 may play a role in regulating sleep maintenance. Furthermore, CLOCK rs3736544 is in complete linkage disequilibrium with rs11735267, which is part of haplotype block of six SNPs (i.e., rs534654, rs2412648, rs4340844, rs11735267, rs6850524, rs7660668) previously associated with insomnia in bipolar disorder (Shi et al., 2008). In the present study, CLOCK rs11735267 was also associated with WASO, but was not reported in Table 5 due to its complete linkage disequilibrium with rs3736544. CLOCK rs6849474 is located in an intron and CLOCK rs2070062 is located in the 5′ untranslated region. Little is known about the functional impacts of these SNPs on the expressions of the gene. The present findings indicate that the number of years since HIV diagnosis may attenuate the relationship between these SNPs and circadian rhythm strength in the HIV population.
Similarly, CRY1, PER1, PER2 and PER3 have been previously associated with diurnal preference (Ojeda et al., 2013; Parsons et al., 2014), sleep quality, and insomnia severity (Brower et al., 2012), although some studies have reported contradictory findings (Barclay et al., 2011; Osland et al., 2011). CRY1, a protein-coding gene, plays a major role in repressing the transcription of CLOCK/BMAL1 and activating the transcription of PER (Ko & Takahashi, 2006; Weger et al., 2011; Hua et al., 2014). An interaction between CRY1 rs10746075 and number of years since HIV diagnosis was associated with circadian quotient. Little is known about the effects of this intronic SNP on sleep-wake rhythms. The findings in the present study suggest that longer HIV exposure may relate to the effects of this SNP on circadian rhythm strength in adults with HIV infection.
A previous study reported an association between PER1 synonymous rs2735611 and diurnal preference (Carpen et al., 2006). In the present study, PER1 SNPs were associated with mesor (i.e., interaction between rs885747 and number of years since HIV diagnosis, interaction between rs2253820 and number of years since HIV diagnosis) and waketime based on self-report (interaction between rs2253820 and viral load). PER1 rs885747 is an intronic SNP associated with risk of more aggressive prostate cancer and autistic disorder (Nicholas et al., 2007; Zhu et al., 2009). PER1 rs2253820 is a synonymous SNP, which has not been associated with sleep-related parameters or other outcomes to date. Our findings suggest that these two SNPs may affect circadian rhythm strength and circadian rhythm timing through interacting with viral load as well as longer HIV exposure. Replication studies focusing on these interactions are warranted.
PER2 has been previously associated with self-reported sleep duration (Parsons et al., 2014) and diurnal preference (Carpen et al., 2005; Lee et al., 2011; Ojeda et al., 2013). Although the present study did not evaluate the same polymorphisms as these prior studies, all three of the intronic SNPs examined in the present study (i.e., rs10198215, rs2304674, rs10462023) were associated with either circadian rhythm strength or phase in adjusted models, often interacting with either viral load or years since HIV diagnosis. PER2 rs10462023 has been associated with depression (Lavebratt et al., 2010) and psychosis (Liu et al., 2015), but to our knowledge, these three SNPs have not been associated with sleep or other circadian-related outcomes. In addition to its links to sleep duration and diurnal preference, PER2 has been associated with reward behavior (Forbes et al., 2012), cocaine addiction (Shumay et al., 2012), and alcohol consumption (Blomeyer et al., 2013). In one study (Comasco et al., 2010), alcohol consumption among adolescent boys was associated with an interaction between a PER2 polymorphism and self-reported sleep problems. Given the prevalence of substance use and sleep disturbance among adults living with HIV, future studies might investigate whether similar associations exist in this population as well.
The PER3 polymorphism most commonly associated with diurnal preference, the coding region variable number tandem repeat (VNTR; rs57875989), was not evaluated in the present study. However, several other PER3 SNPs (i.e., rs228729, rs228682, rs1012477, rs707465, rs4908482, rs2640908) were associated with either circadian rhythm strength or phase. Inhibition of PER3 expression in cell culture results in the inhibition of the early stages of HIV replication (Konig et al., 2008); however, it is unclear if the inhibition is bidirectional. Moreover, whether the variations in PER3 identified herein influence the PER3-mediated inhibition of HIV replication is unknown. A PER2 SNP was associated with self-reported wake time, and the varying associations between PER2 and PER3 across the circadian timing measures may reflect the challenges of comparing self-report and objectively-measured sleep parameters.
