Abstract
Historically, it was thought that the number of X chromosomes plays little role in causing sex differences in traits. Recently, selected mouse models have been used increasingly to compare mice with the same type of gonad but with one versus two copies of the X chromosome. Study of these models demonstrates that mice with one X chromosome can be strikingly different from those with two X chromosomes, when the differences are not attributable to confounding group differences in gonadal hormones. The number of X chromosomes affects adiposity and metabolic disease, cardiovascular ischaemia/reperfusion injury and behaviour. The effects of X chromosome number are likely the result of inherent differences in expression of X genes that escape inactivation, and are therefore expressed from both X chromosomes in XX mice, resulting in a higher level of expression when two X chromosomes are present. The effects of X chromosome number contribute to sex differences in disease phenotypes, and may explain some features of X chromosome aneuploidies such as in Turner and Klinefelter syndromes.
Keywords: sexual differentiation, sex differences, X chromosome, obesity, ischaemia, Klinefelter
1. Introduction
It comes as no surprise to us that males and females are different, because the differences are emphasized and celebrated in our daily conversations from the earliest years of our lives. In everyday discourse, biological sex differences are easily confused with gender differences, i.e. those stemming from cultural attitudes and sex-specific rearing. The study of biological sex differences attempts to identify, categorize and understand the inherent factors that make the two sexes different from each other. These include factors that make every female different from every male (and vice versa). In addition, some factors cause the two sexes to be different, on average, even though some individuals of each sex are similar to individuals of the other sex. Our general goal is to distinguish and understand the separate components causing sex differences.
At the beginning of life, in the zygote, all inherent components must be encoded on the X and Y sex chromosomes, because they are the only genetic factors that are different at that stage. The Y chromosome of mammals encodes several genes that eventually make males different from females, including the testis-determining gene Sry and genes required for spermatogenesis [1,2]. The action of Sry sets up lifelong differences in the levels of gonadal hormones, which act in each sex to make it different from the other sex. Until recently, the X chromosome was thought not to participate significantly in the process of sexual differentiation. That attitude probably stemmed partly from the idea that the process of X inactivation effectively silences most of one X chromosome in XX females, so that they, like XY males, have one active X chromosome in each cell. In the past decade, however, the study of mouse models has provided convincing evidence that cells with two X chromosomes are intrinsically different from those with one X chromosome. Sex differences caused by the number of X chromosomes can have a profound effect on disease. A fundamental understanding of these diseases requires an appreciation of the effects of X chromosome number. The role of the X chromosome implies that specific X genes, at certain levels of expression, protect from disease and therefore might be novel targets for therapy.
2. Mechanisms causing sex differences because of the number of X chromosomes
The X chromosome is one of the most unusual chromosomes in mammals, because it is present in different numbers in males and females. There are numerous ramifications of this inherent imbalance. The inequality in genomic dose of X genes is thought to present a major problem [3], but perhaps for only some gene networks [4]. For some genes, having one or two doses does not make much difference, so that individuals with a single copy of the gene have about the same phenotype as those with two expressed copies. For other X genes, however, the level of expression must be within a limited range, over which the gene product has the optimal balance with its interacting partners, most of which are autosomal [5,6]. These X genes are called ‘dosage-sensitive’ genes. The existence of dosage-sensitive genes on the X chromosome means that cells with two X chromosomes will have too much of some gene products, and/or cells with one X chromosome will have too little, relative to their interacting partners in gene networks. This imbalance creates a selection pressure to increase the expression of the gene in XY cells, which leads to counteracting pressure to decrease expression in XX cells. These selection pressures are thought to have driven the evolution of the current dosage compensation system, X inactivation. The expression of X genes, from a single X chromosome in both sexes, is also upregulated by an unknown mechanism to make X gene expression about on a par with expression of autosomal genes, which are expressed from two copies in most instances [7]. X inactivation is a remarkable process that quite effectively reduces the expected XX>XY sex difference in expression of all but a small minority of X genes [8].
Because X inactivation is a random process in somatic tissues derived from the embryonic epiblast, each XX cell expresses most gene variants and parental imprints from only one of the two X chromosomes. Adult XX tissues and individuals are therefore mosaics of cells that exhibit the effects of either the maternal or paternal X genes. No such mosaicism occurs in XY tissues. The mosaicism of X gene effects has long been recognized as one of the factors that makes individuals with two X chromosomes different from those with one [9,10]. In general, mosaicism is viewed as protective against disease, thus benefitting females. If a deleterious X mutation (or imprint) is inherited from the mother, that mutation is expressed in all cells of XY individuals, because of the male's hemizygous exposure of X alleles, but in only about half of the cells of XX (or XXY) individuals. Thus, X-linked mutations affect males more than females (e.g. as in X-linked developmental disabilities such as Fragile X syndrome). More generally, any genetic variation among X alleles, even those not causing overt disease (e.g. red-green colour blindness), will cause sex differences in traits because the effect of the variant is mitigated in XX tissues by the presence of another variant, but not in XY tissues. The ‘mosaicism buffering effect’ that causes sex differences is relevant only to genetically diverse populations such as humans, but is not a potential explanation of sex differences in traits in inbred laboratory populations such as inbred mice in which the maternal and paternal X alleles are identical.
The sexual inequality of number of X chromosomes also leads to sex differences in traits via at least three mechanisms other than mosaicism: escape from X-inactivation, X imprinting and epigenetic sinks [11]. (i) Escape from X inactivation. X inactivation is not 100% complete, because some genes are insulated from the inactivation process and are thus expressed from both X chromosomes, making expression levels inherently greater in XX (and XXY) cells than in XY (or XO) tissues. The number of ‘X escapees' has been estimated at about 15% of X genes in humans, and 3% in mice [12–15]. These estimates are based on studies of cell lines under artificial conditions in vitro, in which it is possible to detect rigorously even small amounts of expression from the inactive X chromosome. Evidence suggests, however, that many of the putative X escapees do not show the expected XX>XY pattern of expression in whole tissues in vivo [16,17], and the degree of escape from inactivation might be specific to cell types, developmental stages, disease states, environmental conditions, etc. [18]. (ii) Parental imprinting. During production of gametes, each parent methylates DNA in some genes, silencing the allele passed from that parent to its offspring, a process known as imprinting. Sex differences in traits might also arise because of the inherently different pattern of parental imprinting of the X chromosome. Unlike XY tissues, XX tissues are influenced by parent-of-origin effect on X genes. Each parental imprint affects expression in about one-half of the cells because of random X inactivation. For example, a paternal imprint affects only XX cells in which the paternal X chromosome is active. (iii) Epigenetic sinks. The presence of a large inactive and heterochromatic X chromosome in XX cells may attract heterochromatizing factors away from other chromosomes (providing a ‘sink’ for those factors) which could shift the epigenetic status of the genome, and shift gene expression. The ‘epigenetic sink’ hypothesis is still speculative, but has some support [19–23].
