Abstract
Mammalian genomes are densely populated with long duplicated sequences. In this paper, we demonstrate the existence of doublets, short duplications between 25 and 100 bp, distinct from previously described repeats. Each doublet is a pair of exact matches, separated by some distance. The distribution of these intermatch distances is strikingly nonrandom. An unexpectedly high number of doublets have matches either within 100 bp (adjacent) or at distances tightly concentrated ≈1,000 bp apart (nearby). We focus our study on these proximate doublets. First, they tend to have both matches on the same strand. By comparing nearby doublets shared in human and chimpanzee, we can also see that these doublets seem to arise by an insertion event that produces a copy without markedly affecting the surrounding sequence. Most doublets in humans are shared with chimpanzee, but many new pairs arose after the divergence of the species. Doublets found in human but not chimpanzee are most often composed of almost tandem matches, whereas older doublets (found in both species) are more likely to have matches spaced by ≈1 kb, indicating that the nearly tandem doublets may be more dynamic. The spacing of doublets is highly conserved. So far, we have found clearly recognizable doublets in the following genomes: Homo sapiens, Mus musculus, Arabidopsis thaliana, and Caenorhabditis elegans, indicating that the mechanism generating these doublets is widespread. A mechanism that generates short local duplications while conserving polarity could have a profound impact on the evolution of regulatory and proteincoding sequences.
Genome expansion through duplication has been a prominent force in evolution. The human genome in particular is littered with signs of past duplication (1, 2). Transposons (3, 4), processed pseudogenes (5), and segmental duplications (6) are all known classes of repeats found in mammalian genomes. All of these types of duplications play an important role in gene and genome evolution (7), either through gene duplication and subsequent gene specialization or through the creation of unstable genomic regions.
Duplication is also important on a smaller scale. Comparative studies of promoters such as the vertebrate growth hormone gene (8) make it clear that gene regulation often evolves by increasing the number of copies of a given cis regulatory motif. Similarly, protein function can also evolve by the addition of tandem copies of protein domains. These types of short, tandem, or nearly tandem duplication events can have as striking an effect on gene evolution as whole gene or genome duplications. It is clear that once two or more copies of a given sequence are present at a locus, homologous recombination can further increase their number.
In this paper, we present evidence that short unique sequences are being actively duplicated in mammalian genomes. These short duplications occur frequently, have a strong tendency toward proximity and conservation of polarity, and do not fit into any of the well studied classes of interspersed repeats. Studying these short duplications will give us insight into the process by which a unique sequence is duplicated, an important first step in the creation of a tandem array and potentially a key process in the evolution of gene regulation and protein function.
Methods
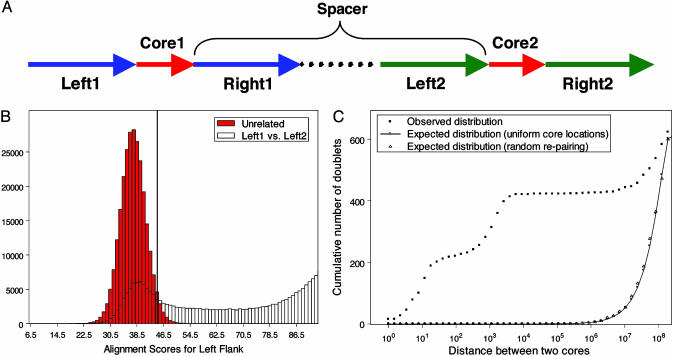
Identifying Doublets. We used the human genome sequence from the April 2003 assembly from University of California, Santa Cruz (9). We first identified cores of at least 25 bp, which occur exactly twice in the genome. Genomic counts (number of occurrences of a substring within the genome) were determined by using the mer-engine method (10). We further required that the 21-bp substrings of the cores occur nowhere else, and that at least one of the cores be flanked on either side by 21 bp of unique sequence. Each core is associated with the 100 bp immediately flanking it to the left and to the right (Fig. 1A). The Needleman–Wunsch global alignment algorithm (11) was used to calculate the alignment scores between the flanks with match, mismatch, and gap scores set to 1, 0, and –1, respectively. Many of the flanks share a large degree of homology (Fig. 1B), indicating that the cores are not independent short exact duplications but small parts of a larger approximate duplication. To eliminate these, we compared the observed alignment score to the distribution of alignment scores between unrelated sequences, determined by aligning one flank of one core and the reverse complement of the corresponding flank of the other core. We calculated the mean plus two standard deviations of the distribution of reverse complemented sequences and used this number as an upper bound on the maximum allowable alignment score. Approximately 86% of paired sequences were eliminated at this stage.
