Abstract
The functionality, substrate specificity, and regiospecificity of enzymes typically evolve by the accumulation of mutations in the catalytic portion of the enzyme until new properties arise. However, emerging evidence suggests enzyme functionality can also be influenced by metabolic context. When the plastidial Arabidopsis 16:0Δ7 desaturase FAD5 (ADS3) was retargeted to the cytoplasm, regiospecificity shifted 70-fold, Δ7 to Δ9. Conversely, retargeting of two related cytoplasmic 16:0Δ9 Arabidopsis desaturases (ADS1 and ADS2) to the plastid, shifted regiospecificity ≈25-fold, Δ9 to Δ7. All three desaturases exhibited Δ9 regiospecificity when expressed in yeast, with desaturated products found predominantly on phosphatidylcholine. Coexpression of each enzyme with cucumber monogalactosyldiacylglycerol (MGDG) synthase in yeast conferred Δ7 desaturation, with 16:1Δ7 accumulating specifically on the plastidial lipid MGDG. Positional analysis is consistent with ADS desaturation of 16:0 on MGDG. The lipid headgroup acts as a molecular switch for desaturase regiospecificity. FAD5 Δ7 regiospecificity is thus attributable to plastidial retargeting of the enzyme by addition of a transit peptide to a cytoplasmic Δ9 desaturase rather than the numerous sequence differences within the catalytic portion of ADS enzymes. The MGDG-dependent desaturase activity enabled plants to synthesize 16:1Δ7 and its abundant metabolite, 16:3Δ7,10,13. Bioinformatics analysis of the Arabidopsis genome identified 239 protein families that contain members predicted to reside in different subcellular compartments, suggesting alternative targeting is widespread. Alternative targeting of bifunctional or multifunctional enzymes can exploit eukaryotic subcellular organization to create metabolic diversity by permitting isozymes to interact with different substrates and thus create different products in alternate compartments.
Keywords: diiron, enzyme evolution, regiospecificity, compartmentation, FAD5
Metabolic diversity in living systems arises primarily from biotransformations catalyzed by enzymes; indeed life itself depends on the specificity of enzymes. The unique functionality, substrate specificity, and regiospecificity of an enzyme typically evolves by the gradual accumulation of changes in the catalytic portion of the enzyme until new properties arise. However, there is an emerging body of evidence suggesting that an enzyme's functional characteristics can also be affected by its metabolic context and that temporal and spatial dynamics of enzyme interactions can be important determinants of enzyme functionality (1). Examples include the “rewiring” of mitogen-activated protein kinase pathways on alternative scaffolds (2); the localization-dependent interactions of transcription factors with RNA polymerase in the nucleus (reviewed in ref. 3); the interchangeable tissue-specific roles of the transcription factors WER and GL1 in plant epidermal hair development (4); the modification of laccase/peroxidase activity in the extracellular matrix by specific dirigent proteins (reviewed in ref. 5); the altered gating properties of the actin regulatory protein N-WASP associated with different “input” domains (6); or the alternative responses to steroid hormones, which depend on the cell-type distribution of various steroid receptors (reviewed in ref. 7). These examples of localization-dependent function involve recruitment of substrates by what has been termed adhesive interactions (1). While characterizing the Arabidopsis desaturase (ADS) enzyme family, we discovered that ADS specificity could be controlled by alternate substrate presentation in different subcellular compartments rather than by changes to the catalytic portion of the enzymes.
The ADS gene family encodes the largest family of fatty acid desaturases identified in the Arabidopsis genome, comprising nine members resembling membrane-bound cyanobacterial acyl-lipid desaturases and mammalian acyl-CoA desaturases (8). We characterized ADS1 (At1g06080) and ADS2 (At2g31360) (8) and a gene we initially termed ADS3 (At3g15850) to gain insights into their metabolic roles. ADS3 differs from other ADS enzymes in that it contains a 71-aa N-terminal extension representing a putative chloroplast transit peptide (9).
A white spruce ADS homolog with a transit peptide was recently characterized as an 18:0Δ9 desaturase (where x:yΔz is a fatty acid containing x carbons and y double bonds in position z counting from the carboxyl end) by heterologous expression in yeast (10). Mekhedov et al. (9) proposed that a mutation in one of two ADS genes (At3g15850 and At3g15870) found on Arabidopsis chromosome III is responsible for the loss of FAD5 activity (11). Loss of FAD5 function results in failure to form 16:1Δ7 on monogalactosyldiacylglycerol (MGDG) in the pathway leading to the production of 16:3Δ7,10,13, a predominant leaf fatty acid involved in cold tolerance (12). The predicted assignment was based on the mapped chromosomal location close to the fad5 locus (13), its similarity to acyl-CoA desaturase genes, the presence of a sequence encoding a transit peptide, and correlation of ESTs in species containing 16:3Δ7,10,13.
At the amino acid level, ADS1 and ADS2 share 76% identity, whereas ADS3 excluding the transit peptide (ADS372-371) is more divergent, with 53% identity to ADS1 and 51% identity to ADS2. The studies on ADS enzyme function in plants presented here show that the regiospecificity exhibited by the ADS enzymes depends on their subcellular targeting to different compartments providing access to 16:0 presented on different head groups. The mechanism controlling ADS regiospecificity thus differs fundamentally from the more widespread mechanism involving changes in the catalytic domains of enzymes that accumulate over the course of evolution.
