Abstract
L55M polymorphism in Paraoxonase 1 (PON1) has been regarded as a risk factor for many cancer types. Nevertheless, the results remain controversial and inconclusive. We therefore performed a meta-analysis of all eligible case–control studies to evaluate the association between L55M polymorphism and cancer risk. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of the associations. Finally, a total of 5,627 cases and 6,390 controls, arising from 21 case–control studies, were enrolled in our study. Significant associations between PON1-L55M polymorphism and overall cancer risk were identified in all genetic models. In the stratified analyses by cancer type, PON1-L55M polymorphism was a risk factor for breast cancer in all genetic models, prostate cancer in the heterozygote model (ML vs LL: OR =1.304, 95% CI =1.049–1.620, Pheterogeneity=0.067), and ovarian cancer in the recessive model (MM vs ML/LL: OR =1.526, 95% CI =1.110–2.097, Pheterogeneity=0.464). Similarly, an increased risk was also identified for the Caucasian population in the heterozygote comparison and homozygote models, and hospital-based controls in all genetic models. To sum up, our study suggests that the PON1-L55M allele increased the risk of cancer. Future well-designed studies with larger sample sizes are warranted to further verify these findings.
Keywords: Paraoxonase 1, L55M, polymorphism, cancer, meta-analysis
Introduction
Cancer is the most lethal factor in developed countries and the second most lethal factor in developing countries.1 According to GLOBOCAN 2012, the number of new cases increased from 12.7 to 14.1 million in 2012, and 8.2 million deaths occurred.1,2 Aging of the population and adoption of cancer-related lifestyle increased the burden of cancer in developing countries. Reducing the incidence of cancer morbidity was the preferred prevention strategy. New and sensitive biomarkers are urgently required for the detection of high-risk populations and as new strategies for early detection. Currently, the underlying mechanisms of carcinogenesis are poorly understood, and research studies have suggested that environmental factors combined with susceptibility genes may play a critical role in the process.3,4 Gene polymorphisms, which can decrease the activity of detoxifying carcinogenic substances, may contribute to the transformation of exposure effects.
PON1 is located on the long arm of chromosome 7. Two important common genetic polymorphisms, PON1-Q192R and PON1-L55M, were identified by the epidemiologic and molecular studies in the coding region of the PON1 gene at positions 192 and 55. Studies revealed that higher PON1 activity and mRNA levels were related to the PON1-55L allele than to PON1-55M,5,6 and a decreased stability of the PON1-55M protein may lead to a lower activity of PON1.7 In addition, the association between the polymorphism and risk of different cancers, such as prostate cancer8 and breast cancer,9 was identified by case–control studies, whereas no significant association was identified between the polymorphism and cancer risk in renal cell carcinoma10 and ovarian cancer.11 Until now, these results remain inconclusive. Therefore, we conducted the present meta-analysis to precisely assess the association between PON1-L55M polymorphism and cancer risk.
Materials and methods
Search strategy
We searched the PubMed, Google Scholar, and Web of Science databases for studies published before November 30, 2015, by adopting keywords “cancer OR malignancy OR carcinoma OR tumor OR neoplasm” AND “polymorphism OR mutation OR SNP OR variant” AND “Paraoxonase 1 OR PON1”. We also conducted a hand search of references of original articles or reviews on this issue for additional studies. All the eligible studies were restricted to humans. And the articles should be presented in English. We extracted data separately when more than one cancer type or ethnicity was involved in one publication. In addition, we enrolled the report with the largest sample size when more than one report published the same data.
Inclusion criteria and exclusion criteria
We selected studies according to the following criteria: 1) reports that assessed the association between PON1 polymorphisms and cancer risk; 2) case–control studies only; and 3) publications that could provide the specific genotype frequency of cases and controls directly or indirectly (can be calculated from the article text). Besides, we excluded studies that were: 1) case reports, case-only studies, or reviews; 2) publications without specific genotype frequency of L55M polymorphism in PON1; 3) animal studies; and 4) duplicate publications.
Data extraction
Two investigators (LC and WL) devoted themselves to the data extraction process, and the following details were captured: the name of the first author, year of publication, ethnicity of each population, cancer type, control source, genotyping method, total number of cases and controls, and P-value of HWE (Hardy–Weinberg equilibrium). We compared the data and reached consensus for all disagreements by the two investigators.
Statistical analysis
We used odds ratio (OR) and 95% confidence interval (CI) to assess the association between PON1-L55M polymorphism and cancer risk. ORs were calculated in five genetic models: allele contrast (M vs L), heterozygote comparison (ML vs LL), homozygote (MM vs LL), recessive (MM vs ML/LL), and dominant (ML/MM vs LL). Between-study heterogeneity was assessed by χ2-test-based Q-statistic test,12 and quantified by I2 values, as well as P-values.13 No significant heterogeneity was observed when I2<50% and P>0.10, and ORs were pooled by a fixed-effects model. Otherwise, the random-effects model was used.14 Besides, stratified analyses by ethnicity, cancer type, genotyping method, and control source were performed. We combined any cancer type with less than two studies into the “other cancers” group. In addition, we also divided these cancer types into solid and hematological malignancies, individually. Sensitivity analysis was performed to assess the stability of these findings by removing one single study from the enrolled studies to reveal the influence of individual data sets on the pooled ORs. In the end, Begg’s funnel plot and Egger’s regression test were performed to assess the publication bias.13,15 We applied STATA software (version 12.0; StataCorp LP, College Station, TX, USA) to conduct all statistical analyses, and P<0.05 for any tests or genetic models was regarded as statistically significant.