Fig. 1.
(A) Anatomy of a doublet. Each doublet has two cores, which are identical (same polarity) or reverse complements (opposite polarity) and are at least 25 bp in length. Each core is associated with 100 bp of flanking sequence on either side (Left1, Right1, Left2, and Right2). These flanking sequences can overlap. The last 21 bp of Left1 and first 21 bp of Right1 must be unique in the human genome. If the two cores are on the same chromosome, the spacer is the sequence between the two cores, and its length is the intercore distance. To be a doublet, the flanks cannot be homologous. (B) Homology between flanks. The hatched histogram shows the distribution of alignment scores from comparing Left1 with Left2 (see A). The red plot shows the distribution of alignment scores from comparing sequences that should be unrelated, Left1 and the reverse complement of Right2. The homology threshold is depicted as a vertical black line. (C) Distribution of distance of doublets on chromosome two. The observed distribution (black squares) is compared with two random models for doublet occurrences: the same number of total doublets, each core occurring uniformly intercore and independently at random across the chromosome (empty circles) and cores occurring at observed locations but randomly repaired into doublets (empty triangles).
We anticipated that a small percentage of the remaining pairs would be the result of processed pseudogenes. If a gene occurs twice in the genome, once with introns and once without, then an exon of the complete gene will be a duplicated sequence immediately flanked by nonhomologous sequence. To exclude this source of paired sequences, we next discarded any pairs in which the matched substring has homology to a sequence in the National Center for Biotechnology Information (NCBI) est_human database (expectation ≤10–4 using megablast with default parameters; http://www.ncbi.nih.gov/blast). Approximately 20% of the pairs were eliminated at this stage.
To find doublets in other genomes, the same procedure was carried out, using matched pairs of genomic sequences and coding sequence databases. Mus musculus sequence is from the National Center for Biotechnology Information (NCBI) Build 30 (12), Caenorhabditis elegans sequence is WS110 from worm-base (13), Drosophila melanogaster sequence is release 3-1 from flybase (14), Plasmodium falciparum sequence is from the Sanger center (15), and Arabidopsis thaliana sequence is from the NCBI (16).
Intercore Distance Distribution. For a doublet with both cores on the same chromosome (intrachromosomal doublets), the intercore distance is the number of base pairs in the spacer (Fig. 1 A) between the two occurrences of the core. For each chromosome, we plotted the distribution of intercore distances of all intrachromosomal doublets on that chromosome (Fig. 1C depicts human chromosome 2). We compared this distribution with two random models that take into account the overall number of intrachromosomal doublets in each chromosome. The first model assumes each core location is independently and uniformly distributed along the chromosome, yielding an expected distance distribution of P(distance< d) = 2d–d2, where d is the intercore distance normalized by chromosome length. If the distribution of core locations along the chromosome is nonuniform (some regions are core-rich and others, core-poor), the distance distribution will deviate from this model. To account for such nonuniformities, the second model uses the true locations of all the intrachromosomal cores on the chromosome but assumes cores are randomly matched up into doublets, independent of their locations. The expected distance distribution based on this model was calculated by Monte Carlo simulation.
Comparison with Chimpanzee. Chimpanzee sequence (Pan troglodytes) is the December 2003 Whitehead Institute for Biomedical Research (Cambridge) assembly from the Chimpanzee Genome Sequencing Consortium (http://ftp.ncbi.nih.gov/genome/Pan_troglodytes). To reduce our chances of finding paralogous rather than orthologous matches, we first screened out doublets in which either core was flanked by nonunique DNA. To do this, we determined the genomic counts for all 21-bp words in each of the flanks. If any of these 21-bp words occurred 10 or more times in the genome, or if the average genomic count was >3, the corresponding doublet was eliminated from the analysis.