3. Methods for detecting differential effects of two versus one X chromosome
Our goal is to test for phenotypic effects of the number of X chromosomes, mirroring the natural difference between females and males, in a manner that will reveal X gene effects involving the molecular mechanisms outlined in §2. We note that some traditional methods of linking genes to phenotypes may not uncover these kinds of X chromosome dosage effects. For example, traditional linkage or association analyses, which establish that variations in the genomic sequence cause phenotypic variation, do not test directly for effects of different doses of genes when there is no difference in genomic DNA sequence. Although variations in genomic sequence might cause changes in gene expression that accidentally mimic sex differences in levels of expression, they do not necessarily do that, and X escapees may have no endogenous differences in DNA sequence. Moreover, many linkage and association studies do not include the X chromosome because of the complexity of analysis of that chromosome [24,25]. However, methods that vary the copy number of the X chromosome to observe its effects in vivo have been informative [25].
We discuss here mouse models for comparing mice with different numbers of the X chromosome [26–28], because this species is the most genetically tractable whole-animal mammalian model of human physiology and disease. An important problem is that groups with different numbers of X chromosomes could conceivably have different levels of gonadal hormones, so that group differences might be caused by gonadal hormones rather than direct effects of X genes on non-gonadal tissues. For example, naturally occurring variation in the number of X chromosomes in humans is associated with changes in adult gonadal hormone levels. Women with Turner syndrome (XO) are infertile and have altered levels of androgens and oestrogens, compared with XX females [29]. Klinefelter syndrome (KS) men (XXY) are also infertile and have lower levels of androgens than XY men [30]. XXY mice similarly are infertile and have lower levels of androgens compared with XY [31–33], but XY and XXY male mice appear to have similar levels of androgens prenatally [34]. The endocrine differences between XO and XX mice are reduced relative to those in humans. XO mice are fertile in some genetic backgrounds, and prenatal levels of androgens appear to be similar to those of XX mice [34]. Nevertheless, one cannot assume that there are no differences in the levels of ovarian hormones. Accordingly, methods must be used to deal with the possible problem that varying X chromosome number could bring changes in the levels of gonadal hormones, which cause differences in non-gonadal traits. Distinguishing hormonal and non-hormonal effects of X chromosome number is a challenge.
One approach to circumventing the issue of confounding gonadal hormone levels is to gonadectomize (GDX) mice as adults (with or without equal hormone replacement) to control hormone levels, so that groups of adult mice can be effectively compared when hormonal levels are the same [35]. Although that method eliminates many possible confounding effects of hormones, it is not sufficient to eliminate all conceivable group differences caused by gonadal hormones. Gonadal hormone effects can be long-lasting, so that group differences may exist even before birth, or before the time of GDX, and can potentially cause phenotypic differences in adulthood [36,37]. (Nevertheless, prenatal differences in gonadal hormone levels, in mice with the same type of gonad, have not been detected in two mouse models discussed here, the Four Core Genotypes (FCG) and XY* models [34,38].) Ultimately, there are almost no methods for keeping gonadal hormones equivalent among groups during the entire lifetime, except in mice that lack gonads entirely [39–41]. However, as we see in §4, it is possible to discover differences in mice with different sex chromosomes that are not explained by effects on gonadal secretions.
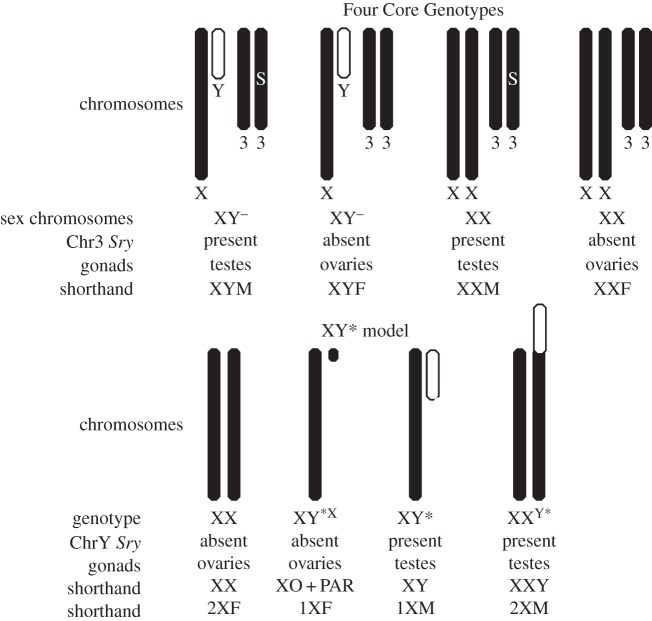
One useful model is the FCG model [28,35,42], which produces XX and XY mice with testes (XXM and XYM) or with ovaries (XXF and XYF) (figure 1). Thus, the XX versus XY comparison can be made when both groups have testes, or have ovaries. The XX and XY groups with the same type of gonad have similar levels of gonadal hormones in adulthood [46–50]. Moreover, differences between XX and XY mice are found after the gonads are removed in adulthood (making their adult gonadal hormone levels zero, and equivalent across groups). In some cases, the XX versus XY difference observed in mice with ovaries is similar to the XX versus XY difference observed in mice with testes. That result suggests that sex chromosome differences can occur under quite different hormonal conditions including during prenatal life (see examples in §4). The FCG model has the major advantage that it detects XX versus XY differences that are independent of type of gonad. The model does not solve whether these sex chromosome effects are caused by the number of X chromosomes (1 versus 2) or the presence of the Y chromosome. For that purpose, the XY* model is useful. It compares groups that have sex chromosomes that are similar to XO, XX, XY and XXY (figure 1). In this model, two comparisons test for different effects of one versus two X chromosomes: XO versus XX, and XY versus XXY (figure 1). The combined use of the FCG and XY* models offers the advantage that a sex chromosome effect can be detected and found to be insensitive to gonadal type in the FCG model, and then confirmed in a completely different genetic model using XY* mice, which also discriminates between effects caused by X or Y chromosome number. The XY* model may be used by itself, to demonstrate differences in the effects of one versus two X chromosomes, in the presence (XY versus XXY) or absence (XO versus XX) of the Y chromosome [51]. The XY* model also tests for the effect of a Y chromosome (XO versus XY or XX versus XXY), but in this model the Y chromosome effects are most likely the result of effects of testicular secretions, because the mice with Y chromosomes have testes. In §4(a–c), we illustrate the use of these and related mouse models to identify mechanisms that contribute to sex differences in traits such as obesity, cardiovascular disease and behaviour.
Figure 1.
Effects of one versus two X chromosomes can be revealed using two mouse models that have various combinations of sex chromosomes and gonad type. In the FCG model, the Y chromosome is deleted for Sry, and designated Y−. An Sry transgene (S) is present on chromosome 3 in some groups. Breeding XYM with XXF produces the four genotypes, XX and XY mice with testes (XXM, XYM), and XX and XY mice with ovaries (XXF, XYF). In the XY* model, breeding an XX mother with XY* father produces the four genotypes, based on the abnormal recombination of the Y* chromosome with the X chromosome [43–45]. Adapted from [27] with permission from Elsevier.