Materials and Methods
cDNA Constructs. For yeast expression, cDNAs for ADS1, ADS2, and ADS372-371 were generated by PCR from Arabidopsis flower cDNA and introduced into the yeast expression plasmid pYES2 (Invitrogen). Details are available in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
For seed expression, ADS1, ADS2, ADS372-371 (i.e., ADS enzymes lacking transit peptides), ADS31-71–ADS1, ADS31-71–ADS2, and ADS3 (i.e., ADS enzymes with transit peptides) were generated by overlap extension PCR from the respective pYES2 constructs and introduced into the binary plasmid pBBV-PHAS.
A Cucumis sativus MGDG synthase cDNA (14) (a gift from H. Ohta, Tokyo Institute of Technology, Tokyo) was introduced into the yeast expression plasmid pESC-His (Stratagene) for coexpression with ADS-pYES2 constructs.
Transformation and Culture. Yeast transformation was carried out according to Gietz and Woods (15). Yeast nonauxotrophic for unsaturated fatty acids (DTY10A) (16) carrying pYES2 constructs were grown at 30°C in synthetic complete medium (SC) without uracil, pH 6, containing 2% (wt/wt) raffinose. For ole1Δ [ole1(HPAΔ::LEU2)] (a gift from C. Martin, Rutgers University, Piscataway, NJ), 0.5 mM each of palmitoleic and oleic acids was added in final 0.1% (wt/wt) tergitol. Solid media contained 1.2% (wt/wt) agar, 18% (wt/wt) sorbitol, and 1% (wt/wt) tergitol. For induction, cells were washed in media consisting of SC without uracil plus 2% (wt/wt) galactose, with no raffinose, palmitoleic, or oleic acid supplements. Media were sterilized by filtration (0.2 μm pore size; Nalgene). Cultures were inoculated at OD ≈0.5. Growth was monitored at 600 nm by using a spectrophotometer (DU640, Beckmann Coulter). For coexpression of MGDG synthase with ADS enzymes, DTY10A cells were transformed simultaneously with MGDG synthase cDNA in pESC-His and with ADS1, ADS2, or ADS372-371 in pYES2. Double transformants were incubated for 5 d at 30°C on solid media (SC without uracil or histidine) containing 2% (wt/wt) glucose and subsequently grown in liquid SC without uracil or histidine plus 2% (wt/wt) galactose for 48 h at 30°C, with shaking at 250 rpm.
An Arabidopsis fab1 fae1 double mutant was constructed by pollinating a fab1 homozygote with a fae1 homozygote. The resulting F1 seed was germinated, and the plants were allowed to self-pollinate. The codominant nature of the fab1 and fae1 mutations enables assignment of genotype on the basis of fatty acid phenotype i.e., >20% 16:0 and trace amounts of fatty acids >18 carbons in length (17). The F2 generation was screened for individuals producing seeds with this phenotype. Of 75 F2 individuals analyzed, three fab1 fae1 double mutants were identified. The phenotype of these individuals was confirmed by test crossing with a fab1 homozygote in which all progeny contained >20% 16:0, homozygosity of the fae1 allele being inferred from the near absence of 20 carbon fatty acids. Arabidopsis plants were grown in soil under continuous exposure to ≈300 microeinsteins of light (1 microeinstein = 1 mol of light) in controlled environment growth chambers. Seven-week-old Arabidopsis plants were transformed according to Clough and Bent (18) by using Agrobacterium tumefaciens strain GV3101. Plants carrying the transgenes were selected for resistance to ammonium glufosinate.
Lipid and Fatty Acid Analysis. Lipids were extracted from Arabidopsis seeds or yeast cultures according to Bligh and Dyer (19). Thin-layer chromatography (TLC) was performed and lipids visualized (20). MGDG was scraped from TLC plates and redissolved in CHCl3/methanol (2:1, vol/vol). Positional analysis of fatty acids esterified to MGDG was performed by using Rhizopus arrhizus lipase (EC.3.1.1.3, Sigma) (21, 22). The resulting lyso-MGDG and free fatty acid fractions were scraped from equivalent unstained TLC plates and redissolved in CHCl3/methanol (2:1, vol/vol). Fatty acids from purified MGDG or lyso-MGDG were directly methylated by using NaOCH3 as described in ref. 23. Free fatty acids were methylated by using 1 ml of 2% (vol/vol) H2SO4 in methanol and incubating for 30 min at 80°C. Fatty acids were extracted from dry cell pellets and methylated by adding 200 μl boron trichloride (BCl3) and incubating for 30 min at 80°C. Fatty acid methyl esters (FAMEs) were reextracted with 2 ml of hexane and dried under N2. FAMEs of single Arabidopsis seeds were prepared (24). FAMEs were analyzed by using an HP5890 gas chromatograph (Hewlett–Packard) fitted with a 60-m × 250-μm SP-2340 capillary column (Supelco). The oven temperature was raised from 100°C to 240°C at a rate of 15°C min–1 with a flow rate of 1.1 ml min–1. Mass spectrometry was performed with an HP5973 mass selective detector (Hewlett–Packard). Double-bond positions of monounsaturated FAMEs were determined (25). Mass spectra, expected fragmentation patterns, and diagnostic ions for the molecular species identified are given in Fig. 4, which is published as supporting information on the PNAS web site.