Results
Study characteristics
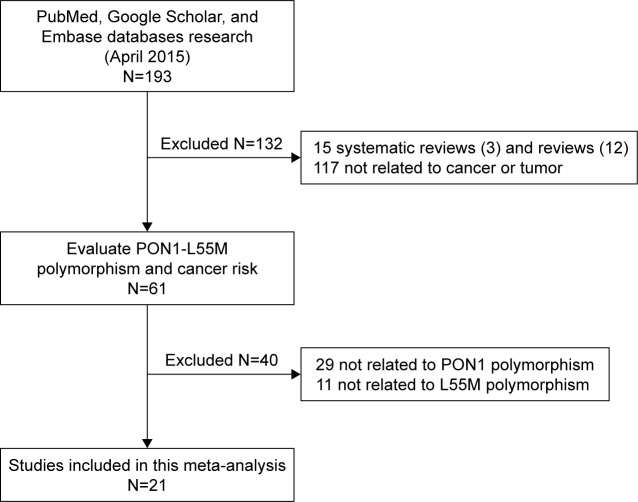
After careful examination according to the inclusion criteria, a total of 21 case–control studies comprising 6,224 cases and 7,014 healthy controls were enrolled in our study (Table 1).8–11,16–32 The flow chart of the study selection process is shown in Figure 1. Among these studies, three studies were performed in Asians, 14 in Caucasians, and four in mixed group. A total of six cancer types were addressed: four studies on breast cancer; three on prostate cancer; two on colorectal cancer, lung cancer, and ovarian cancer; and eight on other cancers (one study each on acute leukemia, brain tumor, embryonal tumor, hepatocellular carcinoma, lymphohematopoietic cancer, osteosarcoma, renal cell carcinoma, and pancreatic cancer). All genotype frequencies were in HWE with the exception of Antognelli et al16 and Ahmed et al,21 and these two studies were excluded from the pooled analyses.
Table 1.
Characteristics of the eligible case–control studies enrolled in the meta-analysis
| First author | Year | Ethnicity | Genotyping method | Control source | Cancer type | Case
|
Control
|
Y or N (HWE) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM | LM | LL | MM | LM | LL | |||||||
| Antognelli et al8 | 2005 | Caucasian | PCR-RFLP | H-B | Prostate cancer | 67 | 197 | 120 | 43 | 169 | 148 | Y |
| Van Der Logt et al18 | 2005 | Caucasian | PCR-RFLP | P-B | Colorectal cancer | 59 | 166 | 139 | 50 | 162 | 140 | Y |
| Stevens et al32 | 2006 | Caucasian | PCR-RFLP | P-B | Breast cancer | 77 | 230 | 176 | 58 | 223 | 202 | Y |
| Stevens et al19 | 2008 | Mixed | TaqMan | P-B | Prostate cancer | 165 | 609 | 481 | 189 | 575 | 498 | Y |
| Lurie et al17 | 2008 | Mixed | TaqMan | P-B | Ovarian cancer | 192 | 65 | 14 | 276 | 145 | 24 | Y |
| Arpaci et al11 | 2009 | Caucasian | PCR-RFLP | H-B | Ovarian cancer | 5 | 19 | 27 | 2 | 27 | 25 | Y |
| Antognelli et al16 | 2009 | Caucasian | PCR-RFLP | P-B | Breast cancer | 325 | 115 | 107 | 231 | 125 | 188 | N |
| Martinez et al31 | 2010 | Caucasian | TaqMan | H-B | Brain tumor | 30 | 32 | 11 | 88 | 94 | 38 | Y |
| Naidu et al23 | 2010 | Asian | PCR-RFLP | P-B | Breast cancer | 50 | 178 | 159 | 17 | 109 | 126 | Y |
| Uyar et al10 | 2011 | Caucasian | PCR-RFLP | P-B | Renal cell carcinoma | 6 | 25 | 29 | 10 | 29 | 21 | Y |
| Ergen et al28 | 2011 | Caucasian | PCR-RFLP | P-B | Osteosarcoma | 3 | 23 | 24 | 9 | 20 | 21 | Y |
| Aksoy-Sagirli et al26 | 2011 | Caucasian | PCR-RFLP | H-B | Lung cancer | 10 | 94 | 119 | 14 | 102 | 118 | Y |
| Hussein et al9 | 2011 | Caucasian | PCR-RFLP | P-B | Breast cancer | 60 | 21 | 19 | 6 | 23 | 35 | Y |
| Vecka et al27 | 2012 | Caucasian | PCR-RFLP | H-B | Pancreatic cancer | 10 | 39 | 24 | 8 | 37 | 28 | Y |
| Kokouva et al30 | 2013 | Caucasian | PCR-RFLP | H-B | Lymphohematopoietic cancers | 60 | 139 | 117 | 50 | 159 | 142 | Y |
| de Aguiar Goncalves et al25 | 2012 | Mixed | TaqMan | H-B | Acute leukemia | 34 | 99 | 104 | 19 | 75 | 131 | Y |
| Wang et al20 | 2012 | Asian | PCR-RFLP | P-B | Lung cancer | 2 | 47 | 307 | 0 | 18 | 166 | Y |
| Akkiz et al22 | 2013 | Caucasian | PCR-RFLP | P-B | Hepatocellular carcinoma | 31 | 81 | 105 | 27 | 89 | 101 | Y |
| Antognelli et al24 | 2013 | Caucasian | PCR-RFLP | H-B | Prostate cancer | 100 | 291 | 180 | 131 | 540 | 497 | Y |
| Vasconcelos et al29 | 2014 | Mixed | TaqMan | P-B | Embryonal tumor | 15 | 56 | 85 | 25 | 134 | 177 | Y |
| Ahmed et al21 | 2015 | Asian | PCR-RFLP | P-B | Colorectal cancer | 2 | 10 | 38 | 16 | 24 | 40 | N |
Abbreviations: PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; HWE, Hardy–Weinberg equilibrium; Y, polymorphisms conformed to HWE in the control group; N, polymorphisms did not conform to HWE in the control group; H-B, hospital based; P-B, population based; L allele, leucine; M allele, methionine.
