Abstract
The completion of whole genome sequencing projects has provided the genetic instructions of life. However, whereas the identification of gene coding regions has progressed, the mapping of transcriptional regulatory motifs has moved more slowly. To understand how distinct expression profiles can be established and maintained, a greater understanding of these sequences and their trans-acting factors is required. Herein we have used a combined in silico and biochemical approach to identify binding sites [repressor element 1/neuron-restrictive silencer element (RE1/NRSE)] and potential target genes of RE1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) within the human, mouse, and Fugu rubripes genomes. We have used this genome-wide analysis to identify 1,892 human, 1,894 mouse, and 554 Fugu RE1/NRSEs and present their location and gene linkages in a searchable database. Furthermore, we identified an in vivo hierarchy in which distinct subsets of RE1/NRSEs interact with endogenous levels of REST/NRSF, whereas others function as bona fide transcriptional control elements only in the presence of elevated levels of REST/NRSF. These data show that individual RE1/NRSE sites interact differentially with REST/NRSF within a particular cell type. This combined bioinformatic and biochemical approach serves to illustrate the selective manner in which a transcription factor interacts with its potential binding sites and regulates target genes. In addition, this approach provides a unique whole-genome map for a given transcription factor-binding site implicated in establishing specific patterns of neuronal gene expression.
Patterns of gene expression in multicellular organisms are established and maintained primarily through interaction of transcription factors with target genes and subsequent transcriptional regulation. However the repertoire of direct target genes of most transcription factors remains unknown. To ultimately understand how the interplay of multiple transcription factors regulates global gene expression, it will be necessary to identify all of the target genes for all transcription factors. This process has primarily relied on individual studies interrogating specific gene promoter sequences for defined regulatory sequence elements. Although such studies yield large volumes of valuable information, they do not provide a complete overview of transcription factor binding sites across a whole genome. The recent successes of genome sequencing projects have provided the field of bioinformatics with the necessary data to achieve this aim, by allowing interrogation of genome sequences for consensus transcription factor binding sites. However, such in silico analyses are hampered by the short and often redundant nature of most transcription factor binding sites, (≈4–8 bp) resulting in the identification of motifs at frequencies many times greater than the small number of bona fide binding sites. Therefore strategies have evolved to make such analyses context-dependent. Searches for clusters of homologous sites (1–4), proximity to other regulatory elements (5–7), or CpG island and core promoters (2, 8) have been successfully used to identify bona fide transcription factor-binding sites but can be computationally intense. However, simple pattern matching algorithms designed to search for large regulatory motifs (which should occur at a low frequency by chance) do not require such context dependency and provide the opportunity to identify a complete set of target genes for a given transcription factor.
Here we have focused on repressor element 1 (RE1), a silencing element [also known as the neuron-restrictive silencer element (NRSE)] that was originally found in the 5′ flanking region of the voltage-gated sodium type II channel (NaV1.2) and superior cervical ganglion 10 (SCG10) genes (9, 10). This 21-bp element has subsequently been identified in the regulatory regions of >30 genes, most of which are neuron-specific (11–20). The essential Kruppel-type zinc-finger transcriptional repressor RE1-silencing transcription factor (REST; also known as the neuron-restrictive silencing factor, or NRSF) interacts with the RE1, and extensive REST expression has been detected in nonneural tissues of several vertebrate species, including Xenopus laevis, Fugu rubripes, chick, mouse, rat, and human (21, 22). REST represses gene expression by recruiting two histone deacetylase-containing corepressor complexes (23–27). The proposed role of REST is that of a transcriptional silencer that restricts neuronal gene expression to the nervous system by silencing expression of these genes in nonneural tissues (21, 22). However, the role of REST in vivo is more ambiguous. REST mRNA is present in adult CNS neurons (28), and its levels can be elevated in response to ischaemic or epileptic insults (28, 29) and more recently, REST protein has been shown to interact with huntingtin, the product of the Huntington's disease gene (30). Against this background, we have undertaken to identify all RE1s and their corresponding target genes across the human, mouse, and Fugu genomes by using a combination of bioinformatic and biochemical approaches. Further, we have used chromatin immunoprecipitation (ChIP) and expression analysis to demonstrate interaction of REST with its endogenous target genes. This study highlights the fact that a single transcription factor can have a highly selective pattern of target gene recruitment within the same cell population.
Materials and Methods
RE1 Database Construction. A search was performed of the human genome GenBank formatted DNA sequence flat files (downloaded from ensembl Version 11.31 and organized as overlapping clones) by using a perl script constrained by a core 17 nucleotide regular expression pattern. This pattern represents an RE1 consensus sequence, derived from alignment of 32 known RE1 sequences containing degeneracies reflecting known variations (similar to ref. 31). The search output and corresponding annotations or external references (swiss-prot Version 40.43 and trembl Version 22.13 protein sequence databases) were used to assign gene description and determination of annotated genes within 100 kb on either strand. The mouse and Fugu genomes were similarly interrogated (by using ensembl Versions 11.3 and 11.2 respectively). Determination of CpG island proximity within the search output used the newcpgreport program (Emboss 2002, www.hgmp.mrc.ac.uk/Software/EMBOSS) with default settings, and a screen to remove search hits in repetitive genomic regions was used. A mySQL relational database (RE1db) of the search results was created (using version 4.10α) and is freely accessible at http://bioinformatics.leeds.ac.uk/group/online/RE1db/re1db_home.htm.