Genomic Analysis and Targeting Predictions. All predicted proteins of the Arabidopsis genome (2003 annotation of The Institute for Genomic Research, Rockville, MD) were clustered with the blastclust program. Details are available in Supporting Materials and Methods.
Three different protein targeting prediction programs were used to estimate putative subcellular locations of proteins within each cluster: ipsort (26), predotar, and targetp (27). Protein families for which all three programs indicated the same location are included in Table 3, which is published as supporting information on the PNAS web site.
Results and Discussion
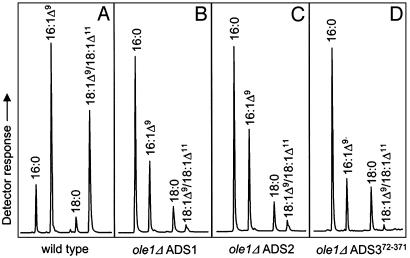
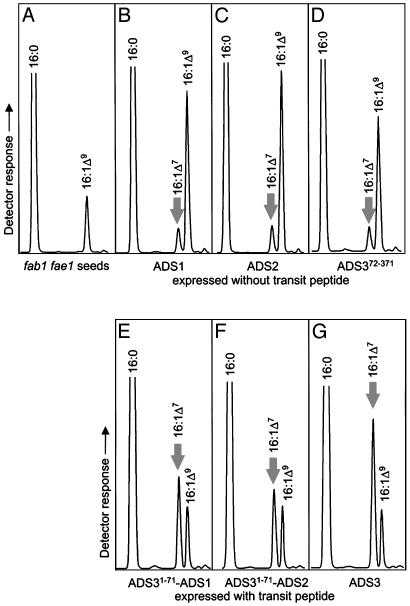
Expression of Arabidopsis ADS1, ADS2, or ADS372-371 in a yeast OLE1 (28) disruption strain restored the ability to grow without unsaturated fatty acid supplementation. Monoenes accumulated to ≈18–25% of the total fatty acids, with palmitoleic, oleic, and vaccenic acids being the only detected unsaturates (Fig. 1; see Fig. 4 for mass spectra). Unsaturated fatty acids were predominantly found on phosphatidylcholine, the most abundant phospholipid, in addition to smaller amounts on phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol (data not shown). Our observation of Δ9, and not Δ7, desaturation in yeast prompted us to ask whether expression in plants might differ from that in yeast, so we further characterized the three ADS genes by expression in Arabidopsis plants under the control of a seed-specific promoter. The fab1fae1 genetic background (17), impaired in both plastidial (FAB1) and endoplasmic reticulum (FAE1) elongase activities, was chosen as a host to provide the desaturases with elevated palmitic acid substrate and to simplify analysis by reducing fatty acid elongation beyond 18-carbons. In contrast to the phenotype observed in yeast, expression of each of the three desaturases, ADS1, ADS2, or ADS372-371, in fab1fae1 Arabidopsis seeds resulted in accumulation of 16:1Δ7 to ≈0.7% of the total fatty acids (Fig. 2 B–D and Table 1; see Fig. 4 for mass spectrum) in addition to an ≈9% increase in 16:1Δ9- and 16:1Δ9-derived vaccenic acid. It was surprising to see an ≈6% increase in the elongation product 18:1Δ11, which must have been formed either in the endoplasmic reticulum after desaturation or by reimportation of the 16:1Δ9 into the plastid. Interestingly, fae1 plants contain some 20-carbon fatty acids (17), suggesting either that the fae1 enzyme possesses residual activity or that there is another FAE1-like activity. Although fab1 plants possess residual plastidial elongation activity, it seems unlikely that the 16:1Δ9 would become esterified to acyl carrier protein in the plastid to become a substrate for fab1. It is possible that the source of this 18-carbon elongation exhibits higher elongation rates with 16-than with 18-carbon substrates. The source of this elongase activity requires further study.
Fig. 1.
Formation of Δ9 unsaturates by ADS enzymes in yeast. GC traces for fatty acids extracted from wild type (DTY10A) (A) and from ole1Δ expressing ADS1 (B), ADS2 (C), or ADS372-371 (D). Fatty acids were identified as indicated.
Fig. 2.
Formation of 16:1Δ7 in Arabidopsis fab1fae1 seeds expressing ADS enzymes. GC traces of fatty acids extracted from nontransformed seeds (A), from seeds expressing ADS1 (B), ADS2 (C), ADS372-371 (D) without transit peptides or ADS31-71-ADS1 (E), ADS31-71-ADS2 (F), or ADS3 (G) with plastidial transit peptides. Only part of each GC trace is shown. Fatty acids were identified as indicated. Arrows mark 16:1Δ7.