Figure 1.
Flow chart presenting the publication selection process.
Quantitative data synthesis
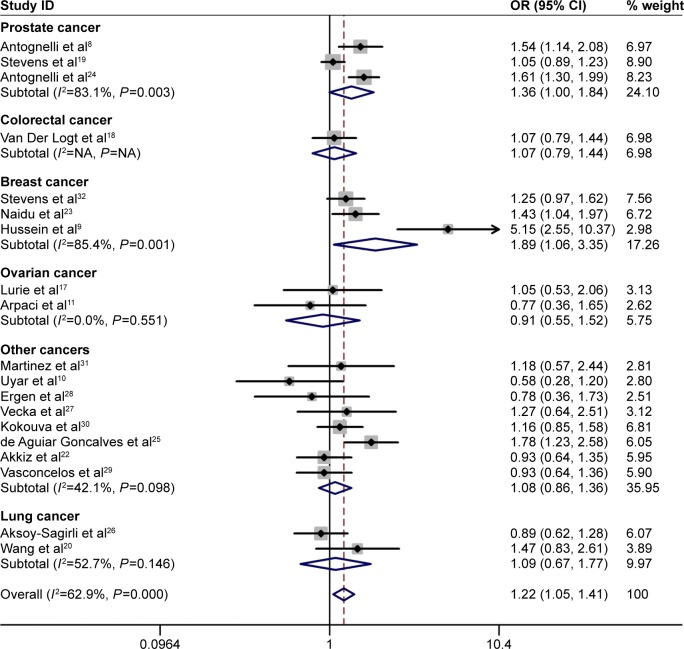
Significant associations between the PON1-L55M polymorphism and cancer risk were identified in the allele contrast (M vs L: OR =1.221, 95% CI =1.066–1.398, Pheterogeneity=0.000), homozygote (MM vs LL: OR =1.463, 95% CI =1.123–1.905, Pheterogeneity=0.000), heterozygote comparison (ML vs LL: OR =1.161, 95% CI =1.069–1.261, Pheterogeneity=0.162), recessive (MM vs ML/LL: OR =1.381, 95% CI =1.107–1.724, Pheterogeneity=0.000), and dominant (MM/ML vs LL: OR =1.218, 95% CI =1.054–1.407, Pheterogeneity=0.000) models (Table 2, Figure 2).
Table 2.