Cell Culture. JTC-19 and U373 cells were cultured in DMEM with 10% FCS containing 6 g/liter penicillin, 10 g/liter streptomycin, and 2 mM l-glutamine at 37°C in 5% CO2.
ChIP. Anti-REST antibody (P-18, Santa Cruz Biotechnology), anti-histone H3 (Upstate Biotechnology, Lake Placid, NY) and normal rabbit IgG (Sigma) were used to carry out a ChIP analysis on U373 cells. ChIPs were performed essentially as described in ref. 27. Purified DNA was analyzed by using real-time PCR (iQ Cycler, Bio-Rad), and 2 μl was used as template in 20-μl real-time PCRs carried out in duplicate. Primer concentration was 300 nM. Individual primer sequences are available as supporting information, which includes Tables 1–4 and Fig. 6 and is published on the PNAS web site.
Adenoviral Construction, Amplification, and Infection. Adenoviral vectors were produced as described in ref. 32. Approximately 109 plaque-forming unit/ml virus particles were used to infect JTC-19 cells or U373 cells. Whole-cell protein, RNA, and chromatin were harvested 48 h later as described in refs. 27 and 32.
Electromobility Shift Assays (EMSA). Klenow fill-in of BglII generated overhangs from pGL3NaII (32), and polyacrylamide electrophoresis purification produced 150-bp, NaV1.2 RE1, [32P]dATP-labeled DNA probes. Nuclear protein was prepared as described in ref. 32, and EMSAs were performed as detailed in ref. 33 (see supporting information for complementary deoxyoligonucleotide sequences used as competitors).
RT-PCR Analysis. Extracted RNA was reverse transcribed by using MMLV RNase H(–) reverse transcriptase (Promega) as described in ref. 32. Two microliters of resultant cDNA was used as template in a 20-μl PCR reaction, with deoxyoligonucleotide primers designed to putative REST/NRSF target genes (see supporting information). PCR products were resolved by electrophoresis on 2% agarose gels and stained with ethidium bromide.
Results
In Silico Identification of RE1 Sites Across Human, Mouse, and Fugu Genomes. Our initial goal was to identify all potential RE1s in the human and mouse genomes by using a bioinformatics approach. However, publication of the Fugu genome (34) prompted us to inspect it for any potential REST homologs, and we subsequently identified a partial gene sequence showing 52% amino acid sequence identity with the DNA-binding domain of human and mouse REST (ENSF00000003748). Consequently, we extended our bioinformatic search to include the Fugu genome. REST homologs have been identified in human, mouse, rat, chick, Xenopus, Danio rerio (subsequently identified as ENS-DARG00000007222), and Fugu; whereas no REST is present in Drosophila, indicating that REST may have first appeared within the last 500 million years, concomitant with vertebrate evolution. A consensus RE1 based on the sequences of 32 known RE1 elements NT(T/C)AG(A/C)(A/G)CCNN(A/G)G(A/C)(G/S)AG was used to search human, mouse, and Fugu ensembl genome sequence databases by using a perl script. The information obtained from these searches has been collated into a searchable online database: RE1db (http://bioinformatics.leeds.ac.uk/group/online/RE1db/re1db_home.htm). The RE1db database includes information on the exact RE1 sequence, position/orientation within a chromosome/contig, and the closest transcriptional units that either overlap or lie within 100 kb of the RE1. Further constraints can be imposed by limiting the search to exonic, intronic, or intergenic sequences (at varying distances from annotated transcriptional start sites) or within regions that either contain CpG islands or are proximal (<500 bp) to them. It is also possible to constrain the output to specific genes by using ensembl gene identifiers, thus establishing whether a particular gene is localized to an RE1. The numbers of putative RE1s identified in the human, mouse, and Fugu genomes were 1,892, 1,894, and 554, respectively, and there are 355, 358, and 416 transcribed units that have RE1s within 10 kb in their 5′ region and 593, 564, and 181 genes that harbor intragenic RE1s (because of the condensed nature of the Fugu genome, more than one transcriptional unit may lie within 10 kb of any individual RE1). Information on the frequency of occurrence of the most common RE1 sequence variations in the human, mouse, and Fugu genomes are given in the supporting information. Comprehensive information can be found online in the RE1db database.