For each of the remaining doublets, we identified the outermost 100-bp flanks of the doublet and used megablast to find regions in the chimpanzee genome with at least 80% identity >90 bp. We then extracted the intervening sequence. We created four different versions of the original doublet from the human or mouse genome: one with flanks, both cores, and the intercore sequence; one with both flanks, a single core, and the intercore sequence; one with both flanks and a single core but no intercore sequence; and one with flanks and intercore sequence but no cores. The needle program from the emboss suite (www.hgmp.mrc.ac.uk/Software/EMBOSS/Apps/needle.html; gap open penalty, 10; gap extend penalty, 0.5; match score, 5; mismatch score, –4) was then used to align the genomic region from the alternate genome to all of these sequences, and the alignment with the best score was used to assign a label to the doublet: two or more cores conserved, one core and spacer conserved, one core and no spacer conserved, or no spacers conserved. The resulting alignments are viewable in Fig. 5, which is published as supporting information on the PNAS web site, and the results are summarized in Table 1.
Table 1. Conservation of human proximate doublets in chimpanzee.
| Two or more cores
|
“Young” one core with or without spacer
|
“Young” with spacer
|
“Young” without spacer
|
|
|---|---|---|---|---|
| Doublet | No. (%) | No. (%) | No. (%) | No. (%) |
| Proximate | 2,283 (100.0) | 306 (100.0) | 27 (8.8) | 279 (91.2) |
| Nearby | 728 (31.9) | 13 (4.2) | 12 (92.3) | 1 (7.7) |
| Adjacent | 1,555 (68.1) | 293 (95.8) | 15 (5.1) | 278 (94.9) |
Proximate doublets are those with intercore distances between 1 and 10 kb. Nearby and adjacent are subclasses of proximate doublets with intercore distance ranges of 101-10,000 and 1-100 bp, respectively. Adjacent doublets are more likely to be “young” than nearby doublets. However, young adjacent doublets are usually missing their intercore spacer sequences in chimpanzee, indicating they are probably either tandem sequences (with an apparent spacer created by point mutation) or the result of a chimpanzee-specific deletion rather than a human-specific insertion. For more detailed alignments, see Fig. 5.
Comparison with Transposons. Alu annotations are from the University of California at Santa Cruz genome browser (9) and consensus sequences are from the Repbase Update database (17). For each of the 51 cases in which a doublet overlaps an annotated Alu, two versions of the annotated sequence were generated, one with the core (as found within the genome) and one with the core removed. Both versions were globally aligned to the consensus by using needle (gap open penalty, 10; gap extend penalty, 0.5; match score, 5; mismatch score, –4), and if the core-excised version had a higher scoring alignment to the consensus, the core was classified as an insertion.
Composition of Intercore Sequences. For each of the 2,020 intercore spacer sequences from nearby human doublets, we downloaded overlapping repeatmasker, segmental duplication, and refgene annotations from the University of California, Santa Cruz, genome browser (9). We did the same for five sets of randomly chosen genomic intervals with the same length distribution as the set of spacers. For each set of sequences and each type of annotation, we counted the number of sequences that overlapped a given type of annotation by at least 50%.
Segmental duplications have been annotated only if they are at least 1 kb in length. To limit any biases introduced by this length threshold, we also looked at uniqueness of the spacer sequences, compared to random genomic intervals and a random sampling of annotated segmental duplications. To determine uniqueness, we annotated each sequence with the number of genomic occurrences of each of its constituent 18 mers (10). We then calculated what percentage of the 18 mers in any given sequence set were unique (found only once in the genome), low count (found between 2 and 5 times in the genome), or high count (found in more than five locations in the genome).
Results and Discussion
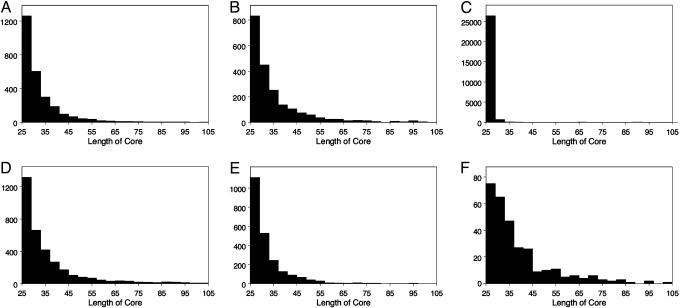
To find instances of short repeats, we searched the human genome for all exact matches (at least 25 bp in length) with dissimilar flanking sequences. We chose to look only at identically matching sequences, or cores, with precisely two copies, to simplify both the definition of the sequences under consideration and the interpretation of the results. We required the flanking sequences to be unrelated to ensure that we were not looking at a small exact patch within a long approximately duplicated region. After filtering out sequences with homologous flanks or with homology to expressed sequences (see Methods), we were left with 32,057 of these paired sequences, or doublets, in the human genome (Fig. 1 A). Although we set no maximum length on the core sequences, 99.9% are <100 bp in length. In fact, over half are 25 bp long, and their length distribution decays rapidly (Fig. 2 A–C).