Results of meta-analysis for PON1-L55M polymorphism and cancer risk
| Variables | Case/control | M vs L
|
MM vs LL
|
ML vs LL
|
ML + MM vs LL
|
MM vs ML + LL
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-valuea | I2 (%) | OR (95% CI) | P-valuea | I2 | OR (95% CI) | P-valuea | I2 | OR (95% CI) | P-valuea | I2 | OR (95% CI) | P-valuea | I2 | ||
| Total | 5,627/6,390 | 1.221 (1.066–1.398)* | 0.000 | 79.5 | 1.463 (1.123–1.905)* | 0.000 | 72.1 | 1.161 (1.069–1.261)* | 0.162 | 24.4 | 1.218 (1.054–1.407)* | 0.000 | 62.9 | 1.381 (1.107–1.724)* | 0.000 | 69.0 |
| Prostate cancer | 2,210/2,790 | 1.244 (0.949–1.631) | 0.000 | 89.6 | 1.521 (0.832–2.778) | 0.000 | 90.3 | 1.258 (1.111–1.423)* | 0.067 | 63.0 | 1.357 (0.999–1.842) | 0.003 | 83.1 | 1.293 (0.803–2.082) | 0.000 | 87.0 |
| Breast cancer | 970/799 | 2.120 (1.066–4.218)* | 0.000 | 94.7 | 3.666 (1.159–11.600)* | 0.000 | 90.3 | 1.252 (1.020–1.536)* | 0.703 | 0.0 | 1.887 (1.064–3.349)* | 0.001 | 85.4 | 3.187 (1.052–9.661)* | 0.000 | 90.7 |
| Ovarian cancer | 322/499 | 1.268 (0.988–1.628) | 0.366 | 0.0 | 1.305 (0.691–2.465) | 0.484 | 0.0 | 0.713 (0.418–1.216) | 0.764 | 0.0 | 0.913 (0.550–1.517) | 0.551 | 0.0 | 1.523 (1.107–2.094)* | 0.464 | 0.0 |
| Lung cancer | 579/418 | 1.095 (0.666–1.801) | 0.104 | 62.2 | 0.781 (0.344–1.771) | 0.404 | 0.0 | 1.047 (0.766–1.433) | 0.215 | 34.8 | 1.089 (0.670–1.767) | 0.146 | 52.7 | 0.806 (0.361–1.799) | 0.432 | 0.0 |
| Other cancers | 1,182/1,532 | 1.072 (0.894–1.285) | 0.035 | 53.5 | 1.249 (0.626–2.492) | 0.124 | 38.3 | 1.062 (0.896–1.258) | 0.268 | 20.3 | 1.082 (0.864–1.356) | 0.098 | 42.1 | 1.219 (0.978–1.519) | 0.265 | 20.7 |
| Cancer type 2 | ||||||||||||||||
| Solid tumor | 5,074/5,814 | 1.201 (1.035–1.395)* | 0.000 | 80.8 | 1.420 (1.055–1.911)* | 0.000 | 74.2 | 1.148 (1.052–1.253)* | 0.208 | 21.1 | 1.191 (1.017–1.394)* | 0.000 | 63.8 | 1.356 (1.058–1.739) | 0.000 | 71.7 |
| Hematological tumor | 553/576 | 1.336 (1.123–1.590)* | 0.089 | 65.3 | 1.717 (1.134–2.600)* | 0.262 | 20.6 | 1.280 (0.992–1.651) | 0.089 | 65.4 | 1.419 (0.928–2.168) | 0.079 | 67.6 | 1.531 (1.092–2.146)* | 0.493 | 0.0 |
| Ethnicity | ||||||||||||||||
| Caucasian | 2,965/3,686 | 1.199 (0.993–1.447) | 0.000 | 82.2 | 1.461 (1.041–2.051)* | 0.000 | 72.9 | 1.170 (1.050–1.303)* | 0.185 | 25.6 | 1.199 (0.981–1.466) | 0.000 | 67.7 | 1.371 (1.025–1.833) | 0.000 | 69.4 |
| Asian | 743/436 | 1.428 (1.143–1.784)* | 0.849 | 0.0 | 2.344 (1.304–4.214)* | 0.925 | 0.0 | 1.324 (0.992–1.767) | 0.797 | 0.0 | 1.443 (1.092–1.907)* | 0.938 | 0.0 | 2.068 (1.175–3.638)* | 0.880 | 0.0 |
| Mixed | 1,919/2,268 | 1.193 (0.938–1.518) | 0.005 | 76.7 | 1.252 (0.818–1.916) | 0.052 | 61.1 | 1.104 (0.842–1.448) | 0.093 | 53.3 | 1.163 (0.878–1.539) | 0.053 | 60.9 | 1.265 (0.859–1.861) | 0.012 | 72.7 |
| Control source | ||||||||||||||||
| Population-based | 3,699/3,705 | 1.221 (0.989–1.509) | 0.000 | 85.2 | 1.374 (0.916–2.061) | 0.000 | 78.6 | 1.082 (0.974–1.203) | 0.574 | 0.0 | 1.162 (0.953–1.417) | 0.001 | 65.1 | 1.376 (0.964–1.966) | 0.000 | 79.3 |
| Hospital-based | 1,928/2,685 | 1.303 (1.194–1.423)* | 0.062 | 48.0 | 1.714 (1.369–2.147)* | 0.303 | 16.2 | 1.293 (1.134–1.474)* | 0.139 | 36.3 | 1.314 (1.083–1.594)* | 0.059 | 48.6 | 1.484 (1.248–1.764)* | 0.510 | 0.0 |
| Genotyping method | ||||||||||||||||
| PCR-RFLP | 3,635/3,902 | 1.239 (1.039–1.478)* | 0.000 | 80.9 | 1.558 (1.119–2.168)* | 0.000 | 71.3 | 1.188 (1.073–1.316)* | 0.208 | 22.6 | 1.235 (1.030–1.482)* | 0.000 | 65.7 | 1.463 (1.097–1.951) | 0.000 | 67.0 |
| TaqMan | 1,992/2,488 | 1.171 (0.956–1.434) | 0.012 | 68.9 | 1.221 (0.861–1.732) | 0.100 | 48.6 | 1.112 (0.967–1.279) | 0.169 | 37.9 | 1.161 (0.912–1.477) | 0.103 | 48.0 | 1.212 (0.886–1.659) | 0.027 | 63.6 |
Notes: I2: 0%–25%, no heterogeneity; 25%–50%, modest heterogeneity; >50%, high heterogeneity;
P-value: P-value of Q test for heterogeneity test;
statistically significant (P<0.05).