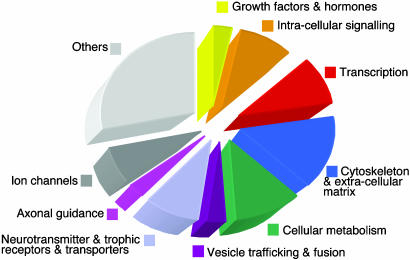
Classification of Genes Found in Silico. We then proceeded to classify potential REST target genes identified with our bioinformatics approach by using ensembl genome annotations (Fig. 1). At least 40% of the genes identified are known to be expressed within the nervous system, consistent with current models of REST function as a silencer or repressor of neuronal gene expression (24, 33, 35, 36). These genes include those encoding neurotransmitter receptors (e.g., M4 muscarinic, D3 dopamine, and γ-aminobutyric acid type β3 receptors), transporters (e.g., γ-aminobutyric acid transporter 4) and neurotrophic receptors [e.g., neurotrophic tyrosine kinase receptor type 3 (NTRK3) and glial cell line-derived neurotrophic factor receptors] and those encoding proteins involved in vesicular trafficking and fusion [e.g., synaptosomal-associated protein, 25 kDa (SNAP25); synaptotagmins IV, V and VII; syntaxin 8; and Rab3), ion channels (e.g., NaV1.3, Kv3.4, and Cavl.3) and axonal guidance [e.g., SCG10, stathmin 3, netrin-2, roundabout, semaphorin 5A, L1 cell-adhesion molecule (L1CAM)]. There are also many genes that encode proteins that do not have obvious neuron-specific functions, such as those involved in cellular metabolic processes (e.g., peroxisome and proteosome components). Additionally, there are genes that specify proteins that perform neuronal functions but are also required in nonneuronal tissues, including those involved in the regulation of cardiovascular tone, such as endothelial nitric oxide synthase, vasoactive intestinal peptide, atrial natriuretic peptide, brain natriuretic peptide, and KCNH2. A comprehensive list can be found online in the RE1db database.
Fig. 1.
Assignment of putative REST target genes within the RE1db database can be assigned to 1 of 10 functional groups. The database includes information on the exact RE1 sequence, position/orientation within a chromosome/contig, and the closest transcriptional units that either overlap or lie within 100 kb of the RE1 and can be accessed at http://bioinformatics.leeds.ac.uk/group/online/RE1db/re1db_home.htm.
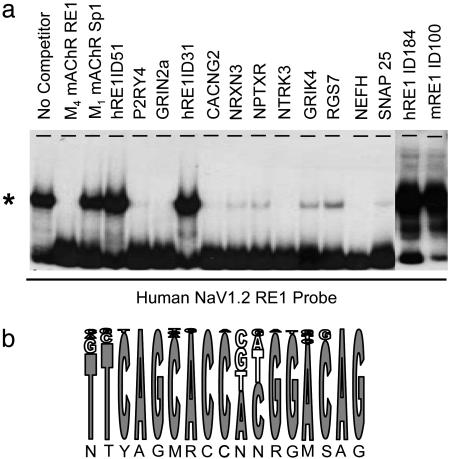
Identification of Bona Fide RE1 Sequences. We next wished to establish which RE1 sequences identified above represented bona fide regulatory sequences, i.e., which sites could bind REST in vitro. To this end we carried out a series of EMSA. The RE1 consensus sequence used in this study is degenerate, allowing a total of 4,096 permutations. Of these, 892, 944, and 291 are found in the human, mouse, and Fugu genomes, respectively. The frequency at which each RE1 variation occurred in the human genome was compared to the frequency in the mouse and Fugu genomes (see Table 1). We reasoned RE1s having higher frequencies are more likely to be functionally important because of their conservation in the regulatory regions of many genes. Accordingly, EMSA analysis was carried out by using 100-fold molar excess of individual RE1s (derived from Table 1) to compete the gel shift produced by incubation of whole-cell protein from JTC19 cells infected with an adenovirus (Ad) carrying a REST DNA-binding domain with a labeled RE1 probe derived from the NaV1.2 gene (Fig. 2a). An RE1 derived from the M4 muscarinic receptor gene was used as a positive control (16, 20). RE1s derived from P2Y4 purinergic receptor (P2RY4), NMDA glutamate receptor 2a (GRIN2a), voltage-gated Ca2+ channel γ2 (CACNG2), glutamate receptor KA1 (GRIK4), and neurofilament triplet H (NEFH) genes all produced a complete inhibition of the gel shift at 100-fold excess; whereas RE1s derived from the neurexin III (NRXN3), neuronal pentraxin receptor (NPTXR), SNAP25, NTRK3, and regulator of G protein signaling 7 (RGs7) genes produced a partial inhibition. We reasoned that this variation in expression may reflect differences in the relative binding affinities of these RE1s for the REST DNA-binding domain used in the EMSA (see below). Sequences that did not compete (hR E1ID51, hR E1ID184, and hRE1ID315) lacked proximity to any annotated genes and occurred exclusively within repetitive regions of the human genome and were not found in either mouse or Fugu. An EMSA analysis showing the inability of hRE1ID51 to bind REST is shown in supporting information. Interestingly, the most prevalent RE1 sequence that was unique to the mouse genome (mRE1ID100) was also unable to bind REST (Fig. 2a). An overall consensus sequence showing the relative occurrence of each base at each position of all functional RE1s identified in our search of the human genome and in previous studies is presented in Fig. 2b.
Fig. 2.