Fig. 2.
Histograms of core length distributions are shown for several different populations of doublets. For each of these populations, a bin size of 4 bp was used to bin the core lengths, and the distribution was plotted. (A) Adjacent human doublets (2,696) with spacer lengths ≤100 bp; (B) 2,077 nearby human doublets with spacer lengths >100 bp and ≤10 kb; (C) 29,013 remote human doublets with spacer lengths >10 kb; (D) 3,430 proximate mouse doublets with spacer lengths ≤10 kb; (E) 2,283 proximate human doublets that are shared between the chimpanzee and human genomes; and (F) 306 proximate young human doublets, where one of the two cores of the doublet is missing in chimpanzee.
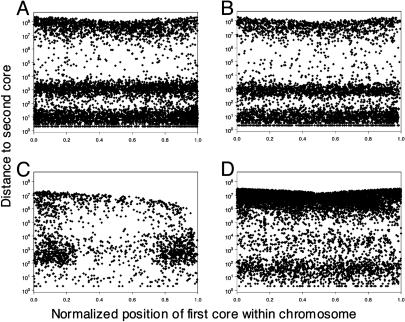
Doublets have several interesting characteristics. First, the distribution of their intercore distances is strikingly nonrandom (Fig. 3A). We observe three populations of doublets: those that are extremely close together (adjacent; cores at most 100 bp apart), those with distances distributed around 1 kb (nearby; cores >100 bp and at most 10 kb apart), and those with cores >10 kb apart or interchromosomal (remote). In addition, there is a bias toward conservation of polarity: the adjacent doublets are always direct repeats, and the nearby doublets have both cores in the same polarity ≈70% of the time. Not surprisingly, the remote doublets show no bias toward conservation of polarity. We made essentially the same observations in mouse (Figs. 3B and 2D).
Fig. 3.
For four different organisms, the distance between the two cores of a doublet is plotted vs. the normalized chromosomal position of one of the cores. Doublets are included only when both cores are on the same chromosome. This graph represents merged data from all of a particular organism's chromosomes. Normalized positions are chromosomal position divided by chromosome length. (A) Homo sapiens.(B) M. musculus.(C) C. elegans.(D) A. thaliana.
The numbers of adjacent and nearby doublets are significantly larger than what can be expected by chance, even considering the biases associated with the overall number of doublets (Fig. 1C). The vast majority of such doublets, which we collectively call proximate, are extremely unlikely to be coincidental matches. However, it is difficult to discern whether the large number of remote doublets is a result of biases in genome sequence composition or represents a more specific phenomenon. We have therefore concentrated our attention on proximate doublets. This decision is supported by the observations that core lengths are shorter among remote than proximate doublets (Fig. 2 A–C) and that remote doublet cores tend to be more AT-rich than proximate (data not shown). Adjacent doublets are comprised of two identical sequences separated by a short spacer sequence of 1–100 bp. Because their polarity is preserved, they can be viewed as a subclass of tandem repeats, loosely defined as direct repeats of approximate matches with little or no spacer. Some of our adjacent doublets are clearly tandem repeats of two units that appear to have a spacer sequence only because the repeat has been partially eroded by point mutations. It is possible that all of our adjacent doublets are variants of this type, and that more of this class would have been found had we loosened our strict ascertainment criteria.