Abbreviations: OR, odds ratio; CI, confidence interval; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; L allele, leucine; M allele, methionine.
Figure 2.
Meta-analysis of the association between PON1-L55M polymorphism and cancer risk in the dominant model (MM/ML vs LL).
Note: Weights are from random effects analysis.
Abbreviations: CI, confidence interval; OR, odds ratio; ID, identification; NA, not available; L allele, leucine; M allele, methionine.
In stratified analyses by cancer type, the PON1-55M allele was a risk factor for breast cancer in all genetic models (allele contrast: M vs L: OR =2.120, 95% CI =1.066–4.218, Pheterogeneity=0.000; homozygote: MM vs LL: OR =3.666, 95% CI =1.159–11.600, Pheterogeneity=0.000; heterozygote comparison: ML vs LL: OR =1.252, 95% CI =1.020–1.536, Pheterogeneity=0.703; recessive: MM vs ML/LL: OR =3.187, 95% CI =1.052–9.661, Pheterogeneity=0.000; and dominant: MM/ML vs LL: OR =1.887, 95% CI =1.064–3.349, Pheterogeneity=0.001), prostate cancer in the heterozygote comparison model (ML vs LL: OR =1.304, 95% CI =1.049–1.620, Pheterogeneity=0.067), and ovarian cancer in the recessive model (MM vs ML/LL: OR =1.526, 95% CI =1.110–2.097, Pheterogeneity=0.464). Similarly, an increased risk was observed in the Caucasian population (homozygote: MM vs LL: OR =1.461, 95% CI =1.041–2.051, Pheterogeneity=0.000; and heterozygote comparison: ML vs LL: OR =1.170, 95% CI =1.050–1.303, Pheterogeneity=0.185) and the Asian population (allele contrast: M vs L: OR =1.428, 95% CI =1.143–1.784, Pheterogeneity=0.849; homozygote: MM vs LL: OR =2.344, 95% CI =1.304–4.214, Pheterogeneity=0.925; recessive: MM vs ML/LL: OR =2.068, 95% CI =1.175–3.638, Pheterogeneity=0.880; MM/ML vs LL: OR =1.443, 95% CI =1.092–1.907, Pheterogeneity=0.938), and hospital-based group (allele contrast: M vs L: OR =1.303, 95% CI =1.194–1.423, Pheterogeneity=0.062; homozygote: MM vs LL: OR =1.714, 95% CI =1.369–2.147, Pheterogeneity=0.303; heterozygote comparison: ML vs LL: OR =1.293, 95% CI =1.134–1.474, Pheterogeneity=0.139; recessive: MM vs ML/LL: OR =1.484, 95% CI =1.248–1.764, Pheterogeneity=0.510; and dominant: MM/ML vs LL: OR =1.314, 95% CI =1.083–1.594, Pheterogeneity=0.059). In addition, we conducted a stratification analysis by genotyping method, and an increased risk for the PCR-RFLP (polymerase chain reaction-restriction fragment length polymorphism) group was identified (Table 2). We divided these tumors into solid and hematological tumor groups, and the results of subgroup analyses were not completely consistent with those of overall cancer analyses (Table 2). We observed an increased risk of solid cancer in the allele contrast, homozygote, heterozygote comparison, and dominant models, and hematological tumor in the allele contrast, homozygote, and recessive models (Table 2).
Publication bias and sensitivity analysis
Each time, one single study was removed from the enrolled assembly to validate the effect of individual studies on the pooled analysis, and no individual study obviously affected the pooled OR observed (Figure 3). Egger’s test and Begg’s funnel plot were performed to assess the publication bias. The shape of the funnel plot was symmetrical (Figure 4). In addition, the results of Egger’s test did not show statistical evidence for bias (PON1-L55M MM vs LL: Egger’s test: t=0.53; P=0.604). Thus, no obvious publication bias was found in our meta-analysis, and our results were credible.
Figure 3.