4. Test cases showing effects of X chromosome number
(a). Sex differences in metabolism and adiposity
Women and men differ in the amount and distribution of fat in the body, and overweight and obesity have differential effects on health of the two sexes [52]. Mice are an important genetic model in research on obesity and metabolic syndrome. As adults, male mice generally weigh more than female mice. In the C57BL/6 strain, the sex difference is approximately 25%. This difference can be seen in gonad-intact FCG mice (figure 2a). By 45 days of age, after puberty, XXM or XYM mice (males with testes) weigh more than XXF and XYF (females with ovaries). To test if the sex difference is caused by adult secretions of gonadal hormones, the gonads are removed (figure 2b). Within a month after GDX, all four groups weigh about the same, because of reduced increase of body weight in males and increase in body weight in females, indicating that the main sex difference in body weight was influenced by both testicular and ovarian sections [43,53,55]. In mice gonadectomized for seven weeks or longer, however, the XX mice gradually gain weight relative to XY mice, leading to a large XX > XY difference in body weight by eight months after GDX. This demonstrates that two X chromosomes are associated with enhanced body weight, a finding that could never be determined by comparing standard XX female and XY male mice, which differ from one another in both gonad type and sex chromosome complement. This sex chromosome effect cannot be explained by group differences in gonadal hormones secreted in adulthood, because the groups had no gonadal hormones for a prolonged period of adulthood. The effect is also not likely to have been caused by group differences in the levels of gonadal hormones secreted before GDX in adulthood, because the XX>XY difference occurs when comparing XX and XY groups that both had either testes or ovaries. The sex chromosome effect therefore occurs robustly under distinctly different hormonal conditions. Moreover, comparison of XX and XY FCG mice with the same type of gonad has uncovered no prenatal or adult XX–XY difference in levels of gonadal hormones [38,46–50].
Figure 2.
The FCG and XY* models are used to demonstrate that the number of X chromosomes contributes to sex differences in body weight and lipoprotein levels. (a) At weaning (age 21 days), the FCG groups have similar body weight, but after puberty (day 45), mice with testes had greater body weight than mice with ovaries (‡p < 0.000001), and mice with XX sex chromosomes weighed slightly more than XY (*p < 0.05). After mice were gonadectomized (GDX) at 75 days of age, and allowed to grow for 10 months, XX mice weighed much more than XY mice, in both gonadal male groups and gonadal female groups (†p < 0.0001), and females were heavier than males (**p < 0.01). The effects of sex chromosome complement interacted significantly with the effects of gonad type (int, *p < 0.05). (b) Growth curves for FCG mice GDX at day 75 (week 0). Before gonadectomy (GDX), mice with ovaries weighed less than mice with testes, and XX mice weighed more than XY mice. The sex difference caused by gonadal secretions acting in adulthood disappeared within four to five weeks after GDX, and thereafter XX mice gained more weight than XY mice. (c) The sex chromosome effect on body weight is confirmed in the XY* model and found to be caused by the number of X chromosomes, using the same GDX design as in a and b. After GDX at day 75, mice with two X chromosomes gained weight more than mice with one X chromosome (p < 0.000001). (d) Metabolic effects of X chromosome number. In FCG mice (left and centre panels) that were gonad-intact or GDX, and in mice eating low-fat lab chow or a high cholesterol diet, XX mice had consistently higher plasma levels of high-density lipoprotein (HDL) cholesterol, independent of their gonadal sex. In the XY* model, mice with two X chromosome had higher HDL levels than mice with one X chromosome. *p < 0.05, **p < 0.01, ***p < 0.001, †p < 0.0001. Adapted from [53,54], with permission from Wolters Kluwer Health, Inc.
The XX chromosome effect on body weight could potentially be caused by the presence of two X chromosomes, or by the absence of a Y chromosome. To distinguish these possibilities, mice from the XY* model were studied. In mice gonadectomized as adults, body weight increased in mice with two X chromosomes (XX and XXY) more than in mice with one X chromosome (XO or XY), but the presence or absence of the Y chromosome had little apparent effect (figure 2c) [53]. Thus, the effect is caused by the inherent sex difference in number of X chromosomes. The results from XY* mice confirm and extend those from FCG mice, using a different genetic model.
The X chromosome effect on body weight is associated with several metabolic changes. One reason for the greater body weight and adiposity of XX mice, relative to XY, could be that they begin their diurnal phase of feeding earlier, and ingest more food during the light phase of the cycle [53,55]. Thus, the time of food intake, rather than total amount eaten, could be affected by the number of X chromosomes. Mice with two X chromosomes also have higher expression of growth hormone mRNA in the arcuate nucleus of the hypothalamus, which likely reflects different activity of hypothalamic circuits regulating feeding [56,57]. Mice with two X chromosomes (relative to those with one) also express higher levels of the gene Pdyn in the striatum [58], which is conceivably related to feeding behaviour [59]. The changes in feeding in XX mice gonadectomized as adults likely contribute to the accrual of nearly double the levels of body fat as XY mice [53]. Moreover, when GDX XX mice are fed a high fat, simple carbohydrate diet, they gain weight faster than XY mice, develop insulin resistance, and have greatly elevated levels of liver fat [53]. In addition, XX mice have altered levels of circulating lipoproteins compared with XY mice, including about 20% higher levels of high-density lipoproteins, both when they are gonad-intact and after GDX, and when fed different diets (figure 2d) [54]. Again, this difference is attributable to the number of X chromosomes.
(b). Sex differences in cardiovascular disease
Men and women differ in their susceptibility to cardiovascular artery disease, which is a leading cause of death [60], but the biological basis of the sex difference is not well understood. The FCG and XY* mouse models have also been studied in a mouse model of ischaemia/reperfusion (I/R) injury in the heart [61] (figure 3) modelling a human heart attack. FCG and XY* mice were GDX and used one month later to remove any possible group differences in levels of gonadal hormones. In an in vivo model, blood flow to some regions of the heart is stopped by ligating one of the cardiac arteries for 30 min, then the ligature is removed to start the blood flow to the heart muscle for 24 h, producing I/R injury (figure 3a–d). In an ex vivo model, in which the heart is removed from the mouse and perfused through the aorta with an oxygenated physiological buffer, the flow is interrupted for 30 min and then restarted for 60 min to simulate I/R injury in humans (figure 3e–h). In both in vivo and ex vivo models, the infarct size was measured at the end of the experiment. In the ex vivo model, haemodynamic parameters were measured throughout the experiment such as rate pressure product (RPP). The infarct size in GDX FCG mice was strikingly greater in XX than XY (approx. 40%) in the in vivo model, independent of the gonadal sex in mice (figure 3b–d). Consistent with larger infarct size in XX mice in vivo, the heart functional recovery after ischaemia in the ex vivo model was significantly lower in XX than XY mice in GDX FCG mice, as indicated by RPP (figure 3f). When the XY* model was subjected to ex vivo I/R injury, RPP was significantly lower and the infarct size was significantly larger in mice with two X chromosomes, relative to mice with one X chromosome, and the presence of the Y chromosome appeared to have no effect (figure 3g,h). These studies of X chromosome effects on cardiovascular disease mirror the studies on metabolism discussed in §4(a), in that two X chromosomes confer a greater disease burden than one X chromosome.
Figure 3.