Putative RE1 sequences linked to potential REST target genes can interact with REST. (a) EMSA analysis was carried out by using 100-fold molar excess of putative RE1s to compete the gel shift produced by incubation of nuclear extracts containing the REST DNA-binding domain with a radiolabeled RE1 derived from the NaV1.2 gene. (b) Consensus RE1 derived from EMSA data showing the relative frequency of occurrence of individual bases at each position of the RE1. The single-letter code is the standard Nomenclature Committee of the International Union of Biochemistry format for incompletely specified bases in nucleic acid sequences (www.chem.qmul.ac.uk/iubmb/misc/naseq.html).
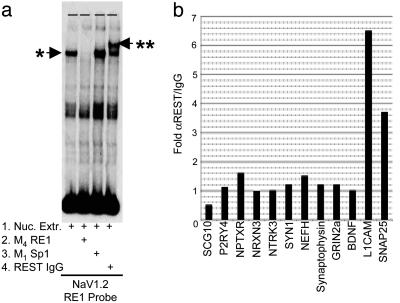
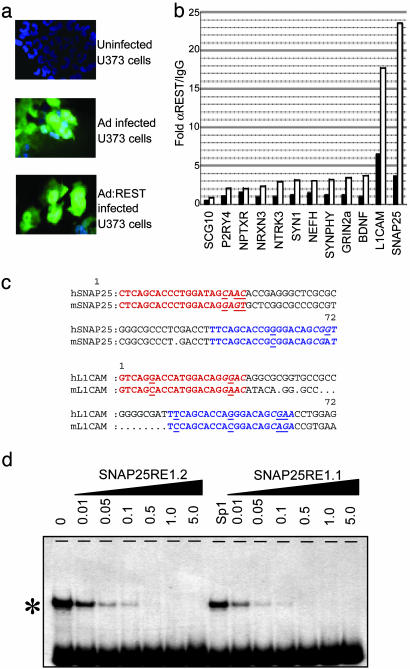
Occupancy of Endogenous Genes by REST. Having identified those RE1s capable of binding REST in vitro, we proceeded to examine interaction of REST with its endogenous target genes. To this end, we carried out ChIP assays in U373 glioma cells. U373 cells were chosen because, although the role played by REST in nonneural cells has been extensively studied, its role within neural cells is poorly understood. We first confirmed that U373 expressed functional REST protein by using an EMSA to show that nuclear protein extracts from U373 produce a specific protein/DNA complex that could be recognized and “supershifted” by an anti-REST antibody (Fig. 3a). Interactions between endogenous U373 REST and the RE1s of published and putative target genes were then probed by ChIP (Fig. 3b). Interestingly, despite the presence of functional U373 REST protein, the RE1s of five of the six known target genes studied, SCG10 (9), GRIN2a (13), synapsin 1 (SYN1) (18), synaptophysin (SYNPHY) (31), and brain-derived neurotrophic factor (BDNF) (19) were not enriched by using an anti-REST antibody. Only the L1CAM gene RE1 (14, 15) was enriched. Six RE1s associated with P2RY4, NPTXR, NRXN3, NTRK3, NEFH, and SNAP25 genes were also tested. Of these, only the RE1 of the SNAP25 gene was enriched. These data suggest that only a subset of RE1s is occupied in U373 cells (or that the L1CAM and SNAP25 gene RE1s show greater REST occupancy). Because all of these RE1 elements are capable of binding REST in vitro (Fig. 2), we considered the possibility that occupancy of the native gene may be limited by endogenous levels of REST. To test this hypothesis, we used an Ad construct (Ad:REST) to drive high levels of REST expression in U373 cells. In all cases, with the exception of the SCG10 RE1, overexpression of REST led to detectable levels of occupancy (Fig. 4b) clearly demonstrating that these sites are all accessible to REST. Further, this experiment shows that apparent lack of endogenous REST recruitment was not caused by the masking of the REST epitope by the chromatin environment. Interestingly, SNAP25 and L1CAM showed greater REST occupancy than the other loci in the presence of either native or overexpressed REST. Closer inspection of the RE1 flanking sequences revealed the presence of an additional RE1 within 30 bp of the original RE1 exclusively in the SNAP25 and L1CAM genes. These second sites deviated from the consensus RE1 used in this study by 1 bp. Nevertheless, subsequent EMSA analysis showed that both of the secondary RE1 sites could bind REST, albeit at a lower affinity than the primary sites (Fig. 4d). Three groups of RE1s could be distinguished. High-affinity sequences were defined as those producing a complete inhibition of the gel shift at 0.5 μM. Low-affinity sequences were those that required 1 μM competitor, whereas those that produced no inhibition, even at 5 μM competitor, were deemed as unable to bind (see Figs. 2 and 4d and supporting information). The existence of tandem RE1s offers a potential explanation for the increased REST occupancy of the SNAP25 and L1CAM genes relative to those genes possessing only a singular RE1.
Fig. 3.
REST/NRSF is selectively recruited to target genes. (a) EMSA analysis of U373 protein extracts incubated with a radiolabeled RE1 derived from the NaV1.2 gene shows a specific “shift” (*) that can be supershifted in the presence of an anti-REST antibody (**), confirming the presence of functional REST protein within U373 cell nuclei. (b) ChIP analysis carried out on U373 cells by using an anti-REST antibody shows detectable levels of U373 REST occupancy only at the RE1s of the SNAP25 and L1CAM genes.