Sleep timing and duration are affected by a homeostatic process and a circadian process (Daan et al., 1984; Borbely, 1998). Circadian process refers to the circadian clock that is regulated by clock genes such as CLOCK, CRY, and PER (Allebrandt et al., 2010; Osland et al., 2011). The association between circadian phase timing and PER3 rs707465 (an intronic SNP) was especially strong in our participants. PER3 rs707465 was associated with acrophase and wake time (habitual and actigraphy) rather than bedtime. These results would suggest that bedtimes are more behaviorally based, while final wake time is genetically influenced by PER3. Given the strong relationship between PER3 rs707465 and circadian phase in the present study, it is possible that this SNP may be in linkage disequilibrium with an unmeasured functional polymorphism elsewhere in the PER3 gene, and further investigation is warranted.
PER3 rs2640908 is a synonymous polymorphism (p.Thr977) associated with circadian rhythm strength (autocorrelation) and circadian phase (objectively measured wake time) in our sample. PER3 rs2640908 is thought to be related to overall survival of hepatocellular carcinoma (Zhao et al., 2012), but little is known about the effects of the SNP on sleep patterns or sleep disturbance in adults with HIV/AIDS.
In this racially/ethnically diverse sample of adults with chronic illness, adjusting for the influence of potentially confounding variables was essential for the identification of genetic associations. Of the 27 unadjusted associations reported, only eleven were significant in adjusted analyses. Furthermore, 17 associations were revealed after adjusting for potential confounders and/or accounting for a possible interaction between genotype and HIV. These findings highlight the importance of identifying and controlling for potentially confounding variables to inform the design of validation studies. Race/ethnicity, waist circumference, use of medications, and smoking were the most common confounders, although their relative influence varied across sleep-wake rhythm parameters. Another recent study reported similar racial differences in sleep disruption and duration as observed in our study and also found that African ancestry is associated with a lower percentage of slow wave sleep (Halder et al., 2015). While our study did not assess slow wave sleep, such racial differences underscore the importance of controlling for genomic estimates of ancestry in genetic association studies. Lifestyle variables, such as smoking and alcohol use, were also relevant covariates for some of the rhythm strength and timing phenotypes. These lifestyle behaviors are likely to be potential efforts to self-manage dysynchrony between genetically-based biological rhythms and societal or cultural rhythms.
We also controlled for HIV-related clinical variables, such as CD4+ T-cell count and viral load, to better isolate genetic associations from disease processes. Although salient HIV clinical variables were associated with some sleep-wake rhythm parameters in bivariate analyses, they were generally not sufficiently significant to warrant being retained as covariates in the adjusted models, suggesting they played a relatively modest role after controlling for other variables. Nonetheless, the majority of the adjusted associations observed did include a significant interaction between genotype and either HIV viral load or the number of years since HIV diagnosis. These two variables were selected for the interactions because they each represent different aspects of HIV exposure: viral load as an estimate of the magnitude of exposure to the virus, and time since HIV diagnosis as an estimate of duration of viral exposure. Because these two HIV variables were generally found to interact with different polymorphisms, our findings suggest that HIV may interact with genotype in different ways. For example, some interactions (e.g., PER3 and viral load) represent direct interactions of gene and virus functioning while other interactions may be due to the indirect impact of poorly controlled viral load on biological functioning (e.g., chronic inflammation and sleep disruption). In addition, evaluation of the beta coefficients for both the genotypes and HIV-genotype interactions would indicate that more intense or longer HIV exposure may attenuate the genotypic effects on phenotype. These interactions were evaluated in light of prior evidence of HIV interactions with both CLOCK and PER3, but the present findings suggest that there may also be interactions between HIV and CRY1, PER1, and PER2. Thus, replication of these interactions in other studies and further exploration of the underlying mechanisms is needed.
This study has a number of limitations that need to be acknowledged. Participants were screened for sleep disorders by self-report rather than formal clinical polysomnography (PSG) assessment. While actigraphy estimates of sleep duration (TST) and disruption (WASO) are not a substitute for PSG estimates, bedtimes and final wake times during actigraphy monitoring are not influenced by PSG electrode monitoring or laboratory environments and are thus more indicative of habitual bedtime and wake time behaviors than PSG. In addition, SNP associations with actigraphy circadian phase estimates of bed times and wake times over 3 weekdays differed in some respects from SNP associations with habitual self-reported bedtimes and wake times. When an individual is asked to indicate a habitual bedtime and final wake time, considering the past month, the response may reflect either a stable and consistent behavior, or represent an average of a fluctuating behavior. Responses with a range of bed and wake times may be indicative of a weak circadian rhythm when not influenced by a daily schedule or routine.