Table 1. Fatty acid patterns of fab1fae1 Arabidopsis seeds expressing ADS constructs.
| Fatty acid | fab1fae1 | ADS1 | ADS2 | ADS372-371 | ADS31-71-ADS1 | ADS31-71-ADS2 | ADS3 |
|---|---|---|---|---|---|---|---|
| 16:0 | 27.2 ± 1.7 | 17.3 ± 1.7 | 17.3 ± 1.5 | 19.5 ± 1.6 | 20.5 ± 1.9 | 20.8 ± 1.9 | 19.4 ± 1.1 |
| 16:1Δ7 | n.d. | 0.8 ± 0.2 | 0.8 ± 0.1 | 0.7 ± 0.4 | 2.6 ± 0.5 | 2.8 ± 0.4 | 4.0 ± 0.5 |
| 16:1Δ9 | 1.9 ± 0.3 | 4.9 ± 0.9 | 4.5 ± 1.0 | 4.1 ± 0.6 | 2.1 ± 0.4 | 2.1 ± 0.4 | 1.9 ± 0.4 |
| 18:0 | 2.5 ± 0.7 | 2.7 ± 0.6 | 2.4 ± 0.8 | 2.3 ± 0.6 | 2.8 ± 0.7 | 2.6 ± 0.5 | 2.9 ± 0.8 |
| 18:1Δ9 | 22.2 ± 1.6 | 10.7 ± 1.4 | 11.1 ± 2.1 | 14.9 ± 1.0 | 19.8 ± 1.6 | 19.8 ± 1.9 | 20.5 ± 1.9 |
| 18:1Δ11 | 2.7 ± 1.0 | 9.4 ± 1.9 | 9.2 ± 1.6 | 8.9 ± 1.0 | 3.9 ± 0.6 | 3.9 ± 0.8 | 3.6 ± 0.5 |
| 18:2 | 24.1 ± 2.1 | 25.2 ± 2.0 | 26.3 ± 1.8 | 22.8 ± 1.8 | 28.3 ± 2.0 | 28.1 ± 1.9 | 28.2 ± 2.1 |
| 18:3 | 19.3 ± 1.6 | 28.0 ± 1.4 | 28.4 ± 1.2 | 26.8 ± 1.9 | 20.0 ± 1.9 | 19.7 ± 2.0 | 19.5 ± 1.3 |
Data are presented as mol% ± SD. n.d., not detectable.
Notably, expression of ADS3, with its plastidial transit peptide intact, in fab1fae1 seeds resulted in the accumulation of ≈3.6% 16:1Δ7 (Fig. 2G), a level 5-fold higher than seen with expression of any ADS enzyme lacking a transit peptide and only an ≈1% increase in Δ9-derived vaccenic acid. This observation raised the question whether targeting of the cytoplasmic ADS1 or ADS2 to the plastid would shift their regiospecificity from Δ9 to Δ7. To address this question, the DNA encoding the ADS3 transit peptide (ADS31-71) was fused in frame to the ADS1 or ADS2 cDNA fragments, respectively. Expression of ADS31-71–ADS1 and ADS31-71–ADS2 in fab1fae1 seeds resulted in patterns similar to those observed with the expression of full-length ADS3 (Fig. 2 E and F) and included increased accumulation of 16:1Δ7 (≈2.5%) in the seeds with only a small increase in 16:1Δ9-derived vaccenic acid (Table 1). The data indicate an overall 25- to 70-fold switch in regiospecificity resulting from alternate targeting, with Δ7:Δ9 product ratios of ≈1:13 (ADS1 and ADS2) to ≈1:14 (ADS372-371) when the desaturases were expressed without a transit peptide and of ≈2:1 (ADS31-71–ADS1 and ADS31-71–ADS2) to ≈5:1 (ADS3) when they were expressed with a transit peptide. From these experiments it appears that, in plants, ADS enzymes are capable of functioning within or outside the plastid; that they are bifunctional for the Δ7 and Δ9 positions of palmitic acid; and that the ratio of accumulating products depends on their expression with or without a transit peptide rather than on the substantial differences in primary sequence of the catalytic portion of ADS1, ADS2, and ADS3.
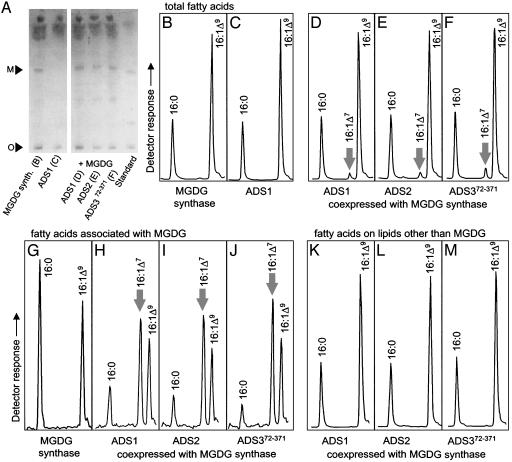
Although the reciprocal targeting experiments reveal changes in regiospecificity, they provide no insight into the molecular mechanism underlying the regiospecificity-switching phenomenon. Because genetic evidence implies FAD5 desaturates palmitic acid on MGDG in the plastid (11), we investigated the possibility that the presentation of palmitic acid on MGDG could impart Δ7 regiospecificity on ADS enzymes. We therefore engineered yeast to accumulate MGDG by introducing a cucumber (Cucumis sativus) MGDG synthase (14) into the yeast strain DTY10A. Expression of the MGDG synthase resulted in the appearance of a compound that comigrated with plant MGDG (Fig. 3A) and accumulated to ≈1–3 mol% of the total lipid. The identity of this compound was confirmed as MGDG by electrospray ionization tandem mass spectrometry (Kansas Lipidomics Research Center, Kansas State University, Manhattan, KS). Fatty acid analysis of transgenic yeast lines indicated that 16:1Δ7 was absent from cultures expressing the MGDG synthase alone (Fig. 3 B and G), from cultures expressing the ADS enzymes alone (e.g., Fig. 3C), and from vector-containing controls (data not shown). However, when ADS1, ADS2, or ADS372-371 was coexpressed with MGDG synthase, 16:1Δ7 accumulated from ≈0.8% (ADS1 and ADS2) to 1.5% (ADS3) of the total yeast fatty acids (Fig. 3D–F). Although the accumulation of 16:1Δ7 was less than the increase in the level of 16:1Δ9-derived vaccenic acid in these yeast strains, it was comparable to the level of accumulation of MGDG itself. Notably, when fatty acids hydrolyzed from the isolated MGDG fraction were analyzed, 16:1Δ7 was enriched ≈15-fold and ≈20-fold (ADS1/ADS2 and ADS372-371, respectively) over that of the total lipid fraction with a concomitant decrease in 16:0 (Table 2 and Fig. 3, compare D–F with H–J and G with H–J). No 16:1Δ7 was detected in total lipid extract after removal of the MGDG fraction (Fig. 3 K–M), suggesting that, within detection limits, 16:1Δ7 occurred exclusively on MGDG.