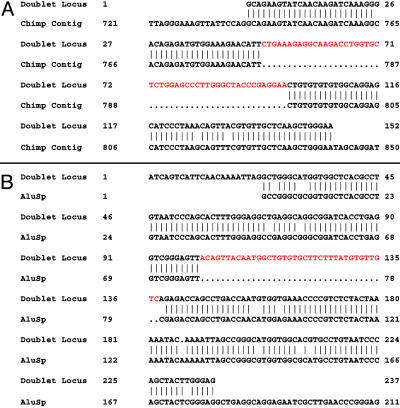
Nearby doublets, with long intercore distances between 100 and 10,000 bp, cannot be classified as degenerate tandem repeats. To study the dynamics of both adjacent and nearby doublets, we compared them to the draft P. troglodytes (chimpanzee) sequence. Of 3,083 doublets with intercore distances ≤10 kb, we found 2,589 in which the outermost flanks have clear homologues in the chimpanzee assembly. In most cases, the cores themselves are also present in chimpanzee. However, in 307 cases, one of the two cores is missing in chimpanzee, implying either a gain of a new copy in the human lineage or a loss in the chimpanzee lineage (Figs. 4A and 5).
Fig. 4.
(A) An alignment between one core of a human doublet and P. troglodytes sequence. The core sequence (highlighted in red) is clearly missing in the orthologous region of chimpanzee. This sequence is polymorphic within human populations; the inserted core has an allele frequency of ≈46%. (B) An alignment between one locus of a different doublet and an AluSp transposon consensus sequence. The core is clearly an insertion relative to the consensus.
In one nearby doublet, we see that both one core and the intercore sequence are missing in chimpanzee relative to human. This particular doublet likely represents a recombination-mediated loss of core and intercore sequence in the chimpanzee lineage rather than a gain in the human lineage (see doublet 643 in Fig. 5 in the supplementary information). However, this example is an exception; in the rest of the nearby doublets, the second core in humans appears to be an insertion relative to chimpanzee.
To unequivocally discriminate between gains and losses of copies, we selected six of the above nearby human doublets for further investigation and used PCR to detect the presence of both cores in chimpanzee, gorilla, orangutan, macaque, spider monkey, and lemur individuals, as well as a set of humans of diverse ethnicity. For each doublet, one core was always missing in non-human primates, whereas the other was always present (data not shown). In one of these six doublets, portrayed in Fig. 4A, we found the variable core to be polymorphic within the human population (data not shown). These data strongly indicate that the nearby doublets seen in human but not in chimpanzee arise by gain of a new copy.
Gains giving rise to nearby doublets in humans are most easily visualized as a simple insertion of a copy of a core into a nearby site with minimal alteration to the surrounding sequence. To look for further examples of these structures, we compared paralogous regions within the human genome. To this end, we identified those doublets that overlap the Alu family of transposons and examined the doublet sequences to determine whether the cores are insertions relative to the Alu consensus sequences. Of 51 nearby doublets with cores that overlap Alu annotations, we found 41 cases where the core appears to be an insertion relative to the transposon (Fig. 4B and Fig. 6, which is published as supporting information on the PNAS web site).
One possible source of nearby doublet generation is segmental duplication. Nearby doublets could be the remnants of old segmental duplications, where only a short exact match remains. Although these should have been eliminated through our filtration process, we used several tests to determine whether they were still a source of doublets. Segmental duplications are preferentially located near centromeres and telomeres in humans (18), so as a first test, we compared the chromosomal distribution of segmental duplications to that of doublets. We did not find any positive correlation between proximate doublet location and these chromosomal structures (Fig. 7, which is published as supporting information on the PNAS web site). Furthermore, we did not observe any clustering of proximate doublets within any 100-kb partitions of the human genome, which strongly argues against their origin as remnants of larger segmental duplications (Fig. 8, which is published as supporting information on the PNAS web site). As a final test, we compared the length distribution of young doublet cores that are found in human but not chimpanzee to the length distribution of conserved cores. If doublets originated in longer sequences, then younger doublets should be longer on average than old ones (Fig. 2 E and F). In fact, the distributions are very similar, with a slight length increase in young doublets, presumably due to the decreased number of point mutations in young sequences.
To search for further clues of the origins of the doublets, particularly the nearby doublets, we compared the content of the intercore sequence to randomly chosen genomic intervals of similar length and also randomly chosen genomic intervals from annotated segmental duplications. We examined these intervals for the uniqueness of their constituent 18 mers and overlap with the following types of genomic annotations: repeatmasker, RefSeq genes, and segmental duplications. With respect to uniqueness, intercore intervals are essentially indistinguishable from random genomic intervals and clearly very distinct from segmental duplications (Table 2, which is published as supporting information on the PNAS web site). Intercore intervals are drastically reduced for annotations as segmental duplications and slightly underrepresented for annotated repeats and genes (Table 2).