Furthermore, circadian sleep-wake rhythm phenotypes were determined from 48–72 consecutive hours on week days, and longer objective monitoring periods (at least 7 days is recommended) would likely have resulted in more reliable estimates, particularly for 24-hour autocorrelation. In addition, by not monitoring weekend sleep/wake behaviors, the potential for weekend sleep rhythm variability could not be evaluated. Although blood draws for CD4+ T-cell counts usually occurred in the morning, blood draws occurring later in the day could have resulted in lower counts related to the circadian nadir (Haus & Smolensky, 1999), thereby limiting the reliability and validity of this measure. Analysis of additional molecular clock genes may result in the identification of additional gene variations associated with these sleep traits. Although this representative sample of adults living with HIV infection in the United States was adequate for our analysis, the sample size was modest for a genetic association study, and larger samples are needed to extend these findings and validate the associations found in our sample. Moreover, this study did not include a comparison group of HIV-seronegative adults, and thus it remains unclear whether the findings are specific to adults with HIV/AIDS or can be generalized to other populations. Findings from this study do, however, contribute to the growing evidence for an association between genes that regulate circadian rhythms and human sleep behavior, regardless of the clinical characteristics of a chronic illness population. Further research is warranted to develop circadian interventions for improving sleep and reducing circadian rhythm disturbances in this patient population. Finally, the PER3 VNTR polymorphism has been strongly linked to circadian rhythmicity and tolerance of sleep debt, but was not evaluated in the present study. Future examination of this polymorphism is also warranted.
Supplementary Material
Footnotes
Declaration of Interest
This research was supported by a grant from the National Institute of Mental Health (NIMH, 5 R01 MH074358). Post-doctoral funding for Eeeseung Byun was provided by the National Institutes of Health/National Institute of Nursing Research (T32 NR007088). Data collection was supported by the General Clinical Research Center in the UCSF CTSA (1 UL RR024131). The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Reference List
- Allebrandt KV, Teder-Laving M, Akyol M, Pichler I, Muller-Myhsok B, Pramstaller P, Merrow M, Meitinger T, Metspalu A, Roenneberg T. CLOCK gene variants associate with sleep duration in two independent populations. Biol Psychiatry. 2010;67:1040–7. doi: 10.1016/j.biopsych.2009.12.026. [DOI] [PubMed] [Google Scholar]
- Ancoli-Israel S, Clopton P, Klauber MR, Fell R, Mason W. Use of wrist activity for monitoring sleep/wake in demented nursing-home patients. Sleep. 1997;20:24–7. doi: 10.1093/sleep/20.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aouizerat BE, Dodd M, Lee K, West C, Paul SM, Cooper BA, Wara W, Swift P, Dunn LB, Miaskowski C. Preliminary evidence of a genetic association between tumor necrosis factor alpha and the severity of sleep disturbance and morning fatigue. Biol Res Nurs. 2009;11:27–41. doi: 10.1177/1099800409333871. [DOI] [PubMed] [Google Scholar]
- Barclay NL, Eley TC, Mill J, Wong CC, Zavos HM, Archer SN, Gregory AM. Sleep quality and diurnal preference in a sample of young adults: associations with 5HTTLPR, PER3, and CLOCK 3111. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:681–90. doi: 10.1002/ajmg.b.31210. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Blomeyer D, Buchmann AF, Lascorz J, Zimmermann US, Esser G, Desrivieres S, Schmidt MH, Banaschewski T, Schumann G, Laucht M. Association of PER2 genotype and stressful life events with alcohol drinking in young adults. PLoS One. 2013;8:e59136. doi: 10.1371/journal.pone.0059136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbely AA. Processes underlying sleep regulation. Horm Res. 1998;49:114–7. doi: 10.1159/000023156. [DOI] [PubMed] [Google Scholar]