Fig. 3.
Formation of 16:1Δ7 on MGDG in yeast coexpressing ADS enzymes with Cucumis sativus MGDG synthase. (A) Thin-layer chromatographic separation of lipids extracted from yeast cultures as labeled. Migration of MGDG (M) and position of the origin (O) and Arabidopsis leaf lipid standard are indicated. (B–F) GC analysis of fatty acids extracted from lipids spotted in A from cultures expressing MGDG synthase alone (B) or ADS1 alone (C) or MGDG synthase plus ADS1 (D), ADS2 (E), or ADS372-371 (F). (G–J) GC analysis of fatty acids associated with MGDG isolated from cultures expressing MGDG synthase alone (G) or MGDG synthase plus ADS1 (H), ADS2 (I), or ADS372-371 (J). (K–M) GC analysis of fatty acids associated with yeast endogenous lipid classes other than MGDG isolated from cultures expressing MGDG synthase plus ADS1 (K), ADS2 (L), or ADS372-371 (M). Only part of each GC trace is shown. Fatty acids were identified as indicated. Arrows mark 16:1Δ7.
Table 2. Positional distribution of fatty acids on MGDG in DTY10A yeast expressing C. sativus MGDG synthase and ADS enzymes as indicated.
| Fatty acid
|
Control without ADS
|
+ADS1
|
+ADS2
|
+ADS372-371
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | sn-1 | sn-2 | Total | sn-1 | sn-2 | Total | sn-1 | sn-2 | Total | sn-1 | sn-2 | |
| 16:0 | 16.4 ± 1.2 | 14.7 ± 0.8 | 1.7 ± 0.3 | 0.8 ± 0.8 | 1.0 ± 0.8 | n.d. | 1.5 ± 0.3 | 1.0 ± 0.7 | n.d. | 1.2 ± 0.3 | 1.4 ± 0.7 | n.d. |
| 16:1Δ7 | n.d. | n.d. | n.d. | 15.7 ± 1.3 | 15.1 ± 1.3 | n.d. | 15.6 ± 2.2 | 14.1 ± 1.6 | 0.6 ± 1.0 | 20.0 ± 0.8 | 18.9 ± 0.5 | n.d. |
| 16:1Δ9 | 14.4 ± 1.0 | 8.4 ± 0.8 | 8.4 ± 0.8 | 15.6 ± 1.1 | 6.4 ± 0.8 | 8.9 ± 1.6 | 16.9 ± 1.0 | 7.5 ± 0.9 | 9.3 ± 0.4 | 16.0 ± 1.3 | 5.3 ± 1.8 | 10.1 ± 0.4 |
| 18:0 | 29.5 ± 1.1 | 26.2 ± 0.8 | 1.7 ± 0.4 | 27.9 ± 0.3 | 26.2 ± 0.8 | 1.6 ± 0.9 | 27.8 ± 1.2 | 25.9 ± 1.8 | 1.3 ± 0.9 | 23.3 ± 1.2 | 22.0 ± 1.8 | 1.1 ± 0.8 |
| 18:1 | 39.7 ± 1.1 | 2.6 ± 0.5 | 38.1 ± 0.5 | 40.1 ± 0.6 | 1.4 ± 0.4 | 39.5 ± 0.8 | 38.2 ± 1.6 | 1.4 ± 0.3 | 38.8 ± 1.2 | 39.4 ± 1.4 | 2.3 ± 1.0 | 38.9 ± 0.7 |
Data are presented as mol% ± SD. n.d., not detectable.
In 16:3 plants such as Arabidopsis and spinach, the successive desaturation of 16:0 to 16:1Δ7 and further to 16:3 occurs almost exclusively on the sn-2 position of MGDG (29). However, in yeast, 16:0 is reported to occur almost exclusively on the sn-1 position of all lipids (30). To determine the position(s) of 16:0 and 16:1Δ7 on the nonnative yeast lipid MGDG we performed positional analysis on MGDG isolated from yeast expressing the MGDG synthase alone and from yeast coexpressing MGDG synthase with ADS1, ADS2, or ADS372-371 (Table 2). In a pattern similar to that reported for native yeast lipid species (30), 16:0 and 18:0 were located almost exclusively on the sn-1 position, 18:1 was found almost exclusively on the sn-2 position, and 16:1Δ9 on both the sn-1 and sn-2 positions of MGDG formed in the transgenic yeast. When ADS1, ADS2, or ADS372-371 was coexpressed with MGDG synthase, the resulting 16:1Δ7 was found almost exclusively on the sn-1 position of MGDG (Table 2).