Use of the FCG model shows that after ischaemia/reperfusion injury, GDX XX mice have worse recovery and larger myocardial infarct area compared with GDX XY mice, irrespective of gonadal type. (a) Experimental protocol in vivo: the left anterior descending artery was occluded in GDX FCG mice for 30 min followed by 24 h of reperfusion. (b) Representative cross sections of heart muscle stained with triphenyl tetrazolium chloride. The white area represents the infarcted area, blue shows the non-infarcted area, red plus white areas show risk area. (c) Percentage of area at risk (AAR) divided by left ventricle area. (d) Infarct size (IS) divided by AAR. **p < 0.01, n = 6–7. (e) Experimental protocol ex vivo: perfusion of the heart is shut off for 30 min, then reperfused for 60 min before measuring heart function. (f) The rate pressure product (RPP), a measure of recovery after injury, was worse in XX than XY mice. **p < 0.01. (g) Use of the XY* model shows that in the ex vivo system, mice with two X chromosomes (XX, XXY) have worse recovery (lower RPP) than mice with one X chromosome (XO, XY). (h) The infarct size as the percentage of total ventricular area in hearts ex vivo. *p < 0.05. Adapted from [61] with permission from Oxford University Press.
(c). Sex differences in behaviour
Studies of mouse behaviour also provide evidence that the number of X chromosomes contributes to sex differences. Fear reactivity in gonad-intact adult mice is greater in XO than XX mice, shown by the reluctance of the mouse to venture into an open area of an elevated plus maze. This difference is not explained by the number of genes in the pseudoautosomal region (PAR), but by the difference in number of non-PAR X genes [62]. In studies of sexual behaviour, when mice of the XY* model are GDX as adults and treated equally with testosterone, then tested with a receptive female, they show male sex behaviour differently depending on the number of X chromosomes [34]. In juvenile mice of the XY* model tested at approximately three weeks of age, mice with one X chromosome show less social behaviour when paired with another mouse, compared with mice with two X chromosomes [51]. Mice with one X chromosome investigated their cage partners more frequently than mice with two X chromosomes, but spent less total time in proximity to or interacting with the partner. When tested for preference for a novel versus familiar mouse, mice with two X chromosomes had greater preference for the unfamiliar mouse. The greater anxiety-like behaviour, found in adult mice with one X chromosome [62], was also found in juvenile mice [51], and may help explain the tendency of mice with one X chromosome to avoid novel mice or social partners, more than mice with two X chromosomes.
(d). Models of sex chromosome aneuploidy
The effects of two versus one X chromosome are not only relevant to the natural difference between the sexes (XX versus XY), but also to two sex chromosome aneuploidies that occur with significant frequency in humans: XO (Turner syndrome, 1/2500 live female births) and XXY (KS, 1/600 live male births). XO females differ from XX females exclusively because of the difference in number of X chromosomes, and XXY males differ from normal XY males for the same reason. XXY males experience inactivation of one of the two X chromosomes, as in XX females [63]. Monosomy of the X chromosome (XO) is usually lethal to human embryos [64], but those who survive to adulthood have multiple phenotypes including ovarian failure with abnormal levels of reproductive hormones (low oestrogen and elevated androgens), short stature, neck webbing, and susceptibility to cardiovascular and metabolic disease [29]. Men with KS have small testes, lowered testosterone levels, and increased height. As a group they show greater incidence of behavioural problems including delayed language development and deficits in social and executive functioning [65]. They experience increased prevalence of several health problems [66,67], some of which are usually more common in women than in men including breast cancer [68,69], osteoporosis [70] and autoimmune diseases including rheumatoid arthritis and systemic lupus erythematosus [71]. KS men have increased body fat, specifically abdominal fat, as well as increased rates of hyperinsulinaemia, insulin resistance, type II diabetes and metabolic syndrome [30].
The loss of one X chromosome in female mice results in a much milder phenotype than in humans. XO mice are fertile at least in outbred genetic backgrounds. The milder phenotype might result from a smaller number of genes in the mouse PAR than human PAR, which are expected to be expressed differently in XO (one PAR) versus XX (two PARs). In addition, fewer X genes escape inactivation in mice than humans, which would reduce the presumed disparity of expression levels of non-PAR X escapees [72], so fewer phenotypes would be affected. Differences in gene expression have been detected in XO mice relative to XX, including in X escapees [73,74]. Although XO mice do not completely model Turner syndrome, numerous genes escape X inactivation in both species, so that the XX versus XO comparison in mice may well model some effects of dosage of those genes in humans.
Several mouse models of KS have been used [33,75–77]. These studies have demonstrated that the second X chromosome in males eliminates sperm production as in KS men [78], reduces testis size, lowers testosterone levels, induces Leydig cell hyperplasia and causes behavioural deficits [33,79]. Moreover, XXY mice have abnormal bone density, and altered sexual partner preferences [80,81]. In most of these studies, gonad-intact mice were studied, so that the lower levels of testosterone in XXY mice, relative to XY mice, could have caused the difference in phenotype, instead of direct (non-gonadal) effects of X genes in mice with one versus two X chromosomes. In some cases, the group differences in adult levels of testosterone were eliminated by castration of adult males, with or without replacement of testosterone [34,80–82]. For some behaviour traits, such as altered social interaction and sex preference, equalizing hormone levels ameliorated group differences [81]. However, differences in male sexual behaviour and in bone architecture persisted despite castration and testosterone replacement [34,80]. These studies ruled out effects of circulating testosterone as the responsible mechanism, but in these studies it is difficult to assess if XY versus XXY differences in levels of testicular hormones before the time of GDX contributed to group differences in phenotype.
Recently, we introduced a novel model of KS, the ‘Sex Chromosome Trisomy’ (SCT) model, which produces XX, XY, XXY and XYY mice (figure 4a) [77,83]. In this model, as in the FCG model, the Sry gene is not on the Y chromosome, but is present on an autosome as a transgene. Thus, each of the four sex chromosome groups is produced with Sry (with testes) or without Sry (with ovaries). The comparison of XXY groups, modelling KS versus normal men, tests for the effects of one versus two X chromosomes when a Y chromosome is present. The model expands the ability to detect effects of the second X chromosome that do not depend on testicular secretions, because the model tests the effects also in mice that do not have testes. In SCT mice GDX as adults and then treated equally with testosterone, XXY mice weigh more than XY mice and have more body fat and less lean mass, compared with XY mice (figure 4b) [77]. The occurrence of these differences in mice that had either testes or ovaries indicates that the group differences do not depend on XXY versus XY differences in the levels of testicular hormones. In tests of sexual partner preference of SCT mice, again GDX and treated with testosterone, XXY male mice also spent less time with a female test mouse than did XY male mice, suggesting a feminizing influence of the second X chromosome on sexual partner preference [83]. This influence appeared to be dependent on the presence of the Y, as the partner preference of XX male mice did not differ from XY males. In the same mice, gene expression was measured in the bed nucleus of the stria terminalis and the striatum. Most genes in XXY males were found to be male-typical in their expression patterns, but a substantial minority of genes in both regions was significantly more female-typical. These genes are candidates for genetic contributors to the KS phenotype
Figure 4.
The Sex Chromosome Trisomy model. (a) The model involves crossing FCG XY ¯(Sry+) male (same as XYM in figure 1) with XXY ¯ female who has the same Y ¯ as in the FCG model (figure 1). The father's Sry is transgenic on chromosome 3. Eight genotypes are produced, XX, XY, XXY and XYY, each with either testes or ovaries. (b) Body weight data from SCT mice shows that after gonadectomy (GDX) in adulthood and treatment with testosterone (T), XXY mice weigh more (b) and have more body fat relative to body weight (c), compared with XY mice. Adapted from [77,83].