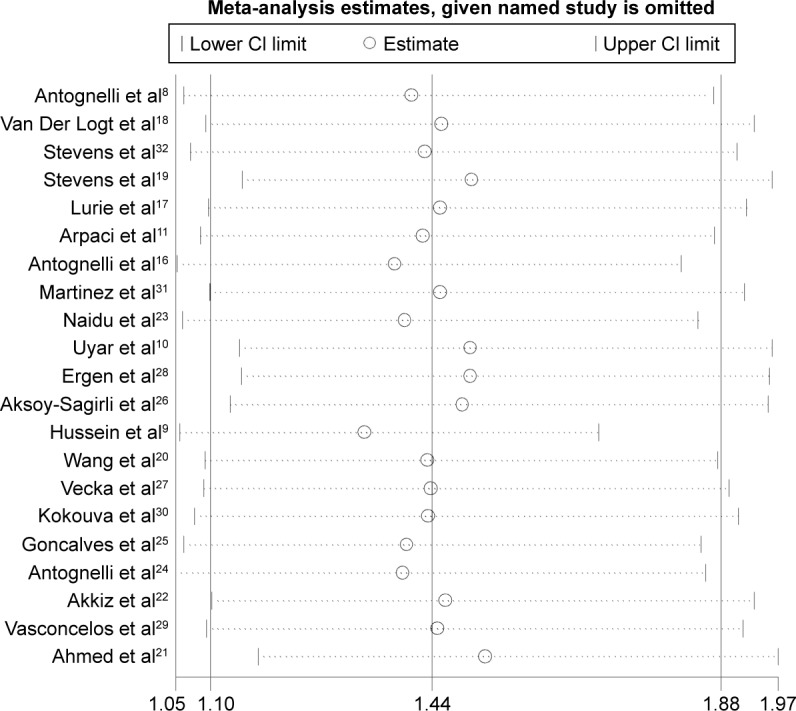
Sensitivity analysis of overall OR coefficients for PON1-L55M (MM vs LL).
Notes: Results were calculated by omitting each study in turn. The two ends of the dotted lines represent the 95% CI.
Abbreviations: CI, confidence interval; OR, odds ratio; L allele, leucine; M allele, methionine.
Figure 4.
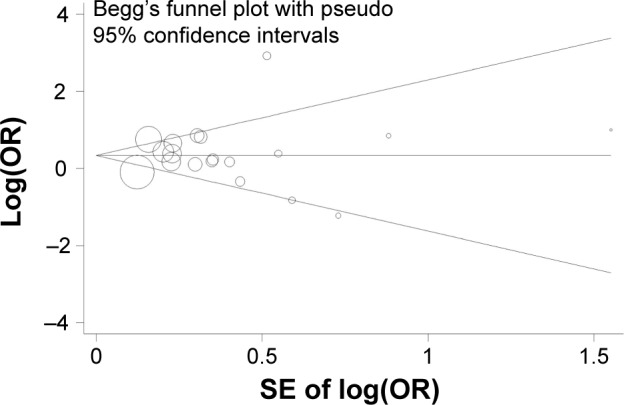
Begg’s funnel plot of publication bias (homozygote model: MM vs LL).
Notes: Each point represents a separate study for the indicated association. Log(OR), natural logarithm of OR; horizontal line, mean effect size.
Abbreviations: OR, odds ratio; SE, standard error of the mean; L allele, leucine; M allele, methionine.
Discussion
Previous studies suggested that lifestyle, estrogens, dietary habits, and oxidative and carbonyl stresses potentially play a critical role in the tumorigenesis and progression of cancers.33–35 There are several enzyme systems in our body that protect against genotoxic damage, either directly or via free-radical detoxification. Moreover, PON1, which is an antioxidant enzyme, may contribute to the disturbance in antioxidant–oxidant balance.36,37 Decreased expression of PON1 was identified in lung cancer and pancreatic cancer by previous studies.38,39 M variants decreased the stability of the PON1 enzyme. Subsequently, the concentration of PON1 in the blood was lowered, which can influence the activity of the enzyme. The LM genotype was identified as having a PON1 activity level between LL and MM genotypes.7
Previous studies suggested that PON1-L55M polymorphism was associated with an increased risk for many cancer types, such as breast and prostate cancers, while a decreased risk was identified in renal cell carcinoma and ovarian cancer. These results were controversial and inconclusive. In our present work, we identified that the PON1-55M allele was associated with an increased risk of cancer. In stratified analyses by cancer type, PON1-L55M polymorphism was a risk factor for breast cancer in all the five genetic models. Previous studies indicated that PON1, which is a part of lipid peroxidation scavenging systems, may affect the cell proliferation and malignant conversion process associated with the development of breast cancer.40 In addition, we also observed an increased risk of prostate cancer in the heterozygote comparison model and ovarian cancer in the recessive model. Similarly, an increased risk was observed in the Caucasian population (homozygote and heterozygote comparison models), the Asian population (allele contrast, homozygote, and recessive and dominant models), and the hospital-based group (all the five genetic models). The controls enrolled in our study were not uniformly defined. Some studies adopted the population-based group as the control source, while others adopted the hospital-based group. As a result, once the polymorphism was considered to influence the risk of other diseases, the control source would not always be representative of the underlying source populations. In addition, we observed an increased risk of solid cancer in the allele contrast, homozygote, heterozygote comparison, and dominant models, and hematological tumor in the allele contrast, homozygote, and recessive models. The cause of these differences may be related to the origin of the tumor.
Although we have presented a comprehensive study of the association between PON1-L55M polymorphism and cancer risk, several limitations should be noted. First, a limited number of publications were enrolled in our study and the sample size of each report was relatively small. Second, most of the enrolled publications were Caucasian, and none of them was African. Third, our results were based on single-factor estimates, which may result in a serious confounding bias, for the reason of lack of original data, without adjustment for age, sex, and other risk factors.
To sum up, our study identified that PON1-L55M polymorphism is a risk factor for cancers, particularly breast cancer, prostate cancer, and ovarian cancer. Further well-designed studies with large sample sizes will be continued on this issue of interest.