In yeast, the correlation of the formation of 16:1Δ7 with the synthesis of MGDG raises the question as to whether 16:1Δ7 formation occurs on MGDG. The following lines of evidence are consistent with desaturation of 16:0 esterified to MGDG: (i) 16:1Δ7 is formed only in yeast expressing ADS enzymes and containing MGDG (Fig. 3, compare C with D–F); (ii) the 16:1Δ7 formed is located exclusively on MGDG and not on other, native yeast lipids; (iii) 16:1Δ7 is restricted to the sn-1 position of MGDG and 16:1Δ7 production is accompanied by a concomitant loss of 16:0 (Fig. 3, compare G with H–J) at that position; and (iv) although highly enriched on MGDG, 16:1Δ7 represents a very minor fraction of the total cellular fatty acid pool (Fig. 3 D–F), and, therefore, the substantial loss of 16:0 specifically from sn-1 of MGDG is most easily explained by 16:0 desaturation directly on the sn-1 position of that lipid. Although it is formally possible that 16:0 desaturation to 16:1Δ7 could take place on CoA, it is difficult to explain why 16:1Δ7 occurrence would strictly depend on the presence of MGDG. Also, if 16:1Δ7 were synthesized on CoA, we would expect it to be transferred to all yeast lipids and occur there on both sn-1 and sn-2 positions based on the distribution of the Δ9 isomer of 16:1, which is presumably formed by the desaturation of 16:0 esterified to CoA by the yeast-endogenous OLE1 acyl-CoA desaturase (28, 31). Furthermore, conversion of CoA-bound 16:0 to 16:1Δ7 would not be expected to cause a substantial concomitant decrease in the global 16:0 pool that would be required to cause the observed 16:0 loss from MGDG (Fig. 3 G–J).
It is perhaps surprising that in yeast, 16:1Δ7 occurs almost exclusively on sn-1 of MGDG, whereas in plants 16:1Δ7-derived 16:3 is found almost exclusively on the sn-2 position of the same lipid. The data suggest that the position on which 16:1Δ7 will be found in MGDG is a consequence of the position the 16:0 substrate takes on the MGDG glycerol backbone (i.e., the sn-1 position in yeast and sn-2 in plants) and that the ADS enzymes do not exhibit sn-positional selectivity. Indeed, Roughan et al. (32) reported that radiolabeled palmitic acid, when supplied to spinach leaves, was incorporated at the sn-1 position of MGDG and that Δ7 16:0 desaturation and formation of 16:3 occurred efficiently on the sn-1 position.
The yeast coexpression experiments show that MGDG is both necessary and sufficient to alter the regiospecificity of palmitic acid desaturation by ADS enzymes from Δ9 to Δ7. Although it has recently been reported that voltage-gated K+ channels can be converted from A-type into delayed rectifiers and vice versa by the nature of their immediate lipid environment (33), the finding that a lipid head group can act as a molecular switch for desaturase regiospecificity is unexpected. Changes in catalytic rate, but not in regiospecificity, were previously reported for the soluble class of desaturases when substrates were presented on different acyl carrier proteins (34). Considering the influence different lipid environments may have, assignments of protein function based solely on heterologous expression (e.g., in yeast) should be viewed with caution. Although no 16:1Δ7 accumulation was observed in yeast with the expression of ADS1, ADS2, or ADS372-371 in the absence of MGDG synthase, the low level of 16:1Δ7 accumulating with extraplastidial targeting of ADS enzymes in fab1fae1 Arabidopsis seeds (Fig. 2 B–D) may be explained by the occurrence of low levels of extraplastidial galactolipids in Arabidopsis discussed by Härtel et al. (20); however, we cannot preclude a fraction of the extraplastidially targeted ADS enzymes acting in the plastid.
Changes to an enzyme's regiospecificity typically require between two and six specific changes at key locations along the amino acid chain that occur over many generations (35, 36). To accumulate mutations at these key sites, many additional mutations also accumulate, which tend to degrade attributes such as stability and turnover of the enzyme (37). In contrast, insertion or deletion of a transit peptide is a single-step process that is potentially instantaneous and does not necessarily result in a degradation of function. We propose that the Δ7 desaturase FAD5 (ADS3) evolved from an ancestral Δ9 desaturase by the addition of a transit peptide, because FAD5 (ADS3) retained Δ9 regiospecificity and product accumulation increased by ≈50% with the removal of the transit peptide. The observation that FAD5 is active in both compartments is perhaps surprising because the environments of the plastidial and cytoplasmic membranes differ markedly in factors including lipid composition, presence of different electron donors (cytochrome b5 in the endoplasmic reticulum versus ferredoxin in the plastid), redox state, and pH. Several lines of evidence support the view that FAD5 evolved from a cytoplasmic ADS enzyme: the widespread occurrence of Δ9 unsaturated fatty acids in nature compared with Δ7 fatty acids, the occurrence of a single gene in Arabidopsis containing the transit peptide versus eight genes lacking one, and that the closest homologs of the ADS enzymes are cyanobacterial desaturases that lack transit peptides. One possible explanation for the efficient functioning of FAD5 in the plastid is that ferredoxin is more electronegative than cytochrome b5, and being a stronger electron donor might overcome less than optimal protein–protein interaction. However, when a cyanobacterial Δ6 desaturase from Synechocystis was expressed in plants, it was found to be equally active when targeted to the plastid, endoplasmic reticulum, or cytoplasm, providing a case in which a ferredoxin-dependent enzyme is presumably functional with endoplasmic reticulum electron donors, such as cytochrome b5 (38). The experimental evidence therefore suggests that for both ADS enzymes and cyanobacterial desaturases, partnering with native electron donors is not essential for function. Although the FAD5 desaturase evidently arose by addition of a plastidial targeting sequence to a member of the multigene ADS family, we note that targeting to different compartments can also occur by alternate mRNA splicing of individual genes (39).