Fig. 4.
Overexpressed REST can interact with RE1s identified in the RE1db in U373 cells. (a) Ad vectors expressing enhanced GFP or enhanced GFP and REST (Ad:REST) were used to effect transgene delivery to U373 cells. Efficiency of gene delivery can be seen from enhanced GFP fluorescence in the photomicrographs, whereas transgene expression can be seen by RT-PCR by using primers directed against REST and GAPDH. Cycle numbers are shown on the left of each panel. (b) ChIP analysis of U373 cells. Chromatin was extracted and precipitated with an anti-REST antibody. Gene-specific primers were used to assess RE1/NRSE occupancy in native U373 (filled bars) and Ad:REST-infected U373 (open bars). (c) Sequence analysis of the SNAP25 and L1CAM gene RE1-flanking regions reveals the presence of secondary RE1/NRSEs conserved between human and mouse. Blue sequences are the originally identified RE1s, and red sequences are the RE1s identified in this study. (d) EMSA analysis of the primary RE1 (SNAP25 RE1.1) and secondary RE1 (SNAP25 RE1.2). Analysis was carried out by using 0.01–5.0 μM unlabeled RE1s to compete the gel shift (*) produced by incubation of nuclear extracts with a radiolabeled RE1 derived from the NaV1.2 gene.
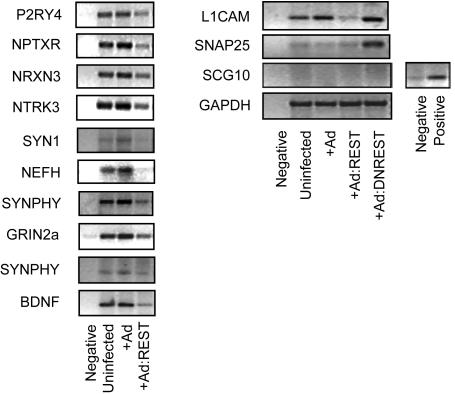
Regulation of Endogenous Gene Expression by REST. Having demonstrated REST/NRSF occupancy by ChIP, we proceeded to use RT-PCR to compare expression levels of each of these genes in uninfected U373 cells with those infected with empty Ad or Ad carrying either REST (Ad:REST) or a dominant negative REST (DNREST) construct (Fig. 5). This latter construct consisted solely of the REST DNA-binding domain and would be expected to lead to derepression of those genes for which presence of REST is required for maintenance of repression or silence. Infection levels using either the Ad:DNREST vector or Ad:REST were identical (Fig. 4a). All target genes except SCG10 are expressed in native U373 cells. Infection with Ad-:REST led to further repression of the majority of genes, including P2RY4, NPTXR, NRXN3, NTRK3, SYN1, NEFH, SYNPHY, GRIN2a, BDNF, and L1CAM. Only SNAP25 and SCG10 gene expression were unaffected. Infection with Ad:DNREST led to derepression of only the SNAP25 gene. The SCG10 gene remained silent under all conditions. Collectively, these results allow four classes of genes to be distinguished. The first group represents the majority of target genes and includes the P2RY4, NPTXR, NRXN3, NTRK3, SYN1, NEFH, SYNPHY, GRIN2a, and BDNF genes. These genes are transcriptionally active and unoccupied by endogenous levels of REST but can be repressed by overexpression of REST. The second group comprises the SNAP25 gene. SNAP25 is also transcribed in U373 cells but is repressed by endogenous levels of REST, and overexpression of REST leads to a greater occupancy but no further repression. The third group is represented by the L1CAM gene. As with SNAP25, L1CAM is transcribed and occupied by endogenous REST, but, unlike SNAP25, L1CAM does not appear to be repressed by endogenous levels of REST. Repression only occurs in the presence of overexpressed REST. The final group comprises the SCG10 gene that is not occupied by REST and remains silent irrespective of the level of REST or DNREST expression. In addition to validating the putative REST target genes, this study highlights the highly selective manner in which endogenous REST is preferentially recruited to specific RE1 sites and is able to differentially regulate target gene expression within the same neural cell.
Fig. 5.
Regulation of gene expression by REST in U373 cells. RT-PCR analysis of mRNA expression levels was carried out on native cells and cells infected with either an Ad vector carrying REST (Ad:REST) or a dominant negative construct comprising the REST DNA-binding domain.
Discussion
This study represents a combined bioinformatic and biochemical approach to the genome-wide identification of RE1 sites and REST target genes. We found 1,892, 1,894, and 554 RE1 sites in the human, mouse, and Fugu genomes, respectively. These numbers are considerably greater than those expected by chance alone (770, 653, and 88, respectively). It is important to note that this study, although comprehensive, does not identify all RE1 sites, as exemplified by the secondary RE1 sites discovered in the L1CAM and SNAP25 genes that diverge from the consensus RE1 (Fig. 4c). However, it was considered important to adopt a conservative consensus sequence for this analysis to minimize identification of false positives. Similarly, adoption of too stringent a consensus could lead to omission of bona fide targets, because individual bases identified as critical in the context of one consensus regulatory element have been shown to be redundant in the context of alternative regulatory elements (31, 37).