5. Candidate X genes that cause sex differences
The inherent sexual imbalance in the number of X chromosomes has now been shown to have unexpectedly large effects on phenotypes, including susceptibility to disease. The next step is to identify the X genes that contribute to this imbalance, and to understand the downstream pathways that affect phenotypes. Although the X chromosome is large and gene-rich, the candidate genes are less numerous, because we can focus on either imprinted genes or genes escaping X inactivation. Tissue-specific parent-of-origin effects on gene expression are just now being reported for an increasing number of X genes [84–86], but it is not clear how many of these will emerge as viable candidates for explaining phenotypic sex differences caused by different number of X chromosomes. By contrast, the number of X escapees resulting in sex differences in expression is relatively small [13], making their analysis more tractable and attractive at present. In mice, several X escapee genes are particularly interesting: Kdm5c, Kdm6a, Eif2s3x and Ddx3x. These are routinely found by numerous laboratories to be expressed at higher levels in XX (or XXY) than XY (or XO) mice in numerous tissues [53,56,61,73,74,87–92]. Each escapes X chromosome inactivation in humans and mice [93–97]. Two of these genes, Kdm5c and Kdm6a, are histone demethylases that are expected to have widespread effects on gene expression throughout the genome, and null mutations of each are implicated in human disease [98,99]. Ddx3x is an RNA helicase involved in several basic cellular processes such as transcription, RNA transport and splicing, and translation, and is implicated in human cancer and intellectual disability [100]. Less is known about Eif2s3x, a translation initiation factor, which is the X-linked paralogue of Eif2s3y, a spermatogonial proliferation factor [101]. Each of these genes has a closely related gene on the Y chromosome, the expression of which in XY males could compensate for the lack of a second copy of the X gene in males. However, the Y genes are unlikely to offset sex differences in effects of X escapees. In some cases, the X and Y copies appear to have diverged in function [102] or pattern of expression [89,90], suggesting that the compensation by the Y paralogue is not complete. Thus, the X escapee genes remain exciting candidates for explaining the effects of two X chromosomes relative to one. Tests of this idea require careful manipulation of the dose of the genes in mouse models, to determine if one versus two copies of the gene causes phenotypic changes similar to the comparison of one versus two copies of the entire X chromosome.
6. Conclusion and prospectus
In the twentieth century, gonadal hormones emerged as the primary proximate factors that act on tissues to cause sex differences in phenotypes. Their effect was so pervasive that they were essentially the only proximate factors incorporated into theories of the origins of sex differences in phenotype. In the past two decades, however, the sexual imbalance of effects of the X and Y chromosomes have been clearly shown to cause sex differences in non-gonadal tissues that are not mediated by gonadal hormones [10,11,35,103–105]. More of these effects have been localized to the X chromosome than to the Y chromosome. Although specific X genes are prime candidates for these effects, we cannot rule out non-genic effects of the X chromosome as reviewed above. These studies are still in their infancy, because we do not know yet which X genes are responsible, and how they act. An important question that is almost completely unstudied is how the multiple sex-biasing factors, hormones and sex chromosome genes, interact with each other in specific instances. Thus, we can expect exciting developments in the near future.
Authors' contributions
A.P.A. drafted the manuscript, which was edited by all other authors.
Competing interests
The authors declare no competing interests.
Funding
Supported by DK083561 (A.P.A., X.C., K.R.), HL119886 (M.E., A.P.A.), HD076125 (A.P.A., E.V.), HL90553 (K.R.), NS043196 (A.P.A.), T32GM007185 (J.C.L.) and T32HD007228 (S.M.W.).
References
- 1.Goodfellow PN, Lovell-Badge R. 1993. SRY and sex determination in mammals. Annu. Rev. Genet. 27, 71–92. ( 10.1146/annurev.ge.27.120193.000443) [DOI] [PubMed] [Google Scholar]
- 2.Burgoyne PS. 1998. The role of Y-encoded genes in mammalian spermatogenesis. Semin. Cell Dev. Biol. 9, 423–432. ( 10.1006/scdb.1998.0228) [DOI] [PubMed] [Google Scholar]
- 3.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 11, 156–166. ( 10.1101/gad.11.2.156) [DOI] [PubMed] [Google Scholar]
- 4.Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B. 2006. Global analysis of X-chromosome dosage compensation. J. Biol. 5, 3 ( 10.1186/jbiol30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oliver B. 2007. Sex, dose, and equality. PLoS Biol. 5, e340 ( 10.1371/journal.pbio.0050340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen ZX, Oliver B. 2015. X chromosome and autosome dosage responses in Drosophila melanogaster heads. G3 (Bethesda) 5, 1057–1063. ( 10.1534/g3.115.017632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen DK, Disteche CM. 2006. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 38, 47–53. ( 10.1038/ng1705) [DOI] [PubMed] [Google Scholar]
- 8.Itoh Y, et al. 2007. Dosage compensation is less effective in birds than in mammals. J. Biol. 6, 2 ( 10.1186/jbiol53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Migeon BR. 2007. Females are mosaic: X inactivation and sex differences in disease. Oxford, UK: Oxford University Press. [DOI] [PubMed] [Google Scholar]
- 10.Arnold AP. 2004. Sex chromosomes and brain gender. Nat. Rev. Neurosci. 5, 701–708. ( 10.1038/nrn1494) [DOI] [PubMed] [Google Scholar]
- 11.Arnold AP. 2011. The end of gonad-centric sex determination in mammals. Trends Genet. 28, 55–61. ( 10.1016/j.tig.2011.10.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrel L, Cottle AA, Goglin KC, Willard HF. 1999. A first-generation X-inactivation profile of the human X chromosome. Proc. Natl Acad. Sci. USA 96, 14 440–14 444. ( 10.1073/pnas.96.25.14440) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang F, Babak T, Shendure J, Disteche CM. 2010. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 20, 614–622. ( 10.1101/gr.103200.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Disteche CM. 2012. Dosage compensation of the sex chromosomes. Annu. Rev. Genet. 46, 537–560. ( 10.1146/annurev-genet-110711-155454) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berletch JB, Yang F, Disteche CM. 2010. Escape from X inactivation in mice and humans. Genome Biol. 11, 213 ( 10.1186/gb-2010-11-6-213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT. 2008. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 4, e9 ( 10.1371/journal.pgen.0040009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng X, Berletch JB, Nguyen DK, Disteche CM. 2014. X chromosome regulation: diverse patterns in development, tissues and disease. Nat. Rev. Genet. 15, 367–378. ( 10.1038/nrg3687) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berletch JB, Ma W, Yang F, Shendure J, Noble WS, Disteche CM, Deng X. 2015. Escape from X inactivation varies in mouse tissues. PLoS Genet. 11, e1005079 ( 10.1371/journal.pgen.1005079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wijchers PJ, Festenstein RJ. 2011. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 27, 132–140. ( 10.1016/j.tig.2011.01.004) [DOI] [PubMed] [Google Scholar]