- Brower KJ, Wojnar M, Sliwerska E, Armitage R, Burmeister M. PER3 polymorphism and insomnia severity in alcohol dependence. Sleep. 2012;35:571–7. doi: 10.5665/sleep.1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney BG, Li JZ, Walsh DM, Stein R, Vawter MP, Cartagena P, Barchas JD, Schatzberg AF, Myers RM, Watson SJ, Akil H, Bunney WE. Circadian dysregulation of clock genes: clues to rapid treatments in major depressive disorder. Mol Psychiatry. 2015;20:48–55. doi: 10.1038/mp.2014.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buysse DJ, Reynolds CF, 3rd, Monk TH, Berman SR, Kupfer DJ. The Pittsburgh Sleep Quality Index: a new instrument for psychiatric practice and research. Psychiatry Res. 1989;28:193–213. doi: 10.1016/0165-1781(89)90047-4. [DOI] [PubMed] [Google Scholar]
- Carpen JD, Archer SN, Skene DJ, Smits M, von Schantz M. A single-nucleotide polymorphism in the 5′-untranslated region of the hPER2 gene is associated with diurnal preference. J Sleep Res. 2005;14:293–7. doi: 10.1111/j.1365-2869.2005.00471.x. [DOI] [PubMed] [Google Scholar]
- Carpen JD, von Schantz M, Smits M, Skene DJ, Archer SN. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J Hum Genet. 2006;51:1122–5. doi: 10.1007/s10038-006-0060-y. [DOI] [PubMed] [Google Scholar]
- Cole RJ, Kripke DF, Gruen W, Mullaney DJ, Gillin JC. Automatic sleep/wake identification from wrist activity. Sleep. 1992;15:461–9. doi: 10.1093/sleep/15.5.461. [DOI] [PubMed] [Google Scholar]
- Comasco E, Nordquist N, Gokturk C, Aslund C, Hallman J, Oreland L, Nilsson KW. The clock gene PER2 and sleep problems: association with alcohol consumption among Swedish adolescents. Ups J Med Sci. 2010;115:41–8. doi: 10.3109/03009731003597127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daan S, Beersma DG, Borbely AA. Timing of human sleep: recovery process gated by a circadian pacemaker. Am J Physiol. 1984;246:R161–83. doi: 10.1152/ajpregu.1984.246.2.R161. [DOI] [PubMed] [Google Scholar]
- Dunlap JC. Molecular bases for circadian clocks. Cell. 1999;96:271–90. doi: 10.1016/s0092-8674(00)80566-8. [DOI] [PubMed] [Google Scholar]
- Forbes EE, Dahl RE, Almeida JR, Ferrell RE, Nimgaonkar VL, Mansour H, Sciarrillo SR, Holm SM, Rodriguez EE, Phillips ML. PER2 rs2304672 polymorphism moderates circadian-relevant reward circuitry activity in adolescents. Biol Psychiatry. 2012;71:451–7. doi: 10.1016/j.biopsych.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamaldo CE, Gamaldo A, Creighton J, Salas RE, Selnes OA, David PM, Mbeo G, Parker BS, Brown A, McArthur JC, Smith MT. Evaluating sleep and cognition in HIV. J Acquir Immune Defic Syndr. 2013a;63:609–16. doi: 10.1097/QAI.0b013e31829d63ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamaldo CE, Spira AP, Hock RS, Salas RE, McArthur JC, David PM, Mbeo G, Smith MT. Sleep, function and HIV: a multi-method assessment. AIDS Behav. 2013b;17:2808–15. doi: 10.1007/s10461-012-0401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–9. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Halder I, Matthews K, Buysse D, Strollo P, Causer V, Reis SE, Hall MH. African Genetic Ancestry is Associated with Sleep Depth in Older African Americans. Sleep. 2015 doi: 10.5665/sleep.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Hum Mutat. 2008;29:648–58. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- Haus E, Smolensky MH. Biologic rhythms in the immune system. Chronobiol Int. 1999;16:581–622. doi: 10.3109/07420529908998730. [DOI] [PubMed] [Google Scholar]
- Hida A, Kitamura S, Katayose Y, Kato M, Ono H, Kadotani H, Uchiyama M, Ebisawa T, Inoue Y, Kamei Y, Okawa M, Takahashi K, Mishima K. Screening of clock gene polymorphisms demonstrates association of a PER3 polymorphism with morningness-eveningness preference and circadian rhythm sleep disorder. Sci Rep. 2014;4:6309. doi: 10.1038/srep06309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne JA, Ostberg O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int J Chronobiol. 1976;4:97–110. [PubMed] [Google Scholar]