Functional diversity of enzymes is commonly probed by feeding a spectrum of potential substrates. Such studies on a fatty acid conjugase/desaturase led Dyer and colleagues (40) to hypothesize that multifunctional enzymes could potentially generate different products if expressed in different metabolic contexts. The current study reports on a natural system in which enzymes evolved distinct regiospecificities by alternate subcellular targeting by means of interaction with different substrates.
Our experimental observation that switching of regiospecificity resulted from the redirection of a plastidial desaturase to the cytoplasm and of cytoplasmic desaturases to the plastid, respectively, prompted us to ask whether this mechanism could have possibly occurred in other protein classes. For changes in enzyme specificity by alternative targeting to occur, several criteria would have to be met. First, individual members of protein families would have to be targeted to different locations. We performed a bioinformatics analysis of the Arabidopsis genome and found 239 encoded protein families with >50% amino acid identity that contain two or more members predicted to localize to different compartments by three independent algorithms. By using these very stringent criteria for inclusion, it is clear that alternative targeting of members of protein families is a wide-spread phenomenon in Arabidopsis. A list of all 239 protein families (Table 3) and a list of their encoding genes (Table 4) are published as supporting information on the PNAS web site. Second, enzymes would have to be capable of accepting two or more alternate substrates for catalysis. A survey of plant lipid-modifying enzymes alone yields many examples of bifunctional enzymes, including desaturases, hydroxylases, and conjugases (36, 40–44), suggesting that plants contain many bifunctional or multifunctional enzymes. Third, compartments would have to contain specific complements of metabolites, a condition that has been experimentally observed for many decades. Our analysis suggests that members of numerous Arabidopsis protein families are exposed to different substrates in alternative subcellular locations, where they may perform different functions. Among the protein families identified as having members in several locations are protein kinases, cytochrome P450s, dehydrogenase/reductases, glycosyl transferases, and lipases: enzymes that can be readily envisaged to exhibit modified functionality in alternate subcellular locations as described in the present work for the ADS enzymes.
Spatial or temporal colocalization of enzymes with pools of distinct substrates, as exemplified by the ADS enzymes, circumvents the barriers between eukaryotic subcellular compartments that separate specific sets of metabolites and enzymes and increases the product diversity resulting from a specific set of enzymes. The mechanism controlling desaturase specificity presented in this study explains the origin of a predominant plant lipid and extends the occurrence of localization-dependent enzyme function to central metabolism.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Sarah L. Zill. We thank Drs. C. Martin and H. Ohta for generous gifts of yeast strains and plasmids and Dr. C. Benning, Dr. J. Setlow, Dr. K. Mayer, E. Whittle, and Dr. J. Broadwater for helpful discussion. This work was supported in part by the Office of Basic Energy Sciences of the U.S. Department of Energy, the Oilseed Engineering Alliance of the Dow Chemical Company and Dow Agrosciences, and a German Science Foundation Emmy Noether Fellowship (to I.H.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ADS, Arabidopsis desaturase; MGDG, monogalactosyldiacylglycerol.
References
- 1.Ptashne, M. & Gann, A. (2002) Genes & Signals (Cold Spring Harbor Lab. Press, Plainview, NY).
- 2.Park, S. H., Zarrinpar, A. & Lim, W. A. (2003) Science 299, 1061–1064. [DOI] [PubMed] [Google Scholar]
- 3.Ptashne, M. & Gann, A. (1998) Curr. Biol. 8, R812–R822. [DOI] [PubMed] [Google Scholar]
- 4.Lee, M. M. & Schiefelbein, J. (2001) Development (Cambridge, U.K.) 128, 1539–1546. [DOI] [PubMed] [Google Scholar]
- 5.Davin, L. B. & Lewis, N. G. (2000) Plant Physiol. 123, 453–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dueber, J. E., Yeh, B. J., Chak, K. & Lim, W. A. (2003) Science 301, 1904–1908. [DOI] [PubMed] [Google Scholar]
- 7.Levin, E. R. (2001) J. Appl. Physiol. 91, 1860–1867. [DOI] [PubMed] [Google Scholar]
- 8.Fukuchi-Mizutani, M., Tasaka, Y., Tanaka, Y., Ashikari, T., Kusumi, T. & Murata, N. (1998) Plant Cell Physiol. 39, 247–253. [DOI] [PubMed] [Google Scholar]
- 9.Mekhedov, S., de Ilarduya, O. M. & Ohlrogge, J. (2000) Plant Physiol. 122, 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marillia, E. F., Giblin, E. M., Covello, P. S. & Taylor, D. C. (2002) FEBS Lett. 526, 49–52. [DOI] [PubMed] [Google Scholar]
- 11.Kunst, L., Browse, J. & Somerville, C. (1989) Plant Physiol. 90, 943–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vijayan, P. & Browse, J. (2002) Plant Physiol. 129, 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hugly, S. & Somerville, C. (1992) Plant Physiol. 99, 197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimojima, M., Ohta, H., Iwamatsu, A., Masuda, T., Shioi, Y. & Takamiya, K. (1997) Proc. Natl. Acad. Sci. USA 94, 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz, R. & Woods, R. (1994) in Molecular Genetics of Yeast: Practical Approaches, ed. Johnston, J. (Oxford Univ. Press, Oxford), pp. 121–134.