- 20.Wijchers PJ, Yandim C, Panousopoulou E, Ahmad M, Harker N, Saveliev A, Burgoyne PS, Festenstein R. 2010. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev. Cell 19, 477–484. ( 10.1016/j.devcel.2010.08.005) [DOI] [PubMed] [Google Scholar]
- 21.Lemos B, Branco AT, Hartl DL. 2010. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc. Natl Acad. Sci. USA 107, 15 826–15 831. ( 10.1073/pnas.1010383107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemos B, Araripe LO, Hartl DL. 2008. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 319, 91–93. ( 10.1126/science.1148861) [DOI] [PubMed] [Google Scholar]
- 23.Silkaitis K, Lemos B. 2014. Sex-biased chromatin and regulatory cross-talk between sex chromosomes, autosomes, and mitochondria. Biol. Sex Differ. 5, 2 ( 10.1186/2042-6410-5-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao F, Chang D, Biddanda A, Ma L, Guo Y, Zhou Z, Keinan A. 2015. XWAS: a software toolset for genetic data analysis and association studies of the X chromosome. J. Hered. 106, 666–671. ( 10.1093/jhered/esv059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Broman KW, Sen S, Owens SE, Manichaikul A, Southard-Smith EM, Churchill GA. 2006. The X chromosome in quantitative trait locus mapping. Genetics 174, 2151–2158. ( 10.1534/genetics.106.061176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold AP. 2009. Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J. Neuroendocrinol. 21, 377–386. ( 10.1111/j.1365-2826.2009.01831.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold AP. 2014. Conceptual frameworks and mouse models for studying sex differences in physiology and disease: why compensation changes the game. Exp. Neurol. 259, 2–9. ( 10.1016/j.expneurol.2014.01.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox KH, Bonthuis PJ, Rissman EF. 2014. Mouse model systems to study sex chromosome genes and behavior: relevance to humans. Front. Neuroendocrinol. 35, 405–419. ( 10.1016/j.yfrne.2013.12.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bondy CA. 2009. Turner syndrome 2008. Horm. Res. 71(Suppl 1), 52–56. ( 10.1159/000178039) [DOI] [PubMed] [Google Scholar]
- 30.Gravholt CH, Jensen AS, Host C, Bojesen A. 2011. Body composition, metabolic syndrome and type 2 diabetes in Klinefelter syndrome. Acta Paediatr. 100, 871–877. ( 10.1111/j.1651-2227.2011.02233.x) [DOI] [PubMed] [Google Scholar]
- 31.Lue YH, Wang C, Liu PY, Erkilla K, Swerdloff RS. 2010. Insights into the pathogenesis of XXY phenotype from comparison of the clinical syndrome with an experimental XXY mouse model. Pediatr. Endocrinol. Rev. 8(Suppl 1), 140–144. [PubMed] [Google Scholar]
- 32.Wistuba J et al. 2010. Male 41, XXY* mice as a model for Klinefelter syndrome: hyperactivation of Leydig cells. Endocrinology 151, 2898–2910. ( 10.1210/en.2009-1396) [DOI] [PubMed] [Google Scholar]
- 33.Wistuba J. 2010. Animal models for Klinefelter's syndrome and their relevance for the clinic. Mol. Hum. Reprod. 16, 375–385. ( 10.1093/molehr/gaq024) [DOI] [PubMed] [Google Scholar]
- 34.Bonthuis PJ, Cox KH, Rissman EF. 2012. X-chromosome dosage affects male sexual behavior. Horm. Behav. 61, 565–572. ( 10.1016/j.yhbeh.2012.02.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Vries GJ, et al. 2002. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J. Neurosci. 22, 9005–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phoenix CH, Goy RW, Gerall AA, Young WC. 1959. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology 65, 369–382. ( 10.1210/endo-65-3-369) [DOI] [PubMed] [Google Scholar]
- 37.Arnold AP. 2009. The organizational-activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm. Behav. 55, 570–578. ( 10.1016/j.yhbeh.2009.03.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itoh Y, Mackie R, Kampf K, Domadia S, Brown JD, O'Neill R, Arnold AP. 2015. Four Core Genotypes mouse model: localization of the Sry transgene and bioassay for testicular hormone levels. BMC Res. Notes 8, 69 ( 10.1186/s13104-015-0986-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Budefeld T, Grgurevic N, Tobet SA, Majdic G. 2008. Sex differences in brain developing in the presence or absence of gonads. Dev. Neurobiol. 68, 981–995. ( 10.1002/dneu.20638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grgurevic N, Budefeld T, Spanic T, Tobet SA, Majdic G. 2012. Evidence that sex chromosome genes affect sexual differentiation of female sexual behavior. Horm. Behav. 61, 719–724. ( 10.1016/j.yhbeh.2012.03.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majdic G, Tobet S. 2011. Cooperation of sex chromosomal genes and endocrine influences for hypothalamic sexual differentiation. Front. Neuroendocrinol. 32, 137–145. ( 10.1016/j.yfrne.2011.02.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arnold AP, Chen X. 2009. What does the ‘four core genotypes’ mouse model tell us about sex differences in the brain and other tissues? Front. Neuroendocrinol. 30, 1–9. ( 10.1016/j.yfrne.2008.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen X, McClusky R, Itoh Y, Reue K, Arnold AP. 2013. X and Y chromosome complement influence adiposity and metabolism in mice. Endocrinology 154, 1092–1104. ( 10.1210/en.2012-2098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eicher EM, Hale DW, Hunt PA, Lee BK, Tucker PK, King TR, Eppig JT, Washburn L. 1991. The mouse Y* chromosome involves a complex rearrangement, including interstitial positioning of the pseudoautosomal region. Cytogenet. Cell Genet. 57, 221–230. ( 10.1159/000133152) [DOI] [PubMed] [Google Scholar]
- 45.Burgoyne PS, Mahadevaiah SK, Perry J, Palmer SJ, Ashworth A. 1998. The Y* rearrangement in mice: new insights into a perplexing PAR. Cytogenet. Cell Genet. 80, 37–40. ( 10.1159/000014954) [DOI] [PubMed] [Google Scholar]
- 46.Gatewood JD, Wills A, Shetty S, Xu J, Arnold AP, Burgoyne PS, Rissman EF. 2006. Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. J. Neurosci. 26, 2335–2342. ( 10.1523/JNEUROSCI.3743-05.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palaszynski KM, Smith DL, Kamrava S, Burgoyne PS, Arnold AP, Voskuhl RR. 2005. A Yin-Yang effect between sex chromosome complement and sex hormones on the immune response. Endocrinology 146, 3280–3285. ( 10.1210/en.2005-0284) [DOI] [PubMed] [Google Scholar]