The Fugu genome contains 38,000 genes (34), a similar number to its mammalian relatives, yet it is largely devoid of repetitive DNA, resulting in a very compact genome of 365 Mb, only 8% of the size of the human genome. On purely stochastic grounds, the RE1 motif should occur 88 times in the Fugu genome; yet it actual occurs 554 times. This finding suggests that the majority of these motifs represent bona fide RE1 sites, indicating a lower estimate for the number of potential REST target genes to be ≈460. This number is likely to be higher in the mammalian genomes, for which the number of RE1s identified was higher. Interrogation of the Drosophila melanogaster genome (that contains no REST homolog) substantiates this observation, given that the occurrence of RE1s is 51 (D. melanogaster genome release, ensembl Version 11.3), a number that is close to the 39 occurrences predicted by chance alone. Consistent occurrence of RE1 consensus sequences at frequencies far greater than their chance occurrence in organisms that express REST suggests that most of these occurrences correspond to genuine RE1s. However, the reduced number of RE1s seen in the Fugu genome suggests that some of the identified sequences in the mammalian genomes are potentially nonfunctional, such as those present within repetitive DNA that are unable to bind REST in vitro (Fig. 2). Accordingly, the RE1db has been modified to filter out RE1/NRSE sequences that appear in known repetitive regions.
The RE1 search presented here substantially expands on a previous limited search conducted before completion of vertebrate genome sequencing projects that used a simple fasta search of the GenBank DNA sequence database to identify 25 candidate genes (31). Another group (38) reported a bioinformatic search for RE1s, but little detail is available on the type of search tool or parameters used, and no details of chromosomal position, species, or RE1 sequence were provided, making comparison difficult. Nevertheless, of the select 75 genes listed in the study, 60 are identified here (as part of the full complement listed in the RE1db). Functional classification of these genes reveals many genes that encode proteins specifically or selectively involved with neuronal functions (e.g., synaptic release and neurotransmission). However some gene products also play roles in nonneuronal tissues, such as atrial natriuretic peptide, brain natriuretic peptide, endothelial nitric oxide synthase, vasoactive intestinal peptide, KCNH2, and voltage-gated potassium type β2 channel, all of which are involved in regulation of cardiovascular or cardiac function (39–41).
In silico and in vitro approaches can identify potential REST-binding sites; however, neither indicates whether endogenous genes are capable of being regulated by REST in vivo. By using ChIP, we have shown that individual RE1s can interact with either endogenous or elevated levels of REST in U373 cells resulting in transcriptional repression. Interestingly, only the SNAP25 and L1CAM genes interact with endogenous REST in U373 cells. The majority of genes do not appear to interact with endogenous levels of U373 REST yet retain the ability to bind REST when expression levels are higher and, as such, appear to be “poised” for repression. The lack of binding at these loci is not attributable to an endogenous REST deficiency, because U373 cells express REST that can bind RE1s in vitro (Fig. 4a), which is detected at the RE1-containing regions of the L1CAM and SNAP25 genes by ChIP (Fig. 4b). How is such a preferential pattern of REST–RE1 interaction established within the same cell nucleus? The existence of tandem RE1 sites in the L1CAM and SNAP25 genes may explain the enhanced occupancy by REST at these two loci. The close proximity of these tandem RE1s is such that ChIP cannot resolve whether there is a preferential occupancy of one site or whether their relative occupancy changes in response to elevated REST levels. However the possibility clearly exists that multiple RE1s have the potential to allow repression to occur at different threshold concentrations or over a greater range of REST concentrations. The fact that the L1CAM gene contains 2 RE1s (Fig. 4c) has important implications for the interpretation of a previous transgenic mouse study in which the L1CAM expression pattern was compared between a reporter gene driven by the L1CAM promoter and one with a mutation in the previously characterized RE1 (14). Removal of the consensus RE1 led to selective derepression of reporter gene expression in mesenchymal derivatives of the neural crest, mesoderm, and ectoderm but not in all tissues where REST is expressed. An implication of the present study is that that mutation of the second RE1 may be required to see ectopic expression in these other tissues.
The observation that REST can selectively bind and regulate target genes within the same cell nucleus resonates with a recent report showing that endogenous c-Myc is detected at only 11% of the targets with which over-expressed c-Myc is capable of binding (42). This association with high-affinity sites seemed to correlate predominantly, although not exclusively, with CpG islands that were characterized by high levels of histone H3 and histone H4 acetylation typical of transcriptionally active chromatin structure. In contrast, overexpressed c-Myc could also associate with low-affinity sites that were characterized by a lower level of basal histone acetylation (42). This model does not appear to offer an analogous explanation for the preferential REST occupancy in U373 cells, because the SCG10 and SNAP25 RE1 sites are both found proximal to CpG islands but only the SNAP25 gene is occupied by endogenous levels of REST. Another report describes selective transcription factor-binding-site occupancy of Ste12p mediated by two different mitogen-activated protein kinases in Saccharomyces cerevisiae (43). Clearly, there is precedent for selective binding of transcription factors, yet the mechanism used by REST to achieve this remains to be elucidated. However, such binding selectivity would lend an extra dimension to REST regulation. For example, REST expression has been shown to be dynamically regulated in the rat forebrain during neurogenesis (21, 22), and such changes in REST levels could lead to various RE1-binding profiles and therefore target gene regulation at different developmental stages. Additionally REST levels have been reported to increase in CA1 pyramidal neurons of the hippocampus in response to ischaemia (29) and across the hippocampus in kainate-induced seizures (28). It is not inconceivable that, as REST levels increase, different classes of RE1 sites can become occupied, thereby establishing a temporally regulated response of REST target gene expression.