Footnotes
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The work by DY and LC was supported by the Department of Urology, the Second Affiliated Hospital of Anhui Medical University. The authors report no other conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer – analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, Weibing Z, Qiong L, et al. XRCC1 R399Q polymorphism and risk of normal tissue injury after radiotherapy in breast cancer patients. Tumour Biol. 2014;35:21–25. doi: 10.1007/s13277-013-0990-x. [DOI] [PubMed] [Google Scholar]
- 5.Li WF, Costa LG, Richter RJ, et al. Catalytic efficiency determines the in-vivo efficacy of PON1 for detoxifying organophosphorus compounds. Pharmacogenetics. 2000;10:767–779. doi: 10.1097/00008571-200012000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Leviev I, Negro F, James RW. Two alleles of the human paraoxonase gene produce different amounts of mRNA. An explanation for differences in serum concentrations of paraoxonase associated with the (Leu-Met54) polymorphism. Arterioscler Thromb Vasc Biol. 1997;17:2935–2939. doi: 10.1161/01.atv.17.11.2935. [DOI] [PubMed] [Google Scholar]
- 7.Leviev I, Deakin S, James RW. Decreased stability of the M54 isoform of paraoxonase as a contributory factor to variations in human serum paraoxonase concentrations. J Lipid Res. 2001;42:528–535. [PubMed] [Google Scholar]
- 8.Antognelli C, Mearini L, Talesa VN, Giannantoni A, Mearini E. Association of CYP17, GSTP1, and PON1 polymorphisms with the risk of prostate cancer. Prostate. 2005;63:240–251. doi: 10.1002/pros.20184. [DOI] [PubMed] [Google Scholar]
- 9.Hussein YM, Gharib AF, Etewa RL, ElSawy WH. Association of L55M and Q192R polymorphisms in paraoxonase 1 (PON1) gene with breast cancer risk and their clinical significance. Mol Cell Biochem. 2011;351:117–123. doi: 10.1007/s11010-011-0718-4. [DOI] [PubMed] [Google Scholar]
- 10.Uyar OA, Kara M, Erol D, Ardicoglu A, Yuce H. Investigating paraoxonase-1 gene Q192R and L55M polymorphism in patients with renal cell cancer. Genet Mol Res. 2011;10:133–139. doi: 10.4238/vol10-1gmr927. [DOI] [PubMed] [Google Scholar]
- 11.Arpaci A, Gormus U, Dalan B, Berkman S, Isbir T. Investigation of PON1 192 and PON1 55 polymorphisms in ovarian cancer patients in Turkish population. In vivo (Athens, Greece) 2009;23:421–424. [PubMed] [Google Scholar]
- 12.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 13.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 14.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 15.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 16.Antognelli C, Del Buono C, Ludovini V, et al. CYP17, GSTP1, PON1 and GLO1 gene polymorphisms as risk factors for breast cancer: an Italian case-control study. BMC Cancer. 2009;9:115. doi: 10.1186/1471-2407-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lurie G, Wilkens LR, Thompson PJ, et al. Genetic polymorphisms in the Paraoxonase 1 gene and risk of ovarian epithelial carcinoma. Cancer Epidemiol Biomarkers Prev. 2008;17:2070–2077. doi: 10.1158/1055-9965.EPI-08-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Der Logt EM, Janssen CH, Van HZ, et al. No association between genetic polymorphisms in NAD(P)H oxidase p22phox and paraoxonase 1 and colorectal cancer risk. Anticancer Res. 2005;25:1465–1470. [PubMed] [Google Scholar]
- 19.Stevens VL, Rodriguez C, Talbot JT, Pavluck AL, Thun MJ, Calle EE. Paraoxonase 1 (PON1) polymorphisms and prostate cancer in the CPS-II Nutrition Cohort. Prostate. 2008;68:1336–1340. doi: 10.1002/pros.20796. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Li L, Ding L, Zhang Z, Pu C. Association of genetic polymorphisms in the paraoxonase 1 gene with the risk and prognosis of non-small cell lung cancer in Chinese Han population. J Investig Med. 2012;60:592–597. doi: 10.2310/JIM.0b013e318245d557. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed NS, Shafik NM, Elraheem OA, Abou-Elnoeman SE. Association of Paraoxonase-1(Q192R and L55M) gene polymorphisms and activity with colorectal cancer and effect of surgical intervention. Asian Pac J Cancer Prev. 2015;16:803–809. doi: 10.7314/apjcp.2015.16.2.803. [DOI] [PubMed] [Google Scholar]