- 48.Sasidhar MV, Itoh N, Gold SM, Lawson GW, Voskuhl RR. 2012. The XX sex chromosome complement in mice is associated with increased spontaneous lupus compared with XY. Ann. Rheum. Dis. 71, 1418–1422. ( 10.1136/annrheumdis-2011-201246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holaskova I, Franko J, Goodman RL, Arnold AP, Schafer R. 2015. The XX sex chromosome complement is required in male and female mice for enhancement of immunity induced by exposure to 3,4-dichloropropionanilide. Am. J. Reprod. Immunol. 74, 136–147. ( 10.1111/aji.12378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corre C, Friedel M, Vousden DA, Metcalf A, Spring S, Qiu LR, Lerch JP, Palmert MR. In press. Separate effects of sex hormones and sex chromosomes on brain structure and function revealed by high-resolution magnetic resonance imaging and spatial navigation assessment of the Four Core Genotype mouse model. Brain Struct. Funct . [DOI] [PubMed] [Google Scholar]
- 51.Cox KH, Quinnies KM, Eschendroeder A, Didrick PM, Eugster EA, Rissman EF. 2015. Number of X-chromosome genes influences social behavior and vasopressin gene expression in mice. Psychoneuroendocrinology 51, 271–281. ( 10.1016/j.psyneuen.2014.10.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karastergiou K, Smith SR, Greenberg AS, Fried SK. 2012. Sex differences in human adipose tissues—the biology of pear shape. Biol. Sex Differ. 3, 13 ( 10.1186/2042-6410-3-13) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen X, McClusky R, Chen J, Beaven SW, Tontonoz P, Arnold AP, Reue K. 2012. The number of X chromosomes causes sex differences in adiposity in mice. PLoS Genet. 8, e1002709 ( 10.1371/journal.pgen.1002709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Link JC, Chen X, Prien C, Borja MS, Hammerson B, Oda MN, Arnold AP, Reue K. 2015. Increased high-density lipoprotein cholesterol levels in mice with XX versus XY sex chromosomes. Arterioscler. Thromb. Vasc. Biol. 35, 1778–1786. ( 10.1161/ATVBAHA.115.305460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen X, Wang L, Loh D, Colwell C, Tache Y, Reue K, Arnold AP. 2015. Sex differences in diurnal rhythms of food intake in mice caused by gonadal hormones and complement of sex chromosomes. Horm. Behav. 75, 55–63. ( 10.1016/j.yhbeh.2015.07.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonthuis PJ, Rissman EF. 2013. Neural growth hormone implicated in body weight sex differences. Endocrinology 154, 3826–3835. ( 10.1210/en.2013-1234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quinnies KM, Bonthuis PJ, Harris EP, Shetty SR, Rissman EF. 2015. Neural growth hormone: regional regulation by estradiol and/or sex chromosome complement in male and female mice. Biol. Sex Differ. 6, 8 ( 10.1186/s13293-015-0026-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X, Grisham W, Arnold AP. 2009. X chromosome number causes sex differences in gene expression in adult mouse striatum. Eur. J. Neurosci. 29, 768–776. ( 10.1111/j.1460-9568.2009.06610.x) [DOI] [PubMed] [Google Scholar]
- 59.Kelley AE, Baldo BA, Pratt WE. 2005. A proposed hypothalamic-thalamic-striatal axis for the integration of energy balance, arousal, and food reward. J. Comp. Neurol. 493, 72–85. ( 10.1002/cne.20769) [DOI] [PubMed] [Google Scholar]
- 60.Barrett-Connor E. 1997. Sex differences in coronary heart disease. Why are women so superior? The 1995 Ancel Keys Lecture. Circulation 95, 252–264. ( 10.1161/01.CIR.95.1.252) [DOI] [PubMed] [Google Scholar]
- 61.Li J, Chen X, McClusky R, Ruiz-Sundstrom M, Itoh Y, Umar S, Arnold AP, Eghbali M. 2014. The number of X chromosomes influences protection from cardiac ischaemia/reperfusion injury in mice: one X is better than two. Cardiovasc. Res. 102, 375–394. ( 10.1093/cvr/cvu064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Isles AR, Davies W, Burrmann D, Burgoyne PS, Wilkinson LS. 2004. Effects on fear reactivity in XO mice are due to haploinsufficiency of a non-PAR X gene: implications for emotional function in Turner's syndrome. Hum. Mol. Genet. 13, 1849–1855. ( 10.1093/hmg/ddh203) [DOI] [PubMed] [Google Scholar]
- 63.Skakkebaek A et al. 2014. Neuropsychology and brain morphology in Klinefelter syndrome—the impact of genetics. Andrology 2, 632–640. ( 10.1111/j.2047-2927.2014.00229.x) [DOI] [PubMed] [Google Scholar]
- 64.Hook EB, Warburton D. 2014. Turner syndrome revisited: review of new data supports the hypothesis that all viable 45,X cases are cryptic mosaics with a rescue cell line, implying an origin by mitotic loss. Hum. Genet. 133, 417–424. ( 10.1007/s00439-014-1420-x) [DOI] [PubMed] [Google Scholar]
- 65.Savic I. 2012. Advances in research on the neurological and neuropsychiatric phenotype of Klinefelter syndrome. Curr. Opin. Neurol. 25, 138–143. ( 10.1097/wco.0b013e32835181a0) [DOI] [PubMed] [Google Scholar]
- 66.Bojesen A, Juul S, Birkebaek NH, Gravholt CH. 2006. Morbidity in Klinefelter syndrome: a Danish register study based on hospital discharge diagnoses. J. Clin. Endocrinol. Metab. 91, 1254–1260. ( 10.1210/jc.2005-0697) [DOI] [PubMed] [Google Scholar]
- 67.Bojesen A, Gravholt CH. 2011. Morbidity and mortality in Klinefelter syndrome (47,XXY). Acta Paediatr. 100, 807–813. ( 10.1111/j.1651-2227.2011.02274.x) [DOI] [PubMed] [Google Scholar]
- 68.Brinton LA. 2011. Breast cancer risk among patients with Klinefelter syndrome. Acta Paediatr. 100, 814–818. ( 10.1111/j.1651-2227.2010.02131.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hultborn R, Hanson C, Kopf I, Verbiene I, Warnhammar E, Weimarck A. 1997. Prevalence of Klinefelter's syndrome in male breast cancer patients. Anticancer Res. 17, 4293–4297. [PubMed] [Google Scholar]
- 70.Ferlin A, Schipilliti M, Foresta C. 2011. Bone density and risk of osteoporosis in Klinefelter syndrome. Acta Paediatr. 100, 878–884. ( 10.1111/j.1651-2227.2010.02138.x) [DOI] [PubMed] [Google Scholar]
- 71.Dillon S, et al. 2011. Klinefelter's syndrome (47,XXY) among men with systemic lupus erythematosus. Acta Paediatr. 100, 819–823. ( 10.1111/j.1651-2227.2011.02185.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. 2011. Genes that escape from X inactivation. Hum. Genet. 130, 237–245. ( 10.1007/s00439-011-1011-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lopes AM, Burgoyne PS, Ojarikre A, Bauer J, Sargent CA, Amorim A, Affara NA. 2010. Transcriptional changes in response to X chromosome dosage in the mouse: implications for X inactivation and the molecular basis of Turner syndrome. BMC Genomics 11, 82 ( 10.1186/1471-2164-11-82) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wolstenholme JT, Rissman EF, Bekiranov S. 2012. Sexual differentiation in the developing mouse brain: contributions of sex chromosome genes. Genes Brain Behav. 12, 166–180. ( 10.1111/gbb.12010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lue Y, Rao PN, Sinha Hikim AP, Im M, Salameh WA, Yen PH, Wang C, Swerdloff RS. 2001. XXY male mice: an experimental model for Klinefelter syndrome. Endocrinology 142, 1461–1470. ( 10.1210/en.142.4.1461) [DOI] [PubMed] [Google Scholar]