For clarity, we have examined a set of precisely defined short exact matches. Another group (Achaz et al., ref. 19) has studied more loosely defined approximate repeats in a range of organisms. For algorithmic simplicity, they too looked only at pairs of duplicated sequences. Although their data presumably encompass segmental duplications and pseudogenes as well as doublets, a bimodal distance relationship similar to what we have observed can be weakly discerned.
Achaz et al. (19) postulate that all of the repeats they found were generated by direct tandem duplications, and that more distantly separated pairs were spread apart by later insertions. We can reject this model, because our sequence comparisons suggest that in many cases, the nearby doublets can be viewed as an insertion of a core copy into an existing target sequence with minimal collateral damage to the target. Furthermore, we have compared the distances between pairs of cores conserved in chimpanzee and human and find that this distance is tightly conserved (Fig. 9, which is published as supporting information on the PNAS web site). Not only is there no evidence of spreading, but also the intercore spacers, if anything, are underrepresented for the agents that might cause spreading, such as transposons and segmental duplications.
Achaz et al. (19) hypothesize that the closest pairs undergo high frequencies of recombination and are consequently unstable. Our data are consistent with this idea. Although there are roughly equal numbers of adjacent and nearby doublets in humans, the doublets that have changed since human–chimpanzee divergence are mainly adjacent. Of the proximate doublets in humans that are conserved in chimpanzee, 68% are adjacent. Of the proximate doublets that are new since chimpanzee, 96% are adjacent (Table 1). This is further supported by the observation that the intercore sequences are often lost in adjacent doublets, implicating a deletion event in the transition from two cores to one.
Much more of the genome may have arisen by this duplication process than is immediately apparent. By requiring exact identity between the two cores, we have missed much older and more divergent short duplications present in the genome. In fact, because only 6% of the proximate doublets are new since the divergence of human and chimpanzee, we expect that most doublets are ancient in origin. Moreover, more than half of exact doublets have cores no longer than our minimum length, so we expect we missed shorter duplications. In fact, smaller sequences, with a minimum length of 21 rather than 25 bp, have a similar intercore distance distribution, and there are at least four times as many of these (Fig. 10, which is published as supporting information on the PNAS web site).
These findings are important because they suggest that the mammalian genome can expand and remodel by local random copying. The genomic forces giving rise to the events we have observed may be responsible for the duplication and shuffling of small functional motifs that have been preserved in vertebrate evolution. Comparisons of doublets orthologous between human and chimpanzee suggest that the short adjacent duplications may be reversible, providing an inexpensive way for the species to rapidly explore the functionality of its local sequence space. In future studies, it might be interesting to relax the requirement of exact identity between cores, to gain further insight into the mutational dynamics of doublets.
As we have already mentioned, the distribution and character of mouse doublets are similar to what we observe in humans. We repeated our analysis in the genomes of C. elegans, D. melanogaster, P. falciparum, and A. thaliana. In D. melanogaster and P. falciparum, we see too few paired matches to conclude whether they have the same characteristics as human doublets. In the other two genomes, we see very significant numbers of doublets (Fig. 3 C and D). In A. thaliana, doublets are mainly adjacent, whereas in C. elegans, doublets are mainly nearby. These observations suggest that the mechanisms that give rise to doublets are fairly widespread among eukaryotic genomes, but that unknown factors alter the relative contribution of these mechanisms to the evolution of different species.
A model involving double-stranded breaks leaving 5′ overhangs, subsequently repaired by filling in followed by nonhomologous recombination, can explain the adjacent doublets. Although we do not know of documented cases of this type of repair, the model seems plausible. The nearby doublets are not so readily explained. Because they too preserve polarity, we may surmise that they too reflect a repair event, but polarity is not absolutely preserved, and different classes of proximate doublets predominate in different genomes, suggesting different types of repair are at play. We offer no mechanism for the much more abundant remote doublets and in fact cannot offer persuasive statistical arguments that remote doublets are not coincidental. A finished assembly of the chimpanzee genome would help resolve this issue. In any case, breakage repair seems a likely mechanism whereby genomes sample and replicate their own composition, which, over a long time, can lead to the amplification and dispersion of small functional motifs.