- 16.Toke, D. A. & Martin, C. E. (1996) J. Biol. Chem. 271, 18413–18422. [DOI] [PubMed] [Google Scholar]
- 17.James, D. & Dooner, H. (1991) Theor. Appl. Genet. 82, 409–412. [DOI] [PubMed] [Google Scholar]
- 18.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- 19.Bligh, E. & Dyer, W. (1959) Can. J. Biochem. Physiol. 37, 911–917. [DOI] [PubMed] [Google Scholar]
- 20.Hartel, H., Dormann, P. & Benning, C. (2000) Proc. Natl. Acad. Sci. USA 97, 10649–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer, W., Heinz, E. & Zeus, M. (1973) Z. Physiol. Chem. (Munich) 354, 1115–1123. [DOI] [PubMed] [Google Scholar]
- 22.Christie, W. W. (2003) Lipid Analysis: Isolation, Separation, Identification and Structural Analysis of Lipids (Barnes, Bridgwater, U.K.).
- 23.Domergue, F., Abbadi, A., Ott, C., Zank, T. K., Zahringer, U. & Heinz, E. (2003) J. Biol. Chem. 278, 35115–35126. [DOI] [PubMed] [Google Scholar]
- 24.Butte, W., Eilers, J. & Kirsch, M. (1982) Anal. Lett. 15, 841–850. [Google Scholar]
- 25.Yamamoto, K., Shibahara, A., Nakayama, T. & Kajimoto, G. (1991) Chem. Phys. Lipids 60, 39–50. [Google Scholar]
- 26.Nakai, K. & Horton, P. (1999) Trends Biochem. Sci. 24, 34–36. [DOI] [PubMed] [Google Scholar]
- 27.Emanuelsson, O., Nielsen, H., Brunak, S. & von Heijne, G. (2000) J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- 28.Stukey, J. E., McDonough, V. M. & Martin, C. E. (1989) J. Biol. Chem. 264, 16537–16544. [PubMed] [Google Scholar]
- 29.Roughan, P. G., Mudd, J. B., McManus, T. T. & Slack, C. R. (1979) Biochem. J. 184, 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner, S. & Paltauf, F. (1994) Yeast 10, 1429–1437. [DOI] [PubMed] [Google Scholar]
- 31.Sperling, P. & Heinz, E. (2001) Eur. J. Lipid Sci. Technol. 103, 158–180. [Google Scholar]
- 32.Roughan, G., Thompson, G. A. & Cho, S. H. (1987) Arch. Biochem. Biophys. 259, 481–496. [DOI] [PubMed] [Google Scholar]
- 33.Oliver, D., Lien, C. C., Soom, M., Baukrowitz, T., Jonas, P. & Fakler, B. (2004) Science 304, 265–270. [DOI] [PubMed] [Google Scholar]
- 34.Suh, M. C., Schultz, D. J. & Ohlrogge, J. B. (1999) Plant J. 17, 679–688. [DOI] [PubMed] [Google Scholar]
- 35.Broadwater, J. A., Whittle, E. & Shanklin, J. (2002) J. Biol. Chem. 277, 15613–15620. [DOI] [PubMed] [Google Scholar]
- 36.Cahoon, E. B., Lindqvist, Y., Schneider, G. & Shanklin, J. (1997) Proc. Natl. Acad. Sci. USA 94, 4872–4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Taverna, D. M. & Goldstein, R. A. (2002) Proteins 46, 105–109. [DOI] [PubMed] [Google Scholar]
- 38.Reddy, A. S. & Thomas, T. L. (1996) Nat. Biotechnol. 14, 639–642. [DOI] [PubMed] [Google Scholar]
- 39.Duchene, A. M., Peeters, N., Dietrich, A., Cosset, A., Small, I. D. & Wintz, H. (2001) J. Biol. Chem. 276, 15275–15283. [DOI] [PubMed] [Google Scholar]
- 40.Dyer, J. M., Chapital, D. C., Kuan, J. C., Mullen, R. T., Turner, C., McKeon, T. A. & Pepperman, A. B. (2002) Plant Physiol. 130, 2027–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van de Loo, F. J., Broun, P., Turner, S. & Somerville, C. (1995) Proc. Natl. Acad. Sci. USA 92, 6743–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Broun, P., Shanklin, J., Whittle, E. & Somerville, C. (1998) Science 282, 1315–1317. [DOI] [PubMed] [Google Scholar]
- 43.Broun, P., Boddupalli, S. & Somerville, C. (1998) Plant J. 13, 201–210. [DOI] [PubMed] [Google Scholar]
- 44.Behrouzian, B., Savile, C. K., Dawson, B., Buist, P. H. & Shanklin, J. (2002) J. Am. Chem. Soc. 124, 3277–3283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.