- Hua P, Liu W, Chen D, Zhao Y, Chen L, Zhang N, Wang C, Guo S, Wang L, Xiao H, Kuo SH. Cry1 and Tef gene polymorphisms are associated with major depressive disorder in the Chinese population. J Affect Disord. 2014;157:100–3. doi: 10.1016/j.jad.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzenberg D, Young T, Finn L, Lin L, King DP, Takahashi JS, Mignot E. A CLOCK polymorphism associated with human diurnal preference. Sleep. 1998;21:569–76. doi: 10.1093/sleep/21.6.569. [DOI] [PubMed] [Google Scholar]
- Kishi T, Kitajima T, Ikeda M, Yamanouchi Y, Kinoshita Y, Kawashima K, Okochi T, Okumura T, Tsunoka T, Ozaki N, Iwata N. CLOCK may predict the response to fluvoxamine treatment in Japanese major depressive disorder patients. Neuromolecular Med. 2009;11:53–7. doi: 10.1007/s12017-009-8060-7. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–7. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, Seidel S, Opaluch AM, Caldwell JS, Weitzman MD, Kuhen KL, Bandyopadhyay S, Ideker T, Orth AP, Miraglia LJ, Bushman FD, Young JA, Chanda SK. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavebratt C, Sjoholm LK, Partonen T, Schalling M, Forsell Y. PER2 variantion is associated with depression vulnerability. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:570–81. doi: 10.1002/ajmg.b.31021. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Kim L, Kang SG, Yoon HK, Choi JE, Park YM, Kim SJ, Kripke DF. PER2 variation is associated with diurnal preference in a Korean young population. Behav Genet. 2011;41:273–7. doi: 10.1007/s10519-010-9396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KA, Gay C, Portillo CJ, Coggins T, Davis H, Pullinger CR, Aouizerat BE. Symptom experience in HIV-infected adults: a function of demographic and clinical characteristics. J Pain Symptom Manage. 2009;38:882–93. doi: 10.1016/j.jpainsymman.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KA, Gay C, Portillo CJ, Coggins T, Davis H, Pullinger CR, Aouizerat BE. Types of sleep problems in adults living with HIV/AIDS. J Clin Sleep Med. 2012;8:67–75. doi: 10.5664/jcsm.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichstein KL, Stone KC, Donaldson J, Nau SD, Soeffing JP, Murray D, Lester KW, Aguillard RN. Actigraphy validation with insomnia. Sleep. 2006;29:232–9. [PubMed] [Google Scholar]
- Lincoln GA, Andersson H, Loudon A. Clock genes in calendar cells as the basis of annual timekeeping in mammals--a unifying hypothesis. J Endocrinol. 2003;179:1–13. doi: 10.1677/joe.0.1790001. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Sudic Hukic D, Forsell Y, Schalling M, Osby U, Lavebratt C. Depression-associated ARNTL and PER2 genetic variants in psychotic disorders. Chronobiol Int. 2015;32:579–84. doi: 10.3109/07420528.2015.1012588. [DOI] [PubMed] [Google Scholar]
- Nicholas B, Rudrasingham V, Nash S, Kirov G, Owen MJ, Wimpory DC. Association of Per1 and Npas2 with autistic disorder: support for the clock genes/social timing hypothesis. Mol Psychiatry. 2007;12:581–92. doi: 10.1038/sj.mp.4001953. [DOI] [PubMed] [Google Scholar]
- Ojeda DA, Perea CS, Nino CL, Gutierrez RM, Lopez-Leon S, Arboleda H, Camargo A, Adan A, Forero DA. A novel association of two non-synonymous polymorphisms in PER2 and PER3 genes with specific diurnal preference subscales. Neurosci Lett. 2013;553:52–6. doi: 10.1016/j.neulet.2013.08.016. [DOI] [PubMed] [Google Scholar]
- Osland TM, Bjorvatn BR, Steen VM, Pallesen S. Association study of a variable-number tandem repeat polymorphism in the clock gene PERIOD3 and chronotype in Norwegian university students. Chronobiol Int. 2011;28:764–70. doi: 10.3109/07420528.2011.607375. [DOI] [PubMed] [Google Scholar]