- 76.Swerdloff RS, Lue Y, Liu PY, Erkkila K, Wang C. 2011. Mouse model for men with Klinefelter syndrome: a multifaceted fit for a complex disorder. Acta Paediatr. 100, 892–899. ( 10.1111/j.1651-2227.2011.02149.x) [DOI] [PubMed] [Google Scholar]
- 77.Chen X, Williams-Burris SM, McClusky R, Ngun TC, Ghahramani N, Barseghyan H, Reue K, Vilain E, Arnold AP. 2013. The sex chromosome trisomy mouse model of XXY and XYY: metabolism and motor performance. Biol. Sex Differ. 4, 15 ( 10.1186/2042-6410-4-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Werler S, Demond H, Damm OS, Ehmcke J, Middendorff R, Gromoll J, Wistuba J. 2014. Germ cell loss is associated with fading Lin28a expression in a mouse model for Klinefelter's syndrome. Reproduction 147, 253–264. ( 10.1530/REP-13-0608) [DOI] [PubMed] [Google Scholar]
- 79.Lewejohann L, Damm OS, Luetjens CM, Hamalainen T, Simoni M, Nieschlag E, Gromoll J, Wistuba J. 2009. Impaired recognition memory in male mice with a supernumerary X chromosome. Physiol. Behav. 96, 23–29. ( 10.1016/j.physbeh.2008.08.007) [DOI] [PubMed] [Google Scholar]
- 80.Liu PY, et al. 2010. Genetic and hormonal control of bone volume, architecture, and remodeling in XXY mice. J. Bone Miner. Res. 25, 2148–2154. ( 10.1002/jbmr.104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu PY, et al. 2010. Genetic, hormonal, and metabolomic influences on social behavior and sex preference of XXY mice. Am. J. Physiol. Endocrinol. Metab. 299, E446–E455. ( 10.1152/ajpendo.00085.2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Park JH, Burns-Cusato M, Dominguez-Salazar E, Riggan A, Shetty S, Arnold AP, Rissman EF. 2008. Effects of sex chromosome aneuploidy on male sexual behavior. Genes Brain Behav. 7, 609–617. ( 10.1111/j.1601-183X.2008.00397.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ngun TC, et al. 2014. Feminized behavior and brain gene expression in a novel mouse model of Klinefelter syndrome. Arch. Sex Behav. 43, 1043–1057. ( 10.1007/s10508-014-0316-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bonthuis PJ, Huang WC, Stacher Horndli CN, Ferris E, Cheng T, Gregg C. 2015. Noncanonical genomic imprinting effects in offspring. Cell Rep. 12, 979–991. ( 10.1016/j.celrep.2015.07.017) [DOI] [PubMed] [Google Scholar]
- 85.Crowley JJ, et al. 2015. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat. Genet. 47, 353–360. ( 10.1038/ng.3222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Babak T, et al. 2015. Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat. Genet. 47, 544–549. ( 10.1038/ng.3274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu J, Burgoyne PS, Arnold AP. 2002. Sex differences in sex chromosome gene expression in mouse brain. Hum. Mol. Genet. 11, 1409–1419. ( 10.1093/hmg/11.12.1409) [DOI] [PubMed] [Google Scholar]
- 88.Xu J, Watkins R, Arnold AP. 2006. Sexually dimorphic expression of the X-linked gene Eif2s3x mRNA but not protein in mouse brain. Gene Expr. Patterns 6, 146–155. ( 10.1016/j.modgep.2005.06.011) [DOI] [PubMed] [Google Scholar]
- 89.Xu J, Deng X, Watkins R, Disteche CM. 2008. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci. 28, 4521–4527. ( 10.1523/JNEUROSCI.5382-07.2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xu J, Deng X, Disteche CM. 2008. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS ONE 3, e2553 ( 10.1371/journal.pone.0002553) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Armoskus C, Moreira D, Bollinger K, Jimenez O, Taniguchi S, Tsai HW. 2014. Identification of sexually dimorphic genes in the neonatal mouse cortex and hippocampus. Brain Res. 1562, 23–38. ( 10.1016/j.brainres.2014.03.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Werler S, Poplinski A, Gromoll J, Wistuba J. 2011. Expression of selected genes escaping from X inactivation in the 41, XXY* mouse model for Klinefelter's syndrome. Acta Paediatr. 100, 885–891. ( 10.1111/j.1651-2227.2010.02112.x) [DOI] [PubMed] [Google Scholar]
- 93.Greenfield A et al. 1998. The UTX gene escapes X inactivation in mice and humans. Hum. Mol. Genet. 7, 737–742. ( 10.1093/hmg/7.4.737) [DOI] [PubMed] [Google Scholar]
- 94.Wu J, Salido EC, Yen PH, Mohandas TK, Heng HH, Tsui LC, Park J, Chapman VM, Shapiro LJ. 1994. The murine Xe169 gene escapes X-inactivation like its human homologue. Nat. Genet. 7, 491–496. ( 10.1038/ng0894-491) [DOI] [PubMed] [Google Scholar]
- 95.Wu J, Ellison J, Salido E, Yen P, Mohandas T, Shapiro LJ. 1994. Isolation and characterization of XE169, a novel human gene that escapes X-inactivation. Hum. Mol. Genet. 3, 153–160. ( 10.1093/hmg/3.1.153) [DOI] [PubMed] [Google Scholar]
- 96.Sheardown S, Norris D, Fisher A, Brockdorff N. 1996. The mouse Smcx gene exhibits developmental and tissue specific variation in degree of escape from X inactivation. Hum. Mol. Genet. 5, 1355–1360. ( 10.1093/hmg/5.9.1355) [DOI] [PubMed] [Google Scholar]
- 97.Carrel L, Willard HF. 2005. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434, 400–404. ( 10.1038/nature03479) [DOI] [PubMed] [Google Scholar]
- 98.Banka S, et al. 2015. Novel KDM6A (UTX) mutations and a clinical and molecular review of the X-linked Kabuki syndrome (KS2). Clin. Genet. 87, 252–258. ( 10.1111/cge.12363) [DOI] [PubMed] [Google Scholar]
- 99.Santos-Reboucas CB, et al. 2015. A novel nonsense mutation in KDM5C/JARID1C gene causing intellectual disability, short stature and speech delay. Neurosci. Lett. 498, 67–71. ( 10.1016/j.neulet.2011.04.065) [DOI] [PubMed] [Google Scholar]
- 100.Snijders Blok L, et al. 2015. Mutations in DDX3X are a common cause of unexplained intellectual disability with gender-specific effects on Wnt signaling. Am. J. Hum. Genet. 97, 343–352. ( 10.1016/j.ajhg.2015.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamauchi Y, Riel JM, Stoytcheva Z, Ward MA. 2014. Two Y genes can replace the entire Y chromosome for assisted reproduction in the mouse. Science 343, 69–72. ( 10.1126/science.1242544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shpargel KB, Sengoku T, Yokoyama S, Magnuson T. 2012. UTX and UTY demonstrate histone demethylase-independent function in mouse embryonic development. PLoS Genet. 8, e1002964 ( 10.1371/journal.pgen.1002964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dewing P, et al. 2006. Direct regulation of adult brain function by the male-specific factor SRY. Curr. Biol. 16, 415–420. ( 10.1016/j.cub.2006.01.017) [DOI] [PubMed] [Google Scholar]
- 104.McCarthy MM, Arnold AP. 2011. Reframing sexual differentiation of the brain. Nat. Neurosci. 14, 677–683. ( 10.1038/nn.2834) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ngun TC, Ghahramani N, Sanchez FJ, Bocklandt S, Vilain E. 2011. The genetics of sex differences in brain and behavior. Front. Neuroendocrinol. 32, 227–246. ( 10.1016/j.yfrne.2010.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]