Supplementary Material
Acknowledgments
We thank Evan Eichler, Mike Zody, Tarjei Mikkelsen, Eric Lander, and Jerzy Jurka for helpful critical reading of our paper; Eric Siggia, Casey Bergman, Izik Pe'er, Dana Pe'er, Guillaume Achaz, and Ira Hall for interesting discussions; and Lakshmi Muthuswamy for help in determining mouse genomic counts. This work was supported by National Institutes of Health and National Cancer Institute Grants 2R01CA078544, 5P30CA45508, 5R01CA81152, and 5R21HG02606 (to M.W.) and New York University/Defense Advanced Research Planning Agency Grant F5239. M.W. is an American Cancer Society Research Professor; E.E.T. is a Farish–Gerry Fellow of the Watson School of Biological Sciences and a Howard Hughes Medical Institute predoctoral fellow; and J.S. is supported by National Institutes of Health Grant 5T32CA09311. This research was also supported by grants to B.M. from the National Science Foundation's Qubic and Information Technology Research programs; the Defense Advanced Research Programming Agency's BioCOMP/BioSPICE program; the U.S. Air Force; and the New York State Office of Science, Technology, and Academic Research (NYSTAR) program.
References
- 1.International Human Genome Sequencing Consortium (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A. & Holt, R. A., et al. (2001) Science 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Deininger, P. L. & Batzer, M. A. (2002) Genome Res. 12, 1455–1465. [DOI] [PubMed] [Google Scholar]
- 4.Prak, E. L. & Kazazian, H. H. (2000) Nat. Rev. Genet. 1, 134–144. [DOI] [PubMed] [Google Scholar]
- 5.Vanin, E. (1985) Annu. Rev. Genet. 19, 253–272. [DOI] [PubMed] [Google Scholar]
- 6.Samonte, R. V. & Eichler, E. E. (2002) Nat. Rev. Genet. 3, 65–72. [DOI] [PubMed] [Google Scholar]
- 7.Ohno, S. (1970) Evolution by Gene and Genome Duplication (Springer, Berlin).
- 8.Chuzhanova, N. A., Krawczak, M, Nemytikova, L. A., Gusev, V. D. & Cooper, D. N. (2000) Gene 254, 9–18. [DOI] [PubMed] [Google Scholar]
- 9.Karolchik, D., Baertsch, R., Diekhans, M., Furey, T. S., Hinrichs, A., Lu, Y. T., Roskin, K. M., Schwartz, M., Sugnet, C. W., Thomas, D. J., et al. (2003) Nucleic Acids Res. 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Healy, J., Thomas, E. E., Schwartz, J. T. & Wigler, M. (2003) Genome Res. 13, 2306–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Needleman, S. B. & Wunsch, C. D. (1970) J. Mol. Biol. 48, 443–453. [DOI] [PubMed] [Google Scholar]
- 12.Mouse Genome Sequencing Consortium (2002) Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 13.The C. elegans Sequencing Consortium (1998) Science 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- 14.Adams, M. D., Celniker, S. E., Holt, R. A., Evans, C. A., Gocayne, J. D., Amanatides, P. G., Scherer, S. E., Li, P. W., Hoskins, R. A., Galle, R. F., et al. (2000) Science 287, 2185–2195. [DOI] [PubMed] [Google Scholar]
- 15.Gardner, M. J., Hall, N., Fung, E., White, O., Berriman, M., Hyman, R. W., Carlton, J. M., Pain, A., Nelson, K. E., Bowman, S., et al. (2002) Nature 419, 498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arabidopsis Genome Initiative (2000) Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 17.Jurka, J. (2000) Trends Genet. 16, 418–420. [DOI] [PubMed] [Google Scholar]
- 18.Bailey, J. A., Gu, Z., Clark, R. A., Reinert, K., Samonte, R. V., Schwartz, S., Adams, M. D., Myers, E. W., Li, P. W. & Eichler, E. E. (2002) Science 297, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 19.Achaz, G., Netter, P. & Coissac, E. (2001) Mol. Biol. Evol. 18, 2280–2288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.