- Parsons MJ, Lester KJ, Barclay NL, Archer SN, Nolan PM, Eley TC, Gregory AM. Polymorphisms in the circadian expressed genes PER3 and ARNTL2 are associated with diurnal preference and GNbeta3 with sleep measures. J Sleep Res. 2014;23:595–604. doi: 10.1111/jsr.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KD, Sowell RL, Boyd M, Dudgeon WD, Hand GA. Sleep quality and health-related quality of life in HIV-infected African-American women of childbearing age. Qual Life Res. 2005;14:959–70. doi: 10.1007/s11136-004-2574-0. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Radloff LS. The CES-D scale: A self-report depression scale for research in the general population. Appl Psych Meas. 1977;1:385–401. [Google Scholar]
- Rubinstein ML, Selwyn PA. High prevalence of insomnia in an outpatient population with HIV infection. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:260–5. doi: 10.1097/00042560-199811010-00008. [DOI] [PubMed] [Google Scholar]
- Serretti A, Cusin C, Benedetti F, Mandelli L, Pirovano A, Zanardi R, Colombo C, Smeraldi E. Insomnia improvement during antidepressant treatment and CLOCK gene polymorphism. Am J Med Genet B Neuropsychiatr Genet. 2005;137B:36–9. doi: 10.1002/ajmg.b.30130. [DOI] [PubMed] [Google Scholar]
- Serretti A, Gaspar-Barba E, Calati R, Cruz-Fuentes CS, Gomez-Sanchez A, Perez-Molina A, De Ronchi D. 3111T/C clock gene polymorphism is not associated with sleep disturbances in untreated depressed patients. Chronobiol Int. 2010;27:265–77. doi: 10.3109/07420521003663785. [DOI] [PubMed] [Google Scholar]
- Shearman LP, Sriram S, Weaver DR, Maywood ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings MH, Reppert SM. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288:1013–9. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- Shi J, Wittke-Thompson JK, Badner JA, Hattori E, Potash JB, Willour VL, McMahon FJ, Gershon ES, Liu C. Clock genes may influence bipolar disorder susceptibility and dysfunctional circadian rhythm. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1047–55. doi: 10.1002/ajmg.b.30714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumay E, Fowler JS, Wang GJ, Logan J, Alia-Klein N, Goldstein RZ, Maloney T, Wong C, Volkow ND. Repeat variation in the human PER2 gene as a new genetic marker associated with cocaine addiction and brain dopamine D2 receptor availability. Transl Psychiatry. 2012;2:e86. doi: 10.1038/tp.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taibi DM, Price C, Voss J. A pilot study of sleep quality and rest-activity patterns in persons living with HIV. J Assoc Nurses AIDS Care. 2013;24:411–21. doi: 10.1016/j.jana.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Hong HK, Ko CH, McDearmon EL. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–50. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger BD, Sahinbas M, Otto GW, Mracek P, Armant O, Dolle D, Lahiri K, Vallone D, Ettwiller L, Geisler R, Foulkes NS, Dickmeis T. The light responsive transcriptome of the zebrafish: function and regulation. PLoS One. 2011;6:e17080. doi: 10.1371/journal.pone.0017080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ptacek LJ, Fu YH. Diversity of human clock genotypes and consequences. Prog Mol Biol Transl Sci. 2013;119:51–81. doi: 10.1016/B978-0-12-396971-2.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Lu J, Yin J, Liu H, Guo X, Yang Y, Ge N, Zhu Y, Zhang H, Xing J. A functional polymorphism in PER3 gene is associated with prognosis in hepatocellular carcinoma. Liver Int. 2012;32:1451–9. doi: 10.1111/j.1478-3231.2012.02849.x. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Stevens RG, Hoffman AE, Fitzgerald LM, Kwon EM, Ostrander EA, Davis S, Zheng T, Stanford JL. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. 2009;69:9315–22. doi: 10.1158/0008-5472.CAN-09-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv-Gal A, Flaws JA, Mahoney MM, Miller SR, Zacur HA, Gallicchio L. Genetic polymorphisms in the aryl hydrocarbon receptor-signaling pathway and sleep disturbances in middle-aged women. Sleep Med. 2013;14:883–7. doi: 10.1016/j.sleep.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.