- 22.Akkiz H, Kuran S, Akgöllü E, et al. Effect of PON1 gene polymorphisms in Turkish patients with hepatocellular carcinoma. Meta Gene. 2013;1:93–101. doi: 10.1016/j.mgene.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naidu R, Har YC, Taib NA. Genetic polymorphisms of Paraoxonase 1 (PON1) gene: association between L55M or Q192R with breast cancer risk and clinico-pathological parameters. Pathol Oncol Res. 2010;16(4):533–540. [Google Scholar]
- 24.Antognelli C, Mezzasoma L, Mearini E, Talesa VN. Glyoxalase 1-419C>A variant is associated with oxidative stress: implications in prostate cancer progression. PloS One. 2013;8:e74014. doi: 10.1371/journal.pone.0074014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Aguiar Goncalves BA, Vasconcelos GM, Thuler LC, et al. NQO1 rs1800566 (C609T), PON1 rs662 (Q192R), and PON1 rs854560 (L55M) polymorphisms segregate the risk of childhood acute leukemias according to age range distribution. Cancer Causes Control. 2012;23:1811–1819. doi: 10.1007/s10552-012-0060-5. [DOI] [PubMed] [Google Scholar]
- 26.Aksoy-Sagirli P, Cakmakoglu B, Isbir T, et al. Paraoxonase-1 192/55 polymorphisms and the risk of lung cancer in a Turkish population. Anticancer Res. 2011;31:2225–2229. [PubMed] [Google Scholar]
- 27.Vecka M, Jáchymová M, Vávrová L, et al. Paraoxonase-1 (PON1) status in pancreatic cancer: relation to clinical parameters. Folia Biol (Praha) 2012;58:231–237. doi: 10.14712/fb2012058060231. [DOI] [PubMed] [Google Scholar]
- 28.Ergen A, Kilicoglu O, Ozger H, Agachan B, Isbir T. Paraoxonase 1 192 and 55 polymorphisms in osteosarcoma. Mol Biol Rep. 2011;38:4181–4184. doi: 10.1007/s11033-010-0538-8. [DOI] [PubMed] [Google Scholar]
- 29.Vasconcelos GM, Gonçalves BA, Montalvão-de-Azevedo R, et al. PON1 Q192R polymorphism (rs662) is associated with childhood embryonal tumors. Mol Biol Rep. 2014;41:6111–6115. doi: 10.1007/s11033-014-3489-7. [DOI] [PubMed] [Google Scholar]
- 30.Kokouva M, Koureas M, Dardiotis E, et al. Relationship between the paraoxonase 1 (PON1) M55L and Q192R polymorphisms and lymphohaematopoietic cancers in a Greek agricultural population. Toxicology. 2013;307:12–16. doi: 10.1016/j.tox.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 31.Martinez C, Molina JA, Alonso-Navarro H, Jiménez-Jiménez FJ, Agúndez JAG, García-Martín E. Two common nonsynonymous paraoxonase 1 (PON1) gene polymorphisms and brain astrocytoma and meningioma. BMC Neurol. 2010;10:71. doi: 10.1186/1471-2377-10-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens VL, Rodriguez C, Pavluck AL, Thun MJ, Calle EE. Association of polymorphisms in the paraoxonase 1 gene with breast cancer incidence in the CPS-II Nutrition Cohort. Cancer Epidemiol Biomarkers Prev. 2006;15:1226–1228. doi: 10.1158/1055-9965.EPI-05-0930. [DOI] [PubMed] [Google Scholar]
- 33.Feigelson HS, Henderson BE. Estrogens and breast cancer. Carcinogenesis. 1996;17:2279–2284. doi: 10.1093/carcin/17.11.2279. [DOI] [PubMed] [Google Scholar]
- 34.Ambrosone CB. Oxidants and antioxidants in breast cancer. Antioxid Redox Signal. 2000;2:903–917. doi: 10.1089/ars.2000.2.4-903. [DOI] [PubMed] [Google Scholar]
- 35.Gonenc A, Erten D, Aslan S, Akinci M, Simşek B, Torun M. Lipid peroxidation and antioxidant status in blood and tissue of malignant breast tumor and benign breast disease. Cell Biol Int. 2006;30:376–380. doi: 10.1016/j.cellbi.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Yu BP. Cellular defenses against damage from reactive oxygen species. Physiol Rev. 1994;74:139–162. doi: 10.1152/physrev.1994.74.1.139. [DOI] [PubMed] [Google Scholar]
- 37.Karaman E, Uzun H, Papila I, et al. Serum paraoxonase activity and oxidative DNA damage in patients with laryngeal squamous cell carcinoma. J Craniofac Surg. 2010;21:1745–1749. doi: 10.1097/SCS.0b013e3181f4040a. [DOI] [PubMed] [Google Scholar]
- 38.Elkiran ET, Mar N, Aygen B, Gursu F, Karaoglu A, Koca S. Serum paraoxonase and arylesterase activities in patients with lung cancer in a Turkish population. BMC Cancer. 2007;7:48. doi: 10.1186/1471-2407-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akcay MN, Polat MF, Yilmaz I, Akcay G. Serum paraoxonase levels in pancreatic cancer. Hepatogastroenterology. 2003;50(Suppl 2):ccxxv–ccxxvii. [PubMed] [Google Scholar]
- 40.Delimaris I, Faviou E, Antonakos G, Stathopoulou E, Zachari A, Dionyssiou-Asteriou A. Oxidized LDL, serum oxidizability and serum lipid levels in patients with breast or ovarian cancer. Clin Biochem. 2007;40:1129–1134. doi: 10.1016/j.clinbiochem.2007.06.007. [DOI] [PubMed] [Google Scholar]