In conclusion, a genome-wide analysis of RE1s and potential REST target genes across three different vertebrate genomes has been undertaken. We have integrated this information into a freely available online database. We have distinguished four groups of target genes based on their occupancy and regulation by REST in U373 glioma cells. We anticipate that membership of these groups will change according to cell type and/or developmental stage.
Supplementary Material
Acknowledgments
The Wellcome Trust and the U.K. Biotechnology and Biological Sciences Research Council (BBSRC) funded this work. A.W.B. is a BBSRC research student.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RE1, repressor element 1; NRSE, neuron-restrictive silencer element; REST, repressor element 1 silencing transcription factor; NRSF, neuron-restrictive silencing factor; ChIP, chromatin immunoprecipitation; EMSA, electromobility shift assay; Ad, adenovirus; DNREST, dominant negative REST.
References
- 1.Davidson, E. (2001) Genomic Regulation Systems: Development and Evolution (Academic, San Diego).
- 2.Kel, A. E., Kel-Margoulis, O. V., Farnham, P. J., Bartley, S. M., Wingender, E. & Zhang, M. Q. (2001) J. Mol. Biol. 309, 99–120. [DOI] [PubMed] [Google Scholar]
- 3.Markstein, M., Markstein, P., Markstein, V. & Levine, M. S. (2002) Proc. Natl. Acad. Sci. USA 99, 763–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rebeiz, M., Reeves, N. L. & Posakony, J. W. (2002) Proc. Natl. Acad. Sci. USA 99, 9888–9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman, B. P., Nibu, Y., Pfeiffer, B. D., Tomancak, P., Celniker, S. E., Levine, M., Rubin, G. M. & Eisen, M. B. (2002) Proc. Natl. Acad. Sci. USA 99, 757–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wagner, A. (1998) Genomics 50, 293–295. [DOI] [PubMed] [Google Scholar]
- 7.Wagner, A. (1998) Pac. Symp. Biocomput. 264–278. [PubMed]
- 8.Weinmann, A. S., Bartley, S. M., Zhang, T., Zhang, M. Q. & Farnham, P. J. (2001) Mol. Cell. Biol. 21, 6820–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mori, N., Schoenherr, C., Vandenbergh, D. J. & Anderson, D. J. (1992) Neuron 9, 45–54. [DOI] [PubMed] [Google Scholar]
- 10.Kraner, S. D., Chong, J. A., Tsay, H. J. & Mandel, G. (1992) Neuron 9, 37–44. [DOI] [PubMed] [Google Scholar]
- 11.Bessis, A., Champtiaux, N., Chatelin, L. & Changeux, J. P. (1997) Proc. Natl. Acad. Sci. USA 94, 5906–5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brene, S., Messer, C., Okado, H., Hartley, M., Heinemann, S. F. & Nestler, E. J. (2000) Eur. J. Neurosci. 12, 1525–1533. [DOI] [PubMed] [Google Scholar]
- 13.Desai, A., Turetsky, D., Vasudevan, K. & Buonanno, A. (2002) J. Biol. Chem. 277, 46374–46384. [DOI] [PubMed] [Google Scholar]
- 14.Kallunki, P., Edelman, G. M. & Jones, F. S. (1997) J. Cell Biol. 138, 1343–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kallunki, P., Edelman, G. M. & Jones, F. S. (1998) Proc. Natl. Acad. Sci. USA 95, 3233–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mieda, M., Haga, T. & Saffen, D. W. (1997) J. Biol. Chem. 272, 5854–5860. [DOI] [PubMed] [Google Scholar]
- 17.Myers, S. J., Peters, J., Huang, Y., Comer, M. B., Barthel, F. & Dingledine, R. (1998) J. Neurosci. 18, 6723–6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoch, S., Cibelli, G. & Thiel, G. (1996) J. Biol. Chem. 271, 3317–3323. [DOI] [PubMed] [Google Scholar]
- 19.Timmusk, T., Palm, K., Lendahl, U. & Metsis, M. (1999) J. Biol. Chem. 274, 1078–1084. [PubMed] [Google Scholar]
- 20.Wood, I. C., Roopra, A. & Buckley, N. J. (1996) J. Biol. Chem. 271, 14221–14225. [DOI] [PubMed] [Google Scholar]
- 21.Chong, J. A., Tapia-Ramirez, J., Kim, S., Toledo-Arai, J. J., Zheng, Y., Boutros, M. C., Altshuller, Y. M., Frohman, M. A., Kraner, S. D. & Mandel, G. (1995) Cell 80, 949–957. [DOI] [PubMed] [Google Scholar]
- 22.Schoenherr, C. J. & Anderson, D. J. (1995) Science 267, 1360–1363. [DOI] [PubMed] [Google Scholar]
- 23.Andres, M. E., Burger, C., Peral-Rubio, M. J., Battaglioli, E., Anderson, M. E., Grimes, J., Dallman, J., Ballas, N. & Mandel, G. (1999) Proc. Natl. Acad. Sci. USA 96, 9873–9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belyaev, N. D., Wood, I. C., Bruce, A. W., Street, M., Trinh, J. B. & Buckley, N. J. (2004) J. Biol. Chem. 279, 556–561. [DOI] [PubMed] [Google Scholar]
- 25.Grimes, J. A., Nielsen, S. J., Battaglioli, E., Miska, E. A., Speh, J. C., Berry, D. L., Atouf, F., Holdener, B. C., Mandel, G. & Kouzarides, T. (2000) J. Biol. Chem. 275, 9461–9467. [DOI] [PubMed] [Google Scholar]
- 26.Huang, Y., Myers, S. J. & Dingledine, R. (1999) Nat. Neurosci. 2, 867–872. [DOI] [PubMed] [Google Scholar]
- 27.Roopra, A., Sharling, L., Wood, I. C., Briggs, T., Bachfischer, U., Paquette, A. J. & Buckley, N. J. (2000) Mol. Cell. Biol. 20, 2147–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palm, K., Belluardo, N., Metsis, M. & Timmusk, T. (1998) J. Neurosci. 18, 1280–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calderone, A., Jover, T., Noh, K. M., Tanaka, H., Yokota, H., Lin, Y., Grooms, S. Y., Regis, R., Bennett, M. V. & Zukin, R. S. (2003) J. Neurosci. 23, 2112–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zuccato, C., Tartari, M., Crotti, A., Goffredo, D., Valenza, M., Conti, L., Cataudella, T., Leavitt, B. R., Hayden, M. R., Timmusk, T., et al. (2003) Nat. Genet. 35, 76–83. [DOI] [PubMed] [Google Scholar]
- 31.Schoenherr, C. J., Paquette, A. J. & Anderson, D. J. (1996) Proc. Natl. Acad. Sci. USA 93, 9881–9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood, I. C., Belyaev, N. D., Bruce, A. W., Jones, C., Mistry, M., Roopra, A. & Buckley, N. J. (2003) J. Mol. Biol. 334, 863–874. [DOI] [PubMed] [Google Scholar]
- 33.Dent, C. L. & Latchman, D. S. (1993) in Transcription Factors: A Practical Approach, ed. Latchman, D. S. (IRL, Oxford), pp. 1–26.
- 34.Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J. M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. (2002) Science 297, 1301–1310. [DOI] [PubMed] [Google Scholar]
- 35.Ballas, N., Battaglioli, E., Atouf, F., Andres, M. E., Chenoweth, J., Anderson, M. E., Burger, C., Moniwa, M., Davie, J. R., Bowers, W. J., et al. (2001) Neuron 31, 353–365. [DOI] [PubMed] [Google Scholar]
- 36.Schoenherr, C. J. & Anderson, D. J. (1995) Curr. Opin. Neurobiol. 5, 566–571. [DOI] [PubMed] [Google Scholar]
- 37.Bulyk, M. L., Johnson, P. L. & Church, G. M. (2002) Nucleic Acids Res. 30, 1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lunyak, V. V., Burgess, R., Prefontaine, G. G., Nelson, C., Sze, S. H., Chenoweth, J., Schwartz, P., Pevzner, P. A., Glass, C., Mandel, G. & Rosenfeld, M. G. (2002) Science 298, 1747–1752. [DOI] [PubMed] [Google Scholar]
- 39.Kuwahara, K., Saito, Y., Takano, M., Arai, Y., Yasuno, S., Nakagawa, Y., Takahashi, N., Adachi, Y., Takemura, G., Horie, M., et al. (2003) EMBO J. 22, 6310–6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuwahara, K., Saito, Y., Ogawa, E., Takahashi, N., Nakagawa, Y., Naruse, Y., Harada, M., Hamanaka, I., Izumi, T., Miyamoto, Y., et al. (2001) Mol. Cell. Biol. 21, 2085–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogawa, E., Saito, Y., Kuwahara, K., Harada, M., Miyamoto, Y., Hamanaka, I., Kajiyama, N., Takahashi, N., Izumi, T., Kawakami, R., et al. (2002) Cardiovasc. Res. 53, 451–459. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez, P. C., Frank, S. R., Wang, L., Schroeder, M., Liu, S., Greene, J., Cocito, A. & Amati, B. (2003) Genes Dev. 17, 1115–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeitlinger, J., Simon, I., Harbison, C. T., Hannett, N. M., Volkert, T. L., Fink, G. R. & Young, R. A. (2003) Cell 113, 